Introduction
Procamallanus (Spirocamallanus) inopinatus Travassos, Artiga & Pereira, Reference Travassos, Artigas and Pereira1928 was originally described infecting the characiform fishes Leporinus copelandii Steindachner, 1855 and Megaleporinus elongatus (Valenciennes, 1850) from River Paraná Basin, Brazil (Travassos et al., Reference Travassos, Artigas and Pereira1928; Kohn and Fernandes, Reference Kohn and Fernandes1987). Since then, the species has been reported in several families of Characiformes, Perciformes and Siluriformes, within a wide geographical range in South America including, besides Brazil, Argentina, Paraguay, Peru and Venezuela (see Moravec, Reference Moravec1998; Iannacone et al., Reference Iannacone, López and Alvariño2000; Chemes and Takemoto, Reference Chemes and Takemoto2011; Luque et al., Reference Luque, Aguiar, Vieira, Gibson and Portes-Santos2011, Reference Luque, Cruces, Chero, Paschoal, Alves, Da Silva, Sanchez and Iannacone2016).
Due to its common occurrence, the general morphology of P. (S.) inopinatus has been well investigated suggesting that the species has homogenous morphology, but quite variable morphometry among component populations (Petter and Dlouhy, Reference Petter and Dlouhy1985; Kohn and Fernandes, Reference Kohn and Fernandes1987; Petter and Thatcher, Reference Petter and Thatcher1988; Moravec et al., Reference Moravec, Kohn and Fernandes1993; Reference Moravec, Prouza and Royero1997). Scanning electron microscopy (SEM) also has been used for observing the ultrastructure of P. (S.) inopinatus, but the detailing level was somewhat limited by the technology available at the time of such studies were performed (Moreira et al., Reference Moreira, Oliveira and Costa1994; Moravec et al., Reference Moravec, Prouza and Royero1997). More recently, Vicentin et al. (Reference Vicentin, Vieira, Tavares, Costa, Takemoto and Paiva2013) investigated P. (S.) inopinatus using SEM, but the approach of the study was not focused on the taxonomy of the species; therefore, results were not detailed.
Due to the lack of genetic data for P. (S.) inopinatus, its phylogenetic position within Camallanidae is unknown and the discussions regarding its wide morphometric variations among different populations remain without this important subsidy. Indeed, the phylogenetic aspects of Camallanidae are far from being elucidated due to the lack of genetic data, which is aggravated by the fact that the morphology-based systematics of the family is problematic and taxonomic labelling of some sequences available in genetic databases (e.g. GenBank) seems to be inaccurate (Černotíková et al., Reference Černotíková, Horák and Moravec2011; Sardella et al., Reference Sardella, Pereira and Luque2017; Ailán-Choke et al., Reference Ailán-Choke, Davies, Tavares and Pereira2019).
Therefore, the present study aimed to compare component populations of P. (S.) inopinatus from two different host species and localities using an integrative taxonomic approach and evaluate the phylogenetic aspects of Camallanidae based on different computational tools (e.g. digital imaging, biostatistics, phylogenetic and species delimitation).
Materials and methods
Sampling and processing of hosts and nematodes
Hosts were sampled as follows: 10 specimens of Anostomoides passionis Santos & Zuanon, 2006 (Characiformes: Anostomidae) from River Xingu (RX), municipality of Altamira, State of Pará, Brazil (3°14′S; 52°06′W) in April 2013, and 12 specimens of M. elongatus (Anostomidae) from River Miranda (RM), municipality of Corumbá, State of Mato Grosso do Sul, Brazil (19°28′S, 57°02′W) in August 2018. Fish were kept alive in small water tanks with oxygen pumps until necropsy, through a ventral longitudinal incision from anus to operculum and extraction of the digestive tract. Organs (oesophagus, stomach, caeca, intestine) were placed individually in Petri dishes with saline and analysed using a stereomicroscope.
Nematodes were found alive, washed in saline, fixed in hot 4% formaldehyde solution and preserved in 70% ethanol. For morphological observations, nematodes were cleared in glycerine. One male of each infrapopulation (i.e. from each infected fish, see also Bush et al., Reference Bush, Lafferty, Lotz and Shostak1997 for details) had the mid-body excised and fixed in molecular-grade 96–99% ethanol for genetic studies; anterior and posterior parts were fixed for morphological identification.
Nomenclature and classification of hosts were updated following Froese and Pauly (Reference Froese and Pauly2019), Frost (Reference Frost2020) and Uetz et al. (Reference Uetz, Freed and Hosek2020).
Morphological procedures
Nematodes were cleared in glycerine and observed in a Leica DM5500 B microscope, with DIC and the software LAS LeicaTM, for morphometric analysis. Measurements are given in micrometres unless otherwise stated. Specimens used for SEM (1 male and 1 female of each infrapopulation) were dehydrated through a graded ethanol series, dried by evaporation with hexamethyldisilazane, coated with gold and observed in a JEOL JSM 6460-LV, at an accelerating voltage of 15 kV. The systematic classification of camallanid nematodes follows the proposal of Moravec and Thatcher (Reference Moravec and Thatcher1997). Voucher specimens were deposited in the Coleção Helmintológica do
Instituto Oswaldo Cruz (accession nos. CHIOC 38939, 38940).
Statistical analysis of morphological data
In order to evaluate significant differences among morphometric features, we performed a series of statistical analyses. First, one sample Kolmogorov–Smirnov (K-S) test was performed to verify the normal distribution of the quantitative variables (Zar, Reference Zar2010). The quantitative variables were the standard length (SL) of fishes, and total body length, maximum width, buccal capsule length and width, distance from the anterior end to nerve ring, deirids and excretory pore, total length of oesophagus, tail length, distance from cephalic end to vulva in females and spicules in males of parasites. All these measurements were also used for calculating their ratio to total body length; the coefficient of variation (CV) for each of the previously mentioned features was also calculated. These morphometric features were chosen because they are used in the specific diagnosis, and ratios were used for verifying if the organ's anatomy (or their locations on the cuticle surface) undergoes modifications according to the parasite growth. Ratios and CV are expressed in percentage (%) and mean values are followed by ± 1 standard deviation unless otherwise stated.
