Introduction
Microfungi have been cultured from a broad range of Antarctic habitats including, soil, moss and permanently frozen lakes (reviewed by Zalar et al. Reference Zalar, Sonjak and Gunde-Cimerman2012). The relatively low diversity of (Fell et al. Reference Fell, Scorzetti, Connell and Craig2006) and potentially high percentage of fungi that are culturable from (Onofri et al. Reference Onofri, Zucconi and Tosi2007) these Antarctic habitats can allow for a more thorough study of community structure than from highly complex lower latitude habitats. Because of the lower diversity and abundance of culturable fungi from each potential sample site, especially in the McMurdo Dry Valleys, identification of each isolate in abundance studies is a viable option.
All methods for the study of soil communities have biases. Identification of the members of a microbial community using culture-independent methods, via ribosomal RNA (rRNA) gene cloning and sequencing techniques, has become standard practice. Yet, despite the data and insights provided, even with second and third generation sequencing methods, a conclusive functional characterization of the community is not yet attainable (Kunin et al. Reference Kunin, Engelbrektson, Ochman and Hugenholtz2010). One important bias for culture-independent methods is the efficiency of cell lysis and nucleic acid extraction from soil (Feinstein et al. Reference Feinstein, Sul and Blackwood2009). The availability of cultured isolates opens the door to numerous microbiological, physiological, functional and other investigations, thereby allowing confirmation and extension of culture-independent data. In addition, species abundance, for culturable species, can only be obtained from culture-based studies. Therefore, complementing microbiological culturing with culture-independent studies is recognized as a strategy to overcome the deficiency of using either method alone.
Fungal taxonomy can be challenging because of intraspecific morphological and physiological plasticity as well as a limited number of morphological markers. Further, the process of identification based on morphology and physiological traits can be time consuming. Sequencing of ribosomal genes (rDNA) and their internal transcribed spacer (ITS) or other variable regions within conserved genes (e.g. actin gene) are often necessary for accurate taxonomic identification. The ITS region of the nuclear rRNA gene has been used as a target for identification of microbial species because it is often variable enough to be used as a species-specific target region (Kurtzman Reference Kurtzman2006). The current cost of traditional sequencing places a substantial limitation on the number of isolates that may be analysed. Frequently in the process of identifying isolates, common species may be sequenced many times demonstrating the need for a lower cost method to screen a large number of isolates. Other molecular methods are available such as arbitrarily primed polymerase chain reaction (apPCR) (Rodriguez et al. Reference Rodriguez, Cullen, Kurtzman, Khachatourians and Hegedua2004), three-primer PCR (Fell Reference Fell1993) and direct hybridization (Kiesling et al. Reference Kiesling, Diaz, Statzell-Tallman and Fell2002). However, direct hybridization and three-primer PCR requires specific primers or probes be developed for each taxa and apPCR is more appropriately used to determine intraspecific variation.
The automated ribosomal intergenic spacer analysis (ARISA) technique is a high-resolution automated capillary electrophoresis DNA fingerprinting method that is based on discriminating taxa by variation in the size of the target gene region. Since its development, along with similar methods such as denaturing gradient gel electrophoresis (DGGE) (Fisher & Lerman Reference Fisher and Lerman1980), ARISA has been applied to a wide range of environmental samples from freshwater bacterial communities to soil fungi (Fisher & Triplett Reference Fisher and Triplett1999, Slabbert et al. Reference Slabbert, van Heerden and Jacobs2010). This technique, when applied to the ITS region, has shown promise as an accurate method for rapidly analysing the diversity and composition of microbial communities at large (reviewed by Popa et al. Reference Popa, Popa, Marshall, Nguyen, Tebo and Brauer2009) by comparing chromatogram peaks among samples. These peaks have been called operational taxonomic units (OTUs). Popa et al. (Reference Popa, Popa, Marshall, Nguyen, Tebo and Brauer2009) proposed the term phylotypes over the use of OTUs as a description of taxa identified because more work still needs to be done to define phylogenetic resolution of data resulting from environmental ARISA. However, the ARISA technique can also be utilized to rapidly and relatively inexpensively determine species (phylotype) using cultured material. For example, ARISA has been used to measure intra-genomic diversity among Shewanella phylotypes (Popa et al. Reference Popa, Popa, Marshall, Nguyen, Tebo and Brauer2009).
