INTRODUCTION
The external surfaces of sea turtles, like many marine organisms, often serve as a substratum for a diverse array of epibionts. The epibiont diversity of various sea turtle species has been characterized in many areas, including the Western Atlantic Ocean (Caine, Reference Caine1986; Frick et al., Reference Frick, Williams and Robinson1998; Pfaller et al., Reference Pfaller, Frick, Reich, Williams and Bjorndal2008), Western Pacific Ocean (Hayashi & Tsuji, Reference Hayashi and Tsuji2008; Hayashi, Reference Hayashi2009) and the Mediterranean Sea (Fuller et al., Reference Fuller, Broderick, Enever, Thorne and Godley2010; Domènech et al., Reference Domènech, Badillo, Tomás, Raga and Aznar2015). Yet the majority of these studies have been largely descriptive (presence/absence data) and focused only on a single sea turtle species at a single location. Consequently, it is not well understood whether epibiont communities are specific to particular host species and what factors influence epibiont community composition.
Frick & Pfaller (Reference Frick, Pfaller, Wyneken, Lohmann and Musick2013) stated that a necessary prerequisite for epibiosis is a geographic and ecological overlap between the host and the epibiont. Thus, it may be expected that sea turtles occupying similar habitats in similar locations would have similar epibiont communities. However, sea turtles are highly migratory and different sea turtle species may only share habitats at certain times of the year (e.g. at nesting areas during the breeding season). As most epibiont surveys are conducted on nesting beaches, the question therefore arises whether epibiont communities of nesting sea turtles are more influenced by the surrounding nesting habitats or the foraging areas that turtles inhabit before their pre-nesting migrations. If epibiont communities are primarily determined by the location of the hosts’ nesting habitats, it might be expected that sympatrically nesting turtles should host similar epibiont communities. In addition, the epibiont communities of non-conspecific, sympatrically nesting turtles should be more similar to each other than to conspecific population nesting in other locations. Alternatively, if epibiont communities are primarily determined by the location of the hosts’ foraging habitats, it might be expected that turtles with similar foraging habitats should host similar epibiont communities. Moreover, turtle species with more diverse foraging behaviour should have more diverse epibiont communities.
Many nesting beaches in the East Pacific Ocean are shared by leatherback Dermochelys coriacea, olive ridley Lepidochelys olivacea and green Chelonia mydas turtles; however, each of these species have distinct foraging habitats. Leatherback turtles, which are obligate gelatinous zooplanktivores (Wallace et al., Reference Wallace, Zolkewitz and James2015), forage in vast open-ocean habitats off the coast of Ecuador, Peru and Chile (Shillinger et al., Reference Shillinger, Palacios, Bailey, Bograd, Swithenbank, Gaspar, Wallace, Spotila, Paladino, Piedra, Eckert and Block2008). Olive ridley turtles, which are opportunistic omnivores (Colman et al., Reference Colman, Sampaio, Weber and Comin De Castilhos2014; Behera et al., Reference Behera, Tripathy, Sivakumar and Choudhury2015), do not appear to migrate to a specific foraging area but instead wander nomadically in search of food (Plotkin et al., Reference Plotkin, Byles, Rostal and Owens1995). Consequently, olive ridley turtles have been recorded inhabiting both oceanic and coastal waters depending on inter-annual patterns in food availability (Plotkin, Reference Plotkin2010). Lastly, East Pacific green turtles (referred to subsequently only as green turtles), which are primarily herbivorous (Seminoff et al., Reference Seminoff, Resendiz and Nichols2002; Amorocho & Reina, Reference Amorocho and Reina2007), forage in shallow coastal habitats, such as gulfs or estuary mouths (Blanco et al., Reference Blanco, Morreale, Bailey, Seminoff, Paladino and Spotila2012; Hart et al., Reference Hart, Blanco, Coyne, Delgado-Trejo, Godley, Jones, Resendiz, Seminoff, Witt and Nichols2015; P. Santidrián Tomillo & N.J. Robinson, unpublished data).