According to the results of K-S test, we performed non-parametric inferential methods and generalized linear models (GLM). As a preliminary analysis, in order to verify possible influences of host-environment attributes and infection burden on the parasite body length, host SL and parasite intensity of infection (see Bush et al., Reference Bush, Lafferty, Lotz and Shostak1997) were tested as predictive of parasite body length using linear regression (Zar, Reference Zar2010). First, host SL and parasite intensity of infection were tested separately against parasite body length using simple linear regression, in order to evaluate which variable was most predictive through the r2 (coefficient of determination). Then a multiple linear regression was performed, inserting the predictive variables into the expected predictive model, in which the most predictive was inserted first followed by the less predictive (Zar, Reference Zar2010). Following the previous methodology, but using logistic regression instead of linear, we tested the effect of host species and parasite sexual maturation (i.e. male, non-gravid and gravid females) on parasite body length, estimating the odds ratio (OR, 0 < OR < 1 indicates antagonistic effect; OR = 1 lack of effect; OR > 1 synergetic effect) and the confidence interval (CI) of each model (Dohoo et al., Reference Dohoo, Martin and Stryhn2003). We also performed a Mann–Whitney (U) to evaluate and prove the differences in SL among the two fish species (Zar, Reference Zar2010).
Following the preliminary analysis, parasites were divided into three categories according to sexual maturation: males, non-gravid females (without larvae in uterus) and gravid females (uterus filled with larvae). All morphometric features previously stated for parasites were tested against their total body length using the Spearman's correlation (rs) (Zar, Reference Zar2010), considering all sexual maturation categories. Differences in morphometric features of parasites among the two-component populations, i.e. from A. passionis and M. elongatus, were evaluated using the U test, within each sexual maturation category and among non-gravid and gravid females (Zar, Reference Zar2010). Additional terminology related to parasite ecology follows the proposal of Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997).
Genetic procedures
Genomic DNA was isolated using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), following the manufacturer's instructions. Three genetic regions were amplified: the 5’ end of the 18S nuclear rDNA, the D2 and D3 domains of the nuclear 28S rDNA and the COI of the mtDNA. All polymerase chain reactions (PCRs), cycling conditions and primers are detailed in online Supplementary Material 1. PCR products were purified through an enzymatic treatment with ExoProStarTM (GE Healthcare) and sent for sequencing at ACTGene (Ludwig Biotec, Rio Grande do Sul, Brazil) with the same primers used in PCRs.
Contiguous sequences were assembled in Geneious (Geneious ver. 9.1.5 created by Biomatters, available from http://www.geneious.com/) and deposited in the GenBank. Preliminary BLAST search on GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) was performed to confirm the genetic proximity between the present sequences and those from representatives of Camallanidae.
Phylogenetic analyses of molecular data
The phylogenetic reconstructions were based on three different datasets: with sequences of the 18S rDNA, with those of COI mtDNA and with sequences of 18S and COI concatenated. Due to the small number of sequences of the 28S rDNA and the high frequency of genetic gaps after aligning them, phylogenies based on this genetic region were not reconstructed; we used this genetic marker only for comparisons between the present samples. Sequences were chosen according to the following criteria: genetic regions congruent with those obtained in the present study and minimum length of 744 bp and 355 bp for 18S and COI, respectively (for details see Table 1). Sequences from samples not identified to species level and clones were excluded. We tried to use as many representatives as possible, including different samples from the same species for species delimitation and validation analyses. The outgroup was chosen according to previous phylogenies of Camallanidae (see Černotíková et al., Reference Černotíková, Horák and Moravec2011). Sequences were aligned using M-Cofee (Notredame et al., Reference Notredame, Higgins and Heringa2000), then evaluated by the transitive consistency score, to verify the reliability of aligned positions and, based on score values, ambiguously aligned positions were trimmed (Chang et al., Reference Chang, Di Tommaso and Notredame2014). Saturation of nucleotide substitution was tested using Xia's method implemented in DAMBE (Xia et al., Reference Xia, Xie and Li2003; Xia, Reference Xia2018).
Table 1. Species whose sequences were obtained from GenBank and used in phylogenetic reconstructions, associated with their hosts (habitat: Freshwater [FW] or Marine [MA]), geographic origin, accession number and genetic regions.

Datasets were subjected to maximum likelihood (ML) and Bayesian inference (BI) analyses, using PHYML and MrBayes, respectively (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001; Guindon and Gascuel, Reference Guindon and Gascuel2003). The model of evolution (nucleotide substitution) and its fixed parameters for each dataset were chosen and estimated under the Akaike information criterion with jModelTest 2 (Guindon and Gascuel, Reference Guindon and Gascuel2003; Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012); in the dataset of 18S + COI the partitions were treated separately and substitution models unlinked (see details in online Supplementary Material 2). Nodal supports for ML were based on 1000 bootstrap non-parametric replications. The same, but for BI posterior probability, were determined after running the Markov chain Monte Carlo (MCMC) (2 runs 4 chains) for 4 × 106 generations, with sampling frequency every 4 × 103 generations and discarding the initial ¼ of sampled trees (1 × 106) as burn-in. In order to check chain convergence, analyses were run in duplicates and inspected using Tracer (Rambaut et al., Reference Rambaut, Drummond, Xie, Baele and Suchard2018).
The genetic markers profile of phylogenetic informativeness was evaluated by PhyDesign (Townsend, Reference Townsend2007) based on the concatenated dataset (18S + COI). For this analysis, an ultrametric tree was generated using BEAST 2.5 (Bouckaert et al., Reference Bouckaert, Vaughan, Barido-Sottani, Duchêne, Fourment, Gavryushkina, Heled, Jones, Kühnert, De Maio, Matschiner, Mendes, Müller, Ogilvie, du Plessis, Popinga, Rambaut, Rasmussen, Siveroni, Suchard, Wu, Xie, Zhang, Stadler and Drummond2019), the best-fit substitution model was chosen according to bModelTest (Bouckaert and Drummond, Reference Bouckaert and Drummond2017), models of each partition were unlinked, clock model was selected based on marginal likelihood estimated (MLE) from stepping stone and path sampling (Baele et al., Reference Baele, Li, Drummond, Suchard and Lemey2012) being 50 path steps, 5 × 105 itinerations and sampling every 500 generations. MCMC chains were run for 5 × 107 generations, sampling every 5 × 103 generations and ¼ of initial runs discarded as burn-in. All analyses were run in duplicates and inspected with Tracer to check convergence. More information can be found in online Supplementary Material 2.