In this study we assessed the ability of ARISA to provide taxonomic identification from cultured fungal material by development of a database consisting of culturable Antarctic fungal ARISA profiles, or ARISA Antarctic fungal fingerprints (ARISA-AFF). This method is also useful as a screening procedure to select isolates that will require sequencing for positive identification or to sort isolates into groups for further analysis. We tested the efficacy of ARISA-AFF in screening 203 isolates (46 species) from samples taken from lakewater, moss, soil and sponge from south Victoria Land, Antarctica. A total of 78% of the isolates in this study were unambiguously identified solely using ARISA-AFF, leaving fewer isolates to be identified by more expensive or time consuming protocols. DNA sequencing following sorting by ARISA-AFF analysis identified the remaining 22% of isolates. In addition, the ARISA-AFF technique helped to recognize several previously unidentified species found in culture. These isolates were morphologically similar to others from the same sample and may have been missed in studies that did not identify each isolate.
Materials and methods
Field sites, collection and fungal isolation
Isolates for this study were from environmental samples collected in south Victoria Land, Antarctica (Table S1, which will be found at http://dx.doi.org/10.1017/S0954102012000879.) during field seasons spanning 2002–10. These samples include lakewater, soil and moss from Taylor Valley, soil from ‘the Labyrinths’ of Wright Valley and from fumarole caves on Mount Erebus and ancient sponge samples from the Ross Ice Shelf.
Wright Valley and Taylor Valley soil samples were collected using a sterile metal scoop and placed in Whirl-Pak bags (Nasco, Fort Atkinson, WI). Fumarole cave soil samples were collected by using a sterile metal scoop to scrape the soil from the bottom of the cave into sterile aluminium foil. Moss was collected by placing c. 20 g into a sterile Whirl-Pak bag using sterile forceps. Sponge samples were obtained from sponges in late Quaternary debris bands on top of the Ross Ice Shelf near Mount Discovery (Kellogg et al. Reference Kellogg, Kellogg and Stuiver1990). Sterile forceps were used to place c. 100 g of sponge material into Whirl-Pak bags. Samples were transported frozen to the continental US (-20°C) then treated as previously described for fungal isolation (Connell et al. Reference Connell, Redman, Craig, Scorzetti, Iszard and Rodriguez2008). Water samples were obtained from various depths of the water column of Lake Fryxell during the 2008–09 field season. Holes were drilled through the ice with a seven-inch steel drill bit attached to a gasoline-powered mechanical Jiffy drill. Water was collected with a 5 l Niskin sampler and transferred to sterile 1 l bottles and stored at 4°C until filtering. Water samples were filtered within five hours on 47 mm, 0.45 μm Metricel Black membrane filters (Pall Corp, Ann Arbor, MI). The filters were transferred to 50 ml tubes, without sides touching and were shipped to the continental US at 4°C. Upon arrival in the US filters were placed on 50 mm petri plates containing one of two media and then incubated at 15°C. The media were yeast peptone dextrose (YPD) (BD, Sparks, MD) or 1/10 YPD, both with antibiotics as described in Connell et al. (Reference Connell, Redman, Craig, Scorzetti, Iszard and Rodriguez2008). Filters were inspected twice weekly for two years and colonies were isolated as previously described for soil (Connell et al. Reference Connell, Redman, Craig and Rodriguez2006).