By combining the hypotheses above concerning the geographic patterns of epibiont community structure with knowledge of sea turtles’ nesting and foraging behaviour, we can make some specific predictions about the epibiont communities of East Pacific sea turtles. If the epibiont communities of nesting turtles are primarily influenced by nesting habitats, we predict that the epibiont communities of sympatrically nesting leatherback, olive ridley and green turtles will be more similar to each other than to conspecific nesting populations at more distant nesting areas. Alternatively, if epibiont communities are more influenced by foraging habitats, we predict that leatherback and green turtles will have different epibiont communities, but both should share epibionts with olive ridley turtles. Furthermore, leatherback turtles should have low epibiont diversity because they inhabit relatively uniform oceanic habitats that are likely to have low diversity in potential epibionts. Olive ridley turtles should have high epibiont diversity because they inhabit a mix of oceanic and coastal environments. Green turtles should have medium to high epibiont diversity because, although this species is primarily coastal, these coastal habitats are often complex and may contain a greater diversity of potential epibionts.
We tested these predictions by characterizing and comparing the epibiont communities of leatherback, olive ridley and green turtles nesting within Parque Nacional Marino Las Baulas on the Pacific Coast of Costa Rica. We also investigated how epibiont community structure differs among these sympatrically nesting species and conspecific populations nesting at other locations in the East Pacific Ocean. This is the first study to statistically compare the epibiont communities of different sea turtle species in the East Pacific Ocean and the first study to investigate the epibiont community structure of leatherback turtles and green turtles on the Pacific coast of Costa Rica.
MATERIALS AND METHODS
Study site
All sampling was conducted on Playa Grande and Playa Ventanas, two sandy beaches that are located within Parque Nacional Marino Las Baulas (PNMB) on the Pacific coastline of the Guanacaste Province in northern Costa Rica (10°20′N 85°51′W).
Sampling protocol
The beaches of PNMB were patrolled nightly between October 2014 and March 2015 to encounter nesting turtles. To avoid interrupting the nesting process, epibiont sampling and tagging were carried out after oviposition had begun. Tagging was achieved using a combination of metal or passive integrated transponder (PIT) tags depending on the species.
After tagging and measuring, each turtle was visually inspected for epibionts. When possible, olive ridley and green turtles were briefly (<5 min) flipped onto their carapace to also inspect ventral surfaces. This was not attempted for leatherback turtles due to their size. All epibionts were removed using a knife or tweezers and preserved in vials of 95% denatured ethanol.
We attempted to collect all visible epibionts from each individual but this was only possible for olive ridley and green turtles. We were not able to exhaustively sample epibionts from leatherback turtles due to their high epibiont loads and the difficulties associated with restraining them.
After collection, all epibionts were counted and identified to the lowest taxonomic level by consulting appropriate literature. The samples were exported to the Yale Peabody Museum of Natural History, USA. On their arrival at the museum, the taxonomy was cross-checked and the samples were catalogued for curation into the Museum's collection. Further information and photos on the samples are available at http://peabody.yale.edu/collections.
Sample size considerations
When characterizing epibiont community structure, there is often a concern that not enough host individuals are sampled to adequately represent the epibiont species richness of the population. To address this issue, we used epibiont presence/absence datasets to estimate the rate at which species richness increased with sample size (termed rarefaction curves) following the Bernoulli product model (Colwell et al., Reference Colwell, Chao, Gotelli, Lin, Mao, Chazdon and Longino2012). Furthermore, as rarefaction curves can be used to reasonably extrapolate species richness up to double or triple the reference sample size (Colwell et al., Reference Colwell, Chao, Gotelli, Lin, Mao, Chazdon and Longino2012), we estimated how many taxa would be found if sample sizes were increased to 40 for each turtle species. Sample-based rarefaction and extrapolation curves as well as their 95% confidence intervals were calculated using the program EstimateS V.9.