Species delimitation
We tested species hypotheses within Camallanidae mainly focused on the present samples. Two different species discovery methods and one species validation method were used to delimit species boundaries. Some analyses required ultrametric trees; these were generated in BEAST 2.5 as previously described and additional details are also in online Supplementary Material 2.
For species discovery, we used the Generalized Mixed Yule Coalescent (GMYC) method and the Poisson Tree Process (PTP) method, which do not require a priori assignments regarding putative species (Pons et al., Reference Pons, Barraclough, Gomez-Zurita, Cardoso, Duran, Hazell, Kamoun, Sumlin and Vogler2006; Zhang et al., Reference Zhang, Kapli, Pavlidis and Stamatakis2013). The GMYC requires an ultrametric guide tree, uses ML to delimit species and estimates a transition point before which all nodes reflects species diversification events and after which all nodes represent population-coalescent process (Pons et al., Reference Pons, Barraclough, Gomez-Zurita, Cardoso, Duran, Hazell, Kamoun, Sumlin and Vogler2006; Razkin et al., Reference Razkin, Sonet, Breugelmans, Madeira, Gómez-Moliner and Backeljau2016). GMYC tests were run in the webserver (http://species.h-its.org/gmyc) under the single-threshold and multiple-threshold models, for each dataset. The PTP does not require ultrametric trees; therefore, the major consensus tree generated from BI of each dataset was used in the analysis; this method tried to identify significant changes in the rate branching of the phylogenetic tree, using the number of substitutions. These analyses were run in the bPTP webserver (http://species.h-its.org/ptp) using the default settings (1 × 105 generations, thinning set to 100, burn-in 1 × 104), which are adequate for datasets with < 50 taxa according to Zhang et al. (Reference Zhang, Kapli, Pavlidis and Stamatakis2013). PTP run both ML and BI to support the delimited species.
To validate the species, we used *BEAST (Heled and Drummond, Reference Heled and Drummond2010) implemented in BEAST 2.5 to generate unrooted (in order to improve species delimitation results; see Zhang et al., Reference Zhang, Kapli, Pavlidis and Stamatakis2013) species trees based on each dataset (minus outgroup sequences). *BEAST uses BI approach to generate phylogenies. We considered samples from the same species as different populations since they are mostly originating from different host individuals (see Table 1). Substitution models were chosen based on bModelTest, molecular clock selected based on MLE from stepping stone and path sampling, Yule process species tree priors and a constant root population size model. MCMC chains were run for 1 × 108 generations, sampling every 2 × 103 generations and ¼ of initial runs discarded as burn-in. Chains were run in duplicates and inspected with Tracer to check convergence. More information can be found in online Supplementary Material 2.
We also used the Automatic Barcode Gap Discovery (ABGD) method (Puillandre et al., Reference Puillandre, Lambert, Brouillet and Achaz2012) to generate pairwise (patristic) distance (P) matrixes, and thus evaluate intra and interspecific divergences among samples according to each genetic marker. The ABGD analyses were run online (http://wwwabi.snv.jussieu.fr/public/abgd/) using Kimura two-parameter (K2P) (Kimura, Reference Kimura1980) as the distance metric, with other parameters set to default. We refrain from the account the partitions generated by ABGD since intraspecific divergence of 18S and COI are poorly known and editing ABGD parameters for intraspecific genetic distance would generate mistaken results.
Results
The prevalence of P. (S.) inopinatus found in the present study was 30% (3 parasitized/10 examined) in A. passionis and 25% (3/12) in M. elongatus. The mean intensity of infection was 7.3 ± 1.5 (ranging from 6 to 9 parasites per infected host) in A. passionis and 8.0 ± 2.6 (0–11 per analysed host) in M. elongatus. Therefore, there were two-component populations (one from each host/locality) each composed of three infrapopulations of the parasite. Procamallanus (S.) inopinatus component population in A. passionis from RX had 9 males, 6 non-gravid and 7 gravid females; that in M. elongatus form RM had 11 males, 5 non-gravid and 8 gravid females. No concurrent infections were observed (i.e. the only helminth found was P. (S.) inopinatus). Specimens of M. elongatus were larger (SL 35.4 ± 1.3 cm; range 34.5–37.3 cm) than those of A. passionis (23.9 ± 1.2 cm; 23–25.5 cm); this difference was statistically significant (p < 0.001). The GLMs indicated no significant effect of host species (P > 0.78), SL (P > 0.37) and parasite intensity of infection (P > 0.59) in parasite body length. However, there was significant effect in parasite body length by sexual maturation (P < 0.001), in which males predicted smaller body length (OR = 0.687; CI 0.599–0.721), non-gravid females intermediate body length (OR = 1.233; CI 1.014–2.357) and gravid females larger body lengths (OR = 3.542; CI 2.987–3.881).
The following diagnostic features were constant and observed in the present specimens using SEM: cephalic end with two median teeth (one dorsal and one ventral) (Fig. 1A, B, online Supplementary Material 3), males with 10 pairs of subventral and sessile caudal papillae (4 pairs pre and 6 pairs prostcloacal) (Fig. 1E, F) and females without distinct terminal appendix on the tail (Fig. 1G, H). Also based on SEM observations, the presence of 6 pore-like structures (2 subdorsal, 2 subventral and 2 lateral) and of small pointed denticles surrounding the oral opening (Fig. 1A–D), as well as the phasmids of females (Figs 1G, H) are reported for the first time. The morphometric variations of the present specimens were within the range reported in previous studies. For these comparative measurements, as well as information on the host and locality of these studies see online Supplementary Material 4.

Fig. 1. Procamallanus (Spirocamallanus) inopinatus, ex. Anostomoides passionis (A, C, G, H) and Megaleporinus elongates (B, D, E, F), SEM micrographs A, B: Cephalic end of female and male, respectively, apical view (dashed lines delimit oral teeth). C, D: Oral opening of female and male, respectively (arrowheads indicate pore-like structures). E, F: posterior end and tail of male, respectively, sublateral views. G, H: tail and posterior end of female, respectively, sublateral views.