Culture and ITS region identification of fungal isolates
Isolates were grown on YPD agar and single colonies selected. Single colonies of yeast and yeast-like isolates were grown in 5 ml YPD broth until an optical density (OD) at 600 nm reached 0.6 (Ultraspec 10; GE Lifesciences, Piscataway, NJ). A cell pellet was produced by centrifugation at 1 x 104 rpm (8624 x g) for 5 min (Eppendorf 5418 microcentrifuge, Eppendorf North America, Hauppauge, NY). Filamentous fungi were scraped off the growth medium into 1.5 ml of MasterPure Yeast DNA Purification Kit lysis buffer (Epicenter, Madison, WI) and then frozen at -80°C for at least 30 min before further processing. DNA was extracted from cell pellets (yeast and yeast-like isolates) or thawed lysate (filamentous fungi) with MasterPure Yeast DNA Purification Kit using the manufacturer's protocol. DNA concentrations were determined using a Nanoview spectrophotometer (GE Lifesciences). Specific ITS region amplicons were produced by PCR completed with 100 ng genomic DNA in 25 ul reactions using Illustra PuReTaq Ready-To-Go™ PCR Beads (GE Lifesciences). PCR primer set ITS5-ITS4 (White et al. Reference White, Bruns, Lee and Taylor1990) were used to target the ITS region for sequencing (Table I). Initial denaturation completed for 2 min at 95°C and 35 cycles with a PTC-200 thermal cycler (MJ Research, Watertown, MA) under the following conditions: 30 sec at 95°C, 30 sec at 52.3°C, 1 min at 72°C with a final 72°C 10 min extension. The resulting PCR products were cleaned using Promega SV Gel and PCR Clean-up System (Promega, Madison, WI) and were visualized using standard gel electrophoresis as previously described (Connell et al. Reference Connell, Redman, Craig, Scorzetti, Iszard and Rodriguez2008). The purified DNA was then used as a double-stranded sequencing template with primers ITS5 and ITS4 in sequencing reactions. Both strands were individually sequenced using Big Dye Terminator Cycle Sequencing in an ABI 377 autosequencer (Applied BioSystems, Foster City, CA). Resulting sequence chromatograms were analysed using Sequencher v 4.9 (GeneCodes, Ann Arbor, MI). Taxonomic identification of each isolate was made by comparison of ITS sequence with GenBank BLASTn (NCBI) data as well as by using physical characteristics of the isolate. Isolate ITS sequences from each species used in this project were submitted to GenBank (Table II).
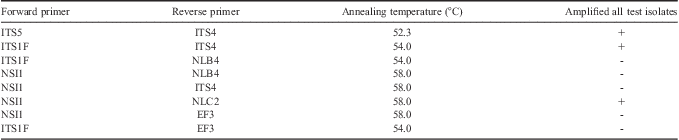
Table I Primer combinations tested for positive PCR amplification on select fungal isolates. Primer sources are as follow: ITS4, ITS5 (White et al. Reference White, Bruns, Lee and Taylor1990), ITS1F (Gardes & Bruns Reference Gardes and Bruns1993), EF3 (Smit et al. Reference Smit, Leeflang, Glandorf, van Elsas and Wernars1999), and NSI1, NLB4, NLC2 (Martin & Rygiewicz Reference Martin and Rygiewicz2005).
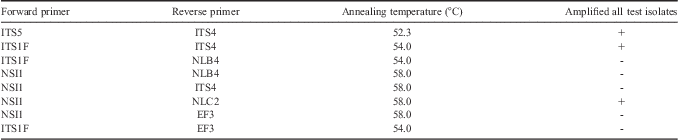
Table II Fungal species used in the development of automated ribosomal intergenic spacer analysis (ARISA) database. Species that cannot be individually resolved using ARISA Antarctic fungal fingerprints (ARISA-AFF) are marked with an asterisk.

ARISA-AFF primer selection and PCR parameters
To determine if selected primer pairs were able to amplify the ITS region of each fungal species in this study, eight sets of primer combinations including ITS4, ITS5 (White et al. Reference White, Bruns, Lee and Taylor1990), ITS1F (Gardes & Bruns Reference Gardes and Bruns1993), EF3 (Smit et al. Reference Smit, Leeflang, Glandorf, van Elsas and Wernars1999), and NSI1, NLB4, NLC2 (Martin & Rygiewicz Reference Martin and Rygiewicz2005) (Table I) were tested to ensure positive amplification with a selection of fungal isolates: Clavispora lusitaniae, Cryptococcus carnescens, Dioszegia cryoxerica, Dioszegia antarctica, Geomyces sp., Glaciozyma sp., Rhodotorula graminis and Thelebolus ellipsoideus. PCR was performed as described above with the exception of annealing temperature, where a 30 sec target annealing temperature for each primer pair was used. PCR products generated were visualized on a standard agarose gel as previously described (Connell et al. Reference Connell, Redman, Craig, Scorzetti, Iszard and Rodriguez2008). When no PCR amplicon was produced the cycling conditions used in the publications listed in Table II were used.
The PCR template concentration and extension time were optimized to maximize the fluorescence recorded from the ABI 3730 DNA Analyser (Applied Biosystems, Carlsbad, CA) without over-saturation of the instruments charged couple device (CCD) digital image sensor. An excess of PCR product will over-saturate the detector, resulting in broad, flat-topped peaks and therefore reduce the precision of size calling. To optimize the sample loading concentration for precision sizing of fragments, eight isolates were tested in duplicate with between 3 ng and 60 ng of PCR product (3 ng, 6 ng, 19 ng, 32 ng, 35 ng, 40 ng, 52 ng, 60 ng). Replicate samples with a final PCR extension step of 30 min compared with 10 min were also compared on ARISA-AFF. Samples were run and analysed as described in the following section.
ARISA-AFF database construction and analysis
Genomic DNA (gDNA) from fungal isolates (Table II) was amplified using the primer set ITS1F-ITS4. In cases where this primer set did not resolve closely related species using ARISA-AFF two other sets were used (ITS5-ITS4 and NSI1-NLB4) to determine if they would resolve the species. The reverse primer of each pair was labelled with 6-FAM carboxyfluorescein, succinimidyl ester and cycling parameters used are as described by Okubo & Sugiyama (Reference Okubo and Sugiyama2009). The resulting PCR products were cleaned using Promega SV Gel and PCR Clean-up System (Promega, Madison, WI) and amplicon concentrations were determined using a NanoVue spectrophotometer. Samples were diluted to 1.5 ng ul-1 with 2 ul placed into duplicate wells of a 96-well, half-skirt plate. Each well contained 0.5 ul of ROX rhodamine labelled standard Mapmarker 1000 (BioVentures, Murfreesboro, TN) and 7.5 ul formamide. One isolate (ANT03-026) required the use of a custom ROX standard that included a 1050 bp fragment (BioVentures) as its size exceeded the largest standard in MapMarker 1000. Sample plates were then heated for 3 min at 90°C and placed on ice. Capillary electrophoresis was applied to samples using a ABI 3730 DNA Analyser run for 40 min at 15 kV, with 1.6 kV injection for 15 sec on a POP7 polymer in a 50 cm array at 60°C. Resultant chromatograms were viewed and analysed using GeneMarker version 1.91 (SoftGenetics, State College, PA) and fragments sized using default analysis parameters with local southern selected as a peak sizing method and large-size fragment algorithm. These parameters included smoothing, corrections for peak saturation, pull-up, baseline subtraction and spike removal. Binning was not used for sizing fragments for identification of fungal isolates. Fragments were sized visually using the peak centre for sizing or for isolates with multiple peaks a mean of the peak sizes was used as an estimate of fragment size. Isolates were considered un-resolvable as distinct peaks when they were overlapping (typically < 1 bp in size difference). Each sample was loaded into three wells per instrument run and three independent PCR reactions were analysed per species. There was very high replication among samples and instrument runs.