Most morphometric features of the present parasite specimens showed CV < 15%, with exception of some measurements related to deirids, excretory pore, tail and distance from the anterior end to vulva in females (see online Supplementary Material 5). The only CV higher than 20%, were observed in the ratio of the excretory pore to total body length (CV = 26.3%) and in the distance of vulva from the anterior end (CV = 24.3%), in gravid females from A. passionis (online Supplementary Material 4). Most morphometric features were correlated with a total body length of the parasites showing moderate to strong association (r2 ranging from |0.594| to |0.928|), except by the length of tail (P = 0.07), length of spicules (P = 0.6), distance of vulva form anterior end (P = 0.9) and its relative position (P = 0.1) in females (online Supplementary Material 5). The size of all anatomic structures (except those previously mentioned) showed a positive correlation with a total body length; conversely, their ratios to total body length were negatively correlated, with exception of that of spicule (online Supplementary Material 5). Gravid females were larger and wider than those non-gravid (both P < 0.001), the tail to body length ratio was higher in non-gravid (P < 0.001) and vulva was more posterior in gravid specimens (P = 0.01); however, the relative position of vulva was similar in both gravid and non-gravid females (P = 0.2) (Table 2, online Supplementary Material 5). Width of buccal capsule and length of oesophagus were also higher in gravid females (P = 0.014 and < 0.001, respectively); distance from the anterior end to deirid and nerve ring to total body length ratio also showed statistical differences according to female ontogeny (Table 2, online Supplementary Material 5).
Table 2. Comparative measurements of Procamallanus (Spirocamallanus) inopinatus in the present study, according to host and locality.

* Measurements not included in the statistical analysis.
Different superscript letters indicate measurements that showed statistical differences based on Mann–Whitney test, these comparisons are within each line of the table, respecting sexual maturation (for more information see online Supplementary Material 5).
Comparing the two-component populations of P. (S.) inopinatus, based on morphometry and according to sex and female ontogeny, most statistical divergences were observed among males. All measurements tended to be higher in males parasitizing A. passionis; however, the significance comparing the body length was in the borderline (i.e., P = 0.05) and the significant differences were found in buccal capsule length (P = 0.003) and width (P = 0.006), in excretory pore location (P = 0.03), as well as in the length of oesophagus (P = 0.003), spicules (P < 0.03) and tail (P = 0.003) (Table 2, online Supplementary Material 4). No statistical differences were observed comparing the morphometry of gravid females from A. passionis and M. elongatus, and regarding the non-gravid specimens from A. passionis showed wider buccal capsule (P = 0.01), more posterior excretory pore (P = 0.03) and larger oesophagus (P = 0.01) (Table 2, online Supplementary Material 5).
After sequencing of the genetic regions, 18S (GenBank accession nos. MT901634; MT901635) and COI (MT898796; MT898796) showed no polymorphisms within samples of the same component population; therefore, we used only one representative of each component population for genetic analyses. Three different genotypes were found for the 28S (MT901636–MT901638), two from parasites of A. passionis and one from M. elongatus, in which the genetic similarity among them was > 98% (sequence lengths were 870 bp, 874 bp and 876 bp).
The partial 18S fragments were 892 bp long and showed only one polymorphism in the position 640 (A/C) according to component populations; their genetic similarity was 99.89% and the patristic distance P = 0.00116. Promacallanus (S.) pintoi (Kohn & Fernandes, 1988) in the freshwater catfish Corydoras atropersonatus Weitzman & Nijssen, 1970 from Peru (DQ442666), was most similar to the present samples (genetic of similarity 94.13% and P = 0.0117 when compared with those parasites of M. elongtus; genetic similarity of 94.24% and P = 0.0105 when compared with those parasites of A. passionis). The partial coding fragments of COI were 388 bp long, the genetic similarity among samples from M. elongtus and A. passionis was 90.72%, the patristic distance P = 0.09901 and no polymorphism resulted in amino acid residue change. These sequences were most similar to that of P. (S.) istiblenni (Noble, 1966) parasitizing the marine snapper Lutjanus kasmira (Forsskål, 1775) from Hawaii (KC517382) (genetic similarity 86.18% and P = 0.1515 from parasites of M. elongtus and 84.75% and P = 0.1700 from parasites of A. passionis). Genetic alignments showed no substantial nucleotide saturation (18S: Iss = 0.068, Iss.c = 0.804, P < 0.001; COI: Iss = 0.2612, Iss.c = 0.6761, P < 0.001). BLAST search indicated that the representative from GenBank database most genetic similar to the present sequences was C. xenopodis Jackson & Tinsle, Reference Jackson and Tinsle1995 [MG947389] (genetic similarity of 64.4%).
The phylogenies using 18S sequences included more representative taxa than those using COI and 18S + COI, because of data availability (Table 1, Figs 2–4). The phylogenetic meaning of the trees reconstructed using ML and BI was the same; therefore, only BI trees were depicted. Monophyly of Camallanidae was strongly supported in all phylogenies (Figs 2, 3); in contrast, its genera and subgenera were not monophyletic (Figs 2, 4). A monophyletic assemblage including sequences with the same origin from India, shaded in the trees of the 18S, made the following species not monophyletic: Camallanus cotti Fujita, 1927, C. oxycephalus Ward & Magarth, 1917 and P. (S.) istiblenni. Procamallanus (S.) rarus Travassos, Artigas & Pereira, 1935 was also non-monophyletic and unrelated to the shaded clade (Figs 2, 4). Samples from the present study formed well-supported monophyletic assemblages in all the phylogenies (Figs 2–4). The phylogenetic informativeness of COI was better for analysing divergences and similarities among inner nodes (e.g. closely related taxa, such as those species) and that of 18S better for evaluating divergence events related to more external nodes (e.g., for comparing genera) (Fig. 3).

Fig. 2. Phylogenetic trees generated using Bayesian Inference from sequences of camallanid nematodes. Nodal supports are indicated on tree nodes, according to the Bayesian posterior probability (BPP) and maximum likelihood (ML) bootstrap replications as follows: full squares (BPP = 1, ML = 100%), full circles (0.96 < BPP < 1, 90% < ML < 100%) and empty circles (0.90 < BPP < 0.96, 95% < ML < 90%). Black polygons next to taxa labels indicate specific entities recognized by the Poisson Tree Process (PTP) and those white by the Generalized Mixed Yule Coalescent (GMYC); superscript numbers make correspondence with information in Table 1. The shaded clade represents atypical phylogenetic behaviour and includes sequences with apparently taxonomic misidentification. Sequences from the present study are in bold and superscriptions are RM = River Miranda, RX = River Xingu.

Fig. 3. Phylogenetic trees generated from concatenated sequences of the 18S rDNA and COI mtDNA of camallanid nematodes using Bayesian Inference. Upper centre tree is the output of Mr. Bayes (gene tree), bottom right is the output of PhyDesign (which does not estimate nodal supports) associated with graphical representation of phylogenetic informativeness for each generic marker, bottom left is a species tree output of *BEAST from 18S and COI concatenated. Nodal supports are indicated on tree nodes, according to the Bayesian posterior probability (BPP) and maximum likelihood (ML) bootstrap replications as follows: full squares (BPP = 1, ML = 100%), full circles (0.96 < BPP < 1, 90% < ML < 100%) and empty circles (0.90 < BPP < 0.96, 95% < ML < 90%). Black polygons next to taxa labels indicate specific entities recognized by the Poisson Tree Process (PTP) and those white by the Generalized Mixed Yule Coalescent (GMYC); superscript numbers make correspondence with information in Table 1. Sequences from the present study are in bold and superscriptions are RM = River Miranda, RX = River Xingu.
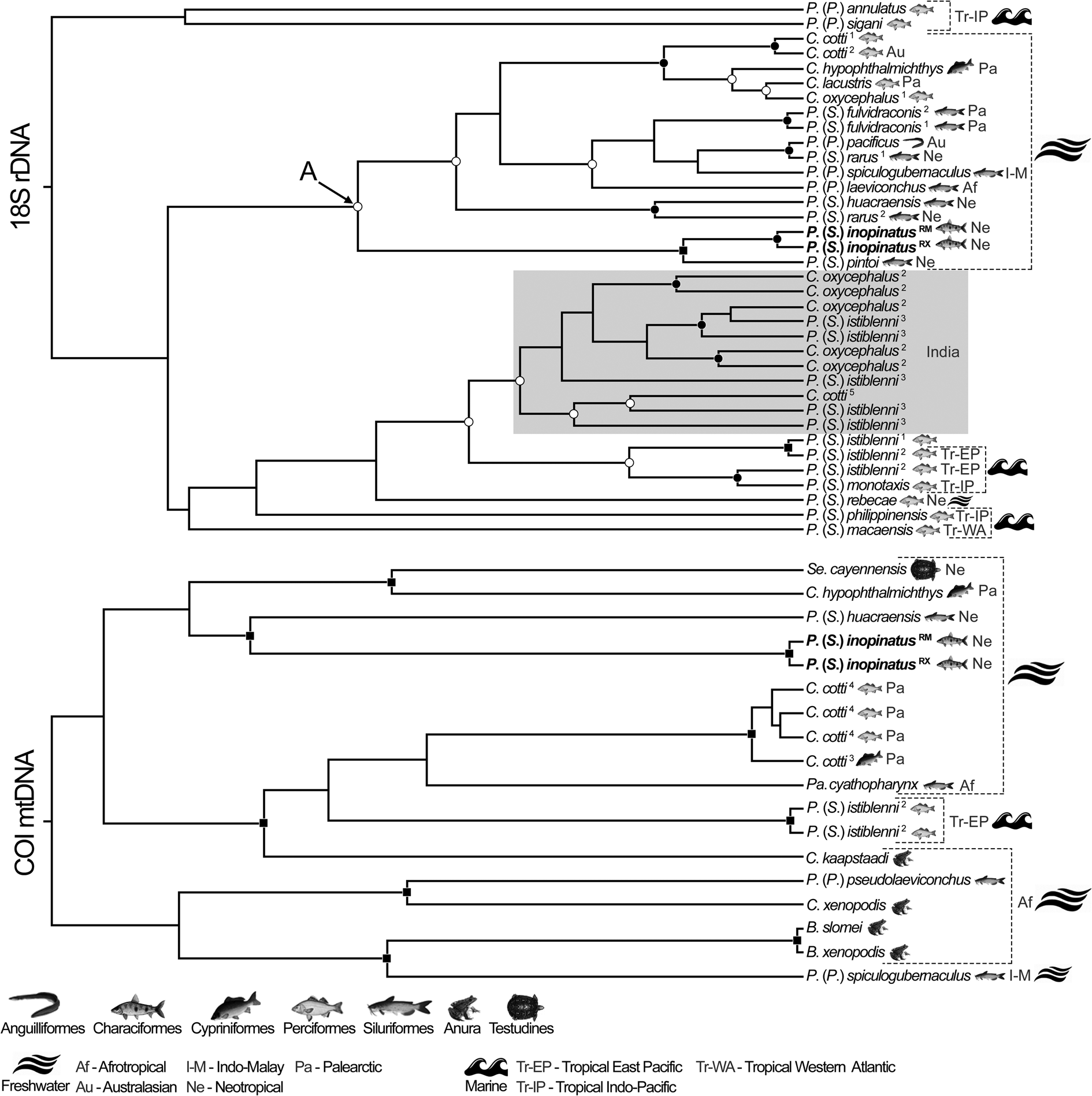
Fig. 4. Species trees based on 18S rDNA and COI mtDNA sequences of camallanid nematodes generated using *BEAST, for species validation. Nodal supports are indicated on tree nodes, according to the Bayesian posterior probability (BPP) as follows: full squares (BPP = 1), full circles (0.96 < BPP < 1) and empty circles (0.90 < BPP < 0.96). Superscript numbers make correspondence with information in Table 1. The shaded clade represents atypical phylogenetic behaviour and includes sequences with apparently taxonomic misidentification. Letter A highlights the clade formed exclusively by species from freshwater hosts. Order of hosts, geographic origin and environment associated with each sequence are depicted. Sequences from the present study are in bold and superscriptions are RM = River Miranda, RX = River Xingu.