Results
ARISA chromatographs for 46 fungal species commonly isolated from south Victoria Land were compared to compile a fingerprint database for rapid identification of fungal isolates. Of these 46 species, ARISA analysis of 34 species (78%) resulted in clearly identifiable chromatograms (Table II). Only three primer combinations amplified all templates (Table I) from test isolates using cycling parameters specified above. Primer pair NSI1-NLC2 yielded fragments that were too large to be sized with the internal lane standard. A commonly used fungal primer, EF3 did not amplify all of the isolates under conditions tested. Fragment sizes using ITS1F-ITS4 ranged from 397.9 bp (Yarrowia lipolytic) to 1006.5 bp (Geomyces ribotype 3) with most clustered between 589 and 690 bp. Signal peaks for many of the isolates showed a single, sharp, diagnostic peak that allowed easy characterization of the isolate as a narrow range of sizes (Table II). Isolates that were not positively identified using ARISA-AFF were identified by isolate morphology and DNA sequencing of the ITS region.
In testing various template concentrations for ARISA-AFF analysis we found 3 ng of gDNA to be adequate for maximizing instrument signal and all subsequent samples were run at this concentration. The effect of longer PCR final extension times on replication of chromatogram peaks and detection of any “false peaks” was tested by running a subset of samples with a 30 min extension. There was no effect with additional extension times in discriminating false peaks within replicates. Therefore, all subsequent samples were processed with the 10 min final extension. Signal “pull-up”, which may appear in one or more dye channels, occurs as a result of strong florescence in a particular dye channel and a sharp peak in the resulting electropherograms. An increased response in the detector, sometimes referred to signal “bleed through”, in the automated sequencer often occurs resulting in a false fluorescent signal in other dye channels. There was more signal pull-up in lane standard dye channel in these samples but the GeneMarker program compensates and corrects this effect. Otherwise, we did not find any effect on accurately sizing fragments (data not shown) and had excellent replication of major diagnostic peaks with little background signal.
This ARISA-AFF technique was successful in discriminating among some closely related taxa (Table II). For example, the four species of Glaciozyma were clearly resolved (Fig. 1). In addition, the Glaciozyma and Dioszegia species in this study were all separated to the species level along with some closely related Tremellales Cryptococcus species (e.g. C. carnescens and C. victoriae). Many of the more distantly related Filobasidiales Cryptococcus species were able to be resolved from one another (e.g. C. magnus, C. satoi and C. vishniacii/C. albidosimilis group) but the C. vishniacii/C. albidosimilis group itself did not separate.

Fig. 1 Combined electropherograms from four species of Glaciozyma demonstrating clear separation of species. The x-axis shows estimated peak size in base pairs (bp).
Heterogeneous ITS sequences within a species may create broad or multiple peaks that showed diagnostic peak sets. Several species in this study exhibited complex and diagnostic peaks using the ITS1F-ITS4 primer set. An example of this is shown in Fig. 2 where chromatograms of individual species (e.g. D. cryoxerica, C. lusitaniae, Y. lipolytica and Cladosporium langeronii) are shown to have more than one peak. For these species the size was estimated as the centre of the group of peak signals.

Fig. 2 Automated ribosomal intergenic spacer analysis (ARISA) profiles of four individual fungal species that demonstrate diagnostic peak patterns using primer pair ITS1F and ITS4. The x-axis shows peak size in base pairs (bp) of each species. a.Clavispora lustaniae, b.Yarrowia lipolytica, c.Cladisporium lageronii, and d.Dioszegia cryoxerica.
Some taxa had peaks that were very similar using the ITS1F-ITS4 primer set (Table II) yet were resolvable using additional primer pairs. For example, the NLB-NSI1 primer set separated Aureobasidium pullulans from Penicillium commune, P. corylophilum and P. dipodomyicola. In addition, Cladosporium sphaerospermum and Tetracladium sp. (isolate ANT CW08-156) were resolved using the ITS5-ITS4 primer set. Other species were not resolved to the species level with any primer set used (e.g. Cryptococcus saitoi and Mrakiella aquaticus) and these isolates were then subjected to direct sequencing and/or morphological/physiological criteria for identification.
This ARISA database also allowed for the identification of unpurified cultures (Fig. 3). Six of the initial isolates from our Lake Fryxell samples were identified as “mixed” by the existence to two or more peaks that corresponded to previously identified taxa. Subsequent ARISA-AFF analysis following re-isolation and separation of the cultures confirmed the identification of each of the species from the mixed sample.