Regarding the species delimitation analyses, PTP was more concordant with the molecular phylogenetic approach than GMYC (Figs 2, 3). ML of the GMYC was significantly higher than the null model, except for the 18S dataset (see online Supplementary Material 6). The entities recognized by the PTP model showed high support values for both ML and Bayesian approaches (see online Supplementary Material 6). The present samples were recognized as a single species by both methods, in all datasets (Figs 2, 3). In the tree of 18S sequences, PTP indicated that P. (S.) istiblenni represented 7 different specific entities, C. oxycephalus 6, C. cotti and P. (S.) rarus 2 each (Fig. 2). In the tree of COI sequences, PTP agreed with taxonomic labelling of all species, except B. slomei and B. xenopodis that were recognized as a single species (Fig. 2), and the K2P distance between these sequences (MG948463/ MN523681) was null (P = 0). The concordance of these methods (mainly PTP) with the taxonomic labelling of the sequences was higher in the dataset of COI and 18S + COI sequences, as expected (Figs 2, 3).
Species tree approach validated the results of phylogenetic and species delimitation methods (mainly of PTP) (Figs 2–4). Samples of P. (S.) inopinatus were validated as a single species, whereas those that appeared as non-monophyletic were not validated as single species, and, as same as in species delimitation approach, B. slomei and B. xenopodis and recognized as a single lineage; nodal supports in these cases were medium to high (Fig. 4). We indicated host, geographic origin and environmental features related to samples in the species tree of 18S and COI (Fig. 4). Thus, most phylogenetic assemblages did not show similarities regarding these characteristics, except the clade A in Fig. 4 that grouped species from freshwater hosts and the shaded one, composed of species from India, with no additional information. These clades showed average nodal supports (Fig. 4). It should be mentioned that P. (S.) inopinatus was phylogenetically close to samples isolated from freshwater fishes of the Neotropical region.
The pairwise distances between samples of P. (S.) inopinatus were P = 0.001 and P = 0.099 in 18S and COI matrices, respectively. Lower than these, were the distances between representative of P. (S.) fulvidraconis Li, 1935 (DQ076689/JF803914) and P. (S.) istiblenni (EF180076/KC505629) (both P = 0) in 18S matrix, representatives of C. cotti (DQ442662/EU598876/EU598845; P < 0.014), C. oxycephalus (EU598879/EU598876/EU598833/EU598845; P < 0.014) of P. (S.) istiblenni (KC517382/KC517383; P = 0.002) and between B. slomei Southwell & Krishner, 1937 (MG948463) and B. xenopodis (MN523681) (P = 0) in COI matrix. Pairwise distances also corroborated previous phylogenetic, species delimitation and validation results, in which representatives of C. cotti, C. oxycephalus and P. (S.) istiblenni forming the shaded clade (Figs 2, 4) showed values of genetic distance relatively high (P > 0.013) when compared with their conspecifics placed out of this clade. Samples of P. (S.) rarus (DQ494195/JF803912) also showed similar results (P = 0.048). Complete distance matrices and comparative tables are available in online Supplementary Material 7, 8, 9.
Discussion
As previously commented, Procamallanus (S.) inopinatus is a nematode parasite with suggestively low host specificity and wide distribution in the freshwaters of South America (from North to South-western Brazil, Venezuela, Peru and Argentina) (see Moravec, Reference Moravec1998; Chemes and Takemoto, Reference Chemes and Takemoto2011; Luque et al., Reference Luque, Aguiar, Vieira, Gibson and Portes-Santos2011, Reference Luque, Cruces, Chero, Paschoal, Alves, Da Silva, Sanchez and Iannacone2016). Some aspects of its taxonomic assessment have been controversial, especially considering the spiral ridges in the buccal capsule (see Moravec et al., Reference Moravec, Kohn and Fernandes1993, Reference Moravec, Prouza and Royero1997). Despite the wide variation in the number of spiral ridges in P. (S.) inopinatus (13–23, see online Supplementary Material 4), they occupied no more than 2/3 of the buccal capsule as shown herein and in other studies (Travassos et al., Reference Travassos, Artigas and Pereira1928; Pereira, Reference Pereira1935; Kloss, Reference Kloss1966; Petter and Thatcher, Reference Petter and Thatcher1988; Moravec et al., Reference Moravec, Kohn and Fernandes1993, Reference Moravec, Prouza and Royero1997; Abdallah et al., Reference Abdallah, Azevedo, Carvalho and Silva2012). Reciprocally, Petter and Dlouhy (Reference Petter and Dlouhy1985) described this structure completely occupied by the spiral ridges and Vicentin et al. (Reference Vicentin, Vieira, Tavares, Costa, Takemoto and Paiva2013) reported the same characteristic but based only on female specimens; both studies analysed characiform fishes from River Paraguay basin (as in the present study). These observations indicate that this characteristic may be a rare intraspecific variation in P. (S.) inopinatus. However, to confirm this assertion, specimens with the whole buccal capsule occupied by spiral ridges need to be genetically characterized and compared to the present data. It should be mentioned that, based on genetic evidence, the morphology of buccal capsule seems to bear some degree of artificiality in the systematics of genera and subgenera of Camallanidae (Wijová et al., Reference Wijová, Moravec, Horák and Lukeš2006; Sardella et al., Reference Sardella, Pereira and Luque2017; Ailán-Choke et al., Reference Ailán-Choke, Davies, Tavares and Pereira2019), which may be also true at the species level, as suggested here.
Moreira et al. (Reference Moreira, Oliveira and Costa1994) performed the first observation of P. (S.) inopinatus using SEM and reported 3 circles of 4 cephalic papillae each in the species. Later, Moravec et. al. (1997) described only 2 circles with 4 papillae each, also using SEM. Based on the present results it was possible to confirm the observations of Moreira et al. (Reference Moreira, Oliveira and Costa1994) through higher resolution micrographs. The present observations also reported for the first time small denticles and pore-like structures surrounding the oral opening internally and externally, respectively. It should be mentioned that Vicentin et al. (Reference Vicentin, Vieira, Tavares, Costa, Takemoto and Paiva2013) reported only 1 circle of 4 cephalic papillae in P. (S.) inopinatus, which is a misinterpretation since in the SEM micrograph provided by the authors it is possible to observe the 2 additional circles of cephalic papillae (see Fig. 1A by Vicentin et al., Reference Vicentin, Vieira, Tavares, Costa, Takemoto and Paiva2013). The presence of two large (dorsal and ventral) teeth (see Petter and Thatcher, Reference Petter and Thatcher1988; Moreira et al., Reference Moreira, Oliveira and Costa1994) was also confirmed and phasmids in female were observed for the first time, contributing to the morphological knowledge of P. (S.) inopinatus.
The present results showed that almost all measurements used for taxonomic diagnosis had a medium to strong correlation with the body length of parasites. Measurements of the organs showed a positive correlation (except the length of spicules and the distance from cephalic and to vulva) whereas the ratio of these measurements to total body length showed negative correlation (see online Supplementary Material 5). It indicates that the growth rates of body and organs are markedly different, in which the first grows much faster than the second (a matter of mathematic reason: if the divisor, i.e., body length, is much higher than the dividend, i.e., the measure of a given organ, the quotient will be low). Moreover, these results indicate general morphometric dependence on body length, except by the spicule length and the relative position of vulva, which are more constant and, consequently, more reliable for intraspecific comparisons, regardless of body length and sexual maturation of P. (S.) inopinatus.
Sexual maturation of females of P. (S.) inopinatus in the present study had a significant effect on their body length, in which gravid specimens were larger than the non-gravid. As a consequence, some ratio measurements, the maximum body and buccal capsule width, oesophagus length, distance from cephalic end to vulva, but not its relative position (vulva to body length ratio), were statistically different (Table 2, online Supplementary Material 5). Therefore, the morphometry of gravid and non-gravid females should be presented separately. However, several studies do not follow this proposal, making the morphometric range of females of P. (S.) inopinatus very wide (see online Supplementary Material 4). Male specimens are evidently smaller than females (present results showed males as predictive of smaller body length) and also exhibit the same wide morphometric ranges, comparing the present results with the previous studies (see online Supplementary Material 4). Thus, researches should be aware when dealing with the morphometry of P. (S.) inopinatus and focus on morphology (see further discussion).
Despite the intensity of infection, host species and SL did not have an effect in the body length of the present parasites; the statistical comparison between specimens of the two-component populations evidenced the morphometric differences from males and non-gravid females, but not from those gravid. It indicates that, once these females reach full sexual maturity, their growth stabilizes (other argument supporting the assertion for separating measurements from non-gravid and gravid specimens). The main differences comparing non-gravid females and males from the two-component populations were in the position of excretory pore and in the length of oesophagus; these variations can be observed in P. (S.) inopinatus from different hosts and river basins (Petter and Dlouhy, Reference Petter and Dlouhy1985; Petter and Thatcher, Reference Petter and Thatcher1988; Moravec et al., Reference Moravec, Kohn and Fernandes1993; Reference Moravec, Prouza and Royero1997). Although the body length is similar between males from the present component populations, specimens parasitizing A. passionis showed larger spicules. The size of these structures is very important in the taxonomy of Procamallanus (Spirocamallanus) spp. (Moravec et al., Reference Moravec, Salgado-Maldonado and Caspeta-Mandujano2000; Moravec and Jirků, Reference Moravec and Jirků2015; Pinheiro et al., Reference Pinheiro, da, Melo, Monks, dos Santos and Giese2018; Moravec and Justine, Reference Moravec and Justine2019). However, in P. (S.) inopinatus the length of spicules seems to be random and not correlated with body length, some of the smallest males have average spicule length (e.g. Petter and Thatcher, Reference Petter and Thatcher1988) whereas some of the larger specimens have them short (e.g. Moravec et al., Reference Moravec, Prouza and Royero1997) (see also online Supplementary Material 4).
The present morphometric heterogeneity observed in P. (S.) inopinatus represents a phenotypic plasticity characteristic of the species, also observed in previous studies (see online Supplementary Material 4). Moreover, although the morphology of P. (S.) inopinatus is almost constant, the number and distribution of spiral ridges on its buccal capsule is highly variable. In fact, these features have been pointed out as weak taxonomic characters within the complicated systematics of Camallanidae (Wijová et al., Reference Wijová, Moravec, Horák and Lukeš2006; Černotíková et al., Reference Černotíková, Horák and Moravec2011; Sardella et al., Reference Sardella, Pereira and Luque2017; Ailán-Choke et al., Reference Ailán-Choke, Davies, Tavares and Pereira2019). Therefore, in order to avoid further taxonomic confusions, we suggest that the specific diagnosis of P. (S.) inopinatus should be based on the presence of two large (one dorsal and one ventral) oral teeth at the cephalic end, which represent the single autapomorphy of the species so far.
The present and first genetic characterization of P. (S.) inopinatus supported the results of morphological analysis, in which the morphology of the specimens was constant among the component populations. It also reaffirms the characteristic wide morphometric variation of the species. The pairwise distances of both 18S and COI among the present sequences were low, as same as observed between representatives of C. cotti (DQ442662/EF180071 from 18S and EU598833/ EU598845 EU598876/EU598879 from COI), P. (S.) istiblenni (KC505629/ KC505630 from 18S and KC517382/ KC517383) and P. (S.) fulvidraconis (DQ076689/ JF803914 from 18S) (for more details see online Supplementary Material 7, 8). The present 28S sequences also supported that the samples are conspecific; their genetic similarity was much higher than that of the most closely related sample from GenBank database (i.e. C. xenopodis). Unfortunately, 28S sequences from GenBank are currently very limited in number and length, giving no robustness for comparing the present intraspecific genetic variations with that from other species. Similarly, species delimitation and validation approaches indicated that P. (S.) inopinatus in A. passionis from River Xingu (Amazon basin) and in M. elongatus from River Miranda (Paraguay basin) are conspecific. It should be highlighted that GMYC was more conservative than PTP, regarding species delimitation and the accuracy of the methods was higher in the databases including COI (see online Supplementary Material 6), because this genetic region was more informative for analysing divergences and similarities among closely related taxa than the 18S (see Fig. 3); this is due to the fact that, in metazoans, mtDNA genes evolve at faster rates than those form nuclear DNA (Allio et al., Reference Allio, Donega, Galtier and Nabholz2017).