Fig. 3 Automated ribosomal intergenic spacer analysis (ARISA) profile of a mixed Lake Fryxell fungal culture generated via PCR amplification with ITS1F-ITS4 primers. The peaks show species names based on Table II. The two yeasts, Glaciozyma watsonii and G. antarctica were co-isolated with Aureobasidium pullulans.
Discussion
In this work we developed an Antarctic fungal ARISA database suitable for rapid identification and sorting into groups of isolates generated from culture-based abundance studies. The use of ARISA-AFF for more cost effective identification of cultured fungal isolates has to be balanced with the consideration that new isolates not in the database may be cultured and therefore will require the same primary analysis before their chromatograms are entered into the ARISA-AFF database. Thus, this technique may be quite useful in initial screening of cultures into higher taxonomic groups for further analysis. In addition, ARISA peaks are viewed in context with isolate morphology. Therefore, gross morphology (e.g. filamentous vs yeast-like) that can clearly be easily discriminated visually help in identification as a second-tier of analysis. The identification of cultures that contain more than one species by comparison with library peaks has been helpful in several respects. First, if these cultures were thought to be single species isolates and were sent directly for sequencing no useable data would have been produced. This method provides identification and the option to re-culture and isolate component species if desired. Second, since many fungi can have similar gross morphology, identification of taxa that are not already in the database will allow for more rapid detection of potential new species. It remains possible that there may be species with very similar morphology and ARISA profiles. In this case, DNA sequencing of some isolates representing specific morphologies may be useful.
Accuracy of this method was higher than anticipated, based on similar studies, with c. 2 bp resolution up to 700 bp and 3 bp for > 700 bp. However, Fisher & Triplett (Reference Fisher and Triplett1999) found a similar level of accuracy in fragment sizing. The higher precision was probably due to a lower baseline resulting from the use of cleaned PCR products that removed non-specific targets (e.g. primer dimers) and excess reagents. Quantitation of individual PCR products and standardization of DNA concentration used for this ARISA technique helped achieve consistent high precision and sharp, unsaturated signals with a high relative fluorescence unit value (RFU). Nonetheless, much of the data analysis was done visually as complications with multiple peaks and peak “shoulders” did not lend to data analysis automation. Regardless, a large number of isolates can be screened and identified to the species level at a lower cost than sequencing.
The complex peaks found in several species (e.g. D. cryoxerica, C. lusitaniae) may be a based on the fact that these species have intraspecific variation of the ITS sequence in each cell (Lachance et al. Reference Lachance, Daniel, Meyer, Prasad, Gautam and Boundy-Mills2003, Connell et al. Reference Connell, Redman, Rodriguez, Barrett, Iszard and Fonesca2010). This variability in ITS does not affect identification of these isolates, in fact the peaks are very distinctive, but it does make future analysis automation more problematic.
The ARISA technique itself may be useful in discriminating fungal communities among treatments or habitats. This Antarctic fungal ARISA-AFF database can be used for identification of species in the traditional ARISA analysis of microbial communities, but it should be done so with some caution. For example, traditional peak calling would identify three phylotypes for D. cryoxerica. However, when environmental samples have few distinctive peaks (< 20) some of the peaks may be identified using this database, especially when combined with information obtained by culturing species from the sample. Therefore, the use of the ARISA-AFF for species identification from environmental samples is of limited utility and not its intended purpose.
Acknowledgements
The authors would like to thank Megan Altenritter, Katie Earle and Ben Segee for laboratory support, Regina Redmond, Rusty Rodriguez, Scott Craig, Amy Chiuchiolo, Tristy Vick, Andrew Barber and Hubert Staudigel for help in field sample collection as well as Raytheon Polar Services and PHI for logistical support. The authors also thank the anonymous reviews that helped improve this manuscript. This work was supported in part by NSF OPP-0125611, NSF ANT 0739696, as well as a University of Maine HEIR grant.
Supplemental material
A supplemental table will be found at http://dx.doi.org/10.1017/S0954102012000879.