The overall phylogeny of Camallanidae was poorly resolved and monophyly was not observed in most genera and subgenera as a consequence of the morphology-based systematics that predominantly uses the buccal capsule structure as a diagnostic feature, which seems to be artificial as commented previously (Wijová et al., Reference Wijová, Moravec, Horák and Lukeš2006; Černotíková et al., Reference Bush, Lafferty, Lotz and Shostak2011; Sardella et al., Reference Sardella, Pereira and Luque2017; Ailán-Choke et al., Reference Ailán-Choke, Davies, Tavares and Pereira2019).
The genus Batrachocamallanus Jackson and Tinsle, Reference Jackson and Tinsle1995 is a particular case; it has been considered a synonym of Procamallanus (Procamallanus) Baylis, 1923 by some authors (Moravec et al., Reference Moravec, Justine, Würtz, Tarachewski and Sasal2006) and valid by others (Jackson and Tinsle, Reference Jackson and Tinsle1995; Svitin et al., Reference Svitin, Schoeman and du Preez2018; Reference Svitin, Truter, Kudlai, Smit and du Preez2019). The buccal capsule structure in both taxa is the same and the characters used in the differential diagnosis of Batrachocamallanus were refuted and proved to be also present in Procamallanus (Procamallanus) (see Moravec et al., Reference Moravec, Justine, Würtz, Tarachewski and Sasal2006). Therefore, we agree with the proposal of Moravec et al. (Reference Moravec, Justine, Würtz, Tarachewski and Sasal2006) and consider Batrachocamallanus junior synonym of Procamallanus (Procamallanus). In this sense, the present genetic analyses support the previous assertion because GMYC considered the two representatives, labelled B. slomei Southwell & Kirshner, 1937 and B. xenopodis (Baylis, 1929), along with P. (P.) spiculocubernaculus Agarwal, 1958 as a single species and in the species tree they were closely related with full nodal support. Furthermore, sequences from B. slomei and B. xenopodis were 100% similar, but we will not assume their synonymy, since data represent only a small and partial fragment of a gene.
In the phylogenetic reconstructions using 18S sequences, representatives of P. (S.) inopinatus formed a fully supported assemblage with P. (S.) pintoi, which was also closely related to P. (S.) rarus and P. (S.) huacraensis Ramallo, 2008, all representatives isolated from freshwater catfishes, in the Amazon (the first two) and la Plata basin (the last). When COI sequences were included in the analysis, the phylogenetic position of P. (S.) inopinatus was somewhat uncertain, but not in the species tree that provides a better genealogy of taxa (Nichols, Reference Nichols2001) and where it formed a fully supported assemblage with P. (S.) huacraensis. The recent ancestrally of the all previously mentioned species is probably the same (or closely related), since the geological and biogeographic history in South America, indicates the close relatedness between the Amazon la and Plata basins (which includes Paraná basin), and the tandem evolution of their fish fauna (Albert and Reis, Reference Albert and Reis2011; Reis et al., Reference Reis, Albert, Di Dario, Mincarones, Petry and Rocha2016; Dagosta and Pinna, Reference Dagosta and Pinna2017). Moreover, P. (S.) inopinatus also parasitizes catfishes (see Moravec, Reference Moravec1998; Iannacone et al., Reference Iannacone, López and Alvariño2000; Chemes and Takemoto, Reference Chemes and Takemoto2011; Luque et al., Reference Luque, Aguiar, Vieira, Gibson and Portes-Santos2011, Reference Luque, Cruces, Chero, Paschoal, Alves, Da Silva, Sanchez and Iannacone2016). These relationships between habitat, host taxa, geographic origin and the phylogenetic aspects of camallanids cannot be generalized and are unclear, since as same as some assemblages were formed by samples isolated from freshwater hosts, they were from very different geographic origins and host taxa; other assemblages were formed independently from these characteristics. Similar results were previously observed (Wijová et al., Reference Wijová, Moravec, Horák and Lukeš2006; Ailán-Choke et al., Reference Ailán-Choke, Davies, Tavares and Pereira2019).
The conspecific-labelled sequences referring to C. cotti, C. oxycephalus, P. (S.) istiblenni and P. (S.) rarus were not monophyletic. The main responsible for this result was the shaded clade in Figs 2, 4, composed by sequences with the same origin (location and authors), with poorly detailed information and unpublished in scientific papers. The usage of these sequences generated misleading results, which were kept here with the intent of highlighting the importance of a careful analysis when dealing with the genetic database of camallanids, since these data have considerable taxonomic inaccuracies also observed in other studies (Černotíková et al., Reference Černotíková, Horák and Moravec2011; Ailán-Choke et al., Reference Ailán-Choke, Davies, Tavares and Pereira2019).
The present approach confirmed that the general morphometry of P. (S.) inopinatus is markedly variable, whereas the general morphological features are constant, especially the following that should be used for species diagnosis: the presence of two large (dorsal and ventral) oral teeth (only synapomorphy of the species), the relative position of vulva (on the second third of body) and 10 then pairs of subventral caudal papillae in males (4 pairs pre and 6 pair post cloacal). The present genetic characterization and the phylogenetic analyses supported the morphological analyses and that the component populations of P. (S.) inopinatus from A. passionis and M. elongatus are conspecific, their genetic structure differ, but this difference is minimal regarding all genetic markers analysed. The data provided here improve the current scarce genetic database of camallanid nematodes and represents the first step for a better understanding of the genetic population structure of P. (S.) inopinatus.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182020001687
Acknowledgements
The authors would like to thank Dr Elias Nogueira de Aguiar from the Laboratório Multiusuário de Análises de Materiais do Instituto de Física (MULTILAM-INFI), Universidade Federal de Mato Grosso do Sul (UFMS), for help with scanning electron microscopical procedures. Dr Carina Elisei from the Universidade Católica Dom Bosco provided the facilities for molecular studies. Dr Philippe Vieira Alves for valuable conversations that inspired the approach of the present manuscript and Lennon de Souza Malta for his assistance in field collections and laboratorial procedures.
Financial support
This work was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES)-Finance code 001 and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico – Brazil (Proc. 474077/2011-0).
Conflict of interest
None.
Ethical standards
All procedures involving animal manipulation were permitted by Sistema de Informação e Autorização em Biodiversidade (license number 54895) and were in strict accordance with the recommendations of the Colégio Brasileiro de Experimentação Animal.