Comparing epibiont community structure
For each sea turtle species, we calculated four metrics that characterized the epibiont community structure. (1) The total number of unique epibiont taxa recorded on each sea turtle species. (2) The mean abundance of epibionts on each sea turtle host. (3) The Dominance Index (or 1 – Simpson's Index) – this is a measure of the probability that two epibionts selected from a single host turtle will belong to the same taxon. With this index, 0 represents a 100% probability that both epibionts are from the same taxon meaning there is low diversity in the sample, while numbers converging on 1 represent a decreasing probability that both epibionts are from the same taxon and thus means there is high diversity in the sample. (4) The Shannon–Wiener Index – a diversity index that takes into account both the number of individuals and the number of taxa. The value of this diversity index increases when epibiont species richness increases and when evenness in abundance among taxa increases.
To compare the epibiont community structure of different sea turtle species we used a one-way ANOSIM (ANalysis Of SIMilarities) using the Bray–Curtis index of similarity. Although it is possible to use the Bray–Curtis indices of similarity to compare raw abundance data, we used frequency of presence/absence data as it was not possible to exhaustively collect epibiont samples from most leatherback turtles. Significance was computed through permutation by group membership with 9999 replicates. We also examined the contribution of individual epibiont taxa to any separation identified between sea turtle species in the ANOSIM by following the SIMPER procedure.
We examined the similarity between the epibiont communities of sea turtles in PNMB and conspecific sea turtle populations in East Pacific Ocean. Specifically, we compared the data collected in this study to samples collected from olive ridley turtles nesting at Playa Ostional, Costa Rica (see Majewska et al., Reference Majewska, Santoro, Bolaños, Chaves and De Stafano2015) and olive ridley and green turtles nesting at Jalisco, Mexico (see Lazo-Wasem et al., Reference Lazo-Wasem, Pinou, Peña de Niz and Feuerstein2011), which is about 2300 km north-east of PNMB (Figure 1). The frequency of epibiont occurrence was compared between species and locations using hierarchical clustering analysis and Ward's method (Ward, Reference Ward1963). Each model was bootstrapped through 20,000 random permutations to determine the bootstrap probability (percentage of replicates that support each node) of each separation. Although Lazo-Wasem et al. (Reference Lazo-Wasem, Pinou, Peña de Niz and Feuerstein2011) and Majewska et al. (Reference Majewska, Santoro, Bolaños, Chaves and De Stafano2015) are not the only studies to have categorized sea turtle epibiont communities in the East Pacific Ocean (see Hernández-Vásquez & Valadez-González, Reference Hernández-Vásquez and Valadez-González1998; Gámez Vivaldo et al., Reference Gámez Vivaldo, Ororio Sarabia, Peñaflores Salazar, García Hernández and Ramírez Lezema2006; Angulo-Lozano et al., Reference Angulo-Lozano, Nava-Duran and Frick2007), these other studies only indicated the presence or absence of different epibiont taxa and did not indicate the frequency at which these epibionts were found. Consequently, only data from Lazo-Wasem et al. (Reference Lazo-Wasem, Pinou, Peña de Niz and Feuerstein2011) and Majewska et al. (Reference Majewska, Santoro, Bolaños, Chaves and De Stafano2015) were used for statistical comparisons. Leatherback turtles were also not included in these analyses due to a lack of appropriate regional comparisons.
Fig. 1. Map of the Pacific coast of Central America highlighting the primary sampling location (Parque Nacional Marino Las Baulas) as well as other nearby locations (Teopa Beach & Playa Ostional) where published recorded of epibiont communities are available.
RESULTS
Characterizing epibiont communities
Epibionts were present on each of the 43 turtles sampled (18 leatherback turtles, 19 olive ridley turtles and six green turtles) in PNMB. In total, we collected 2571 epibionts belonging to 18 unique taxa. For a full list of these taxa see Table 1.
Table 1. Individual epibiont identification, abundance and frequency per host species of leatherback (Dermochelys coriacea), olive ridley (Lepidochelys olivacea), and green (Chelonia mydas) sea turtles nesting on Parque Nacional Marino Las Baulas, Costa Rica.

a Taxon represents presence only. Individual counts were not undertaken.
b The count only includes ‘adults’ and not complemental males.
c Only the stalks of these lepadomorphs were present and so it was not possible to determine the species.
d It is not certain that the Caecum sp. were epibionts or were in the sand that was thrown onto the carapace while the turtle was nesting.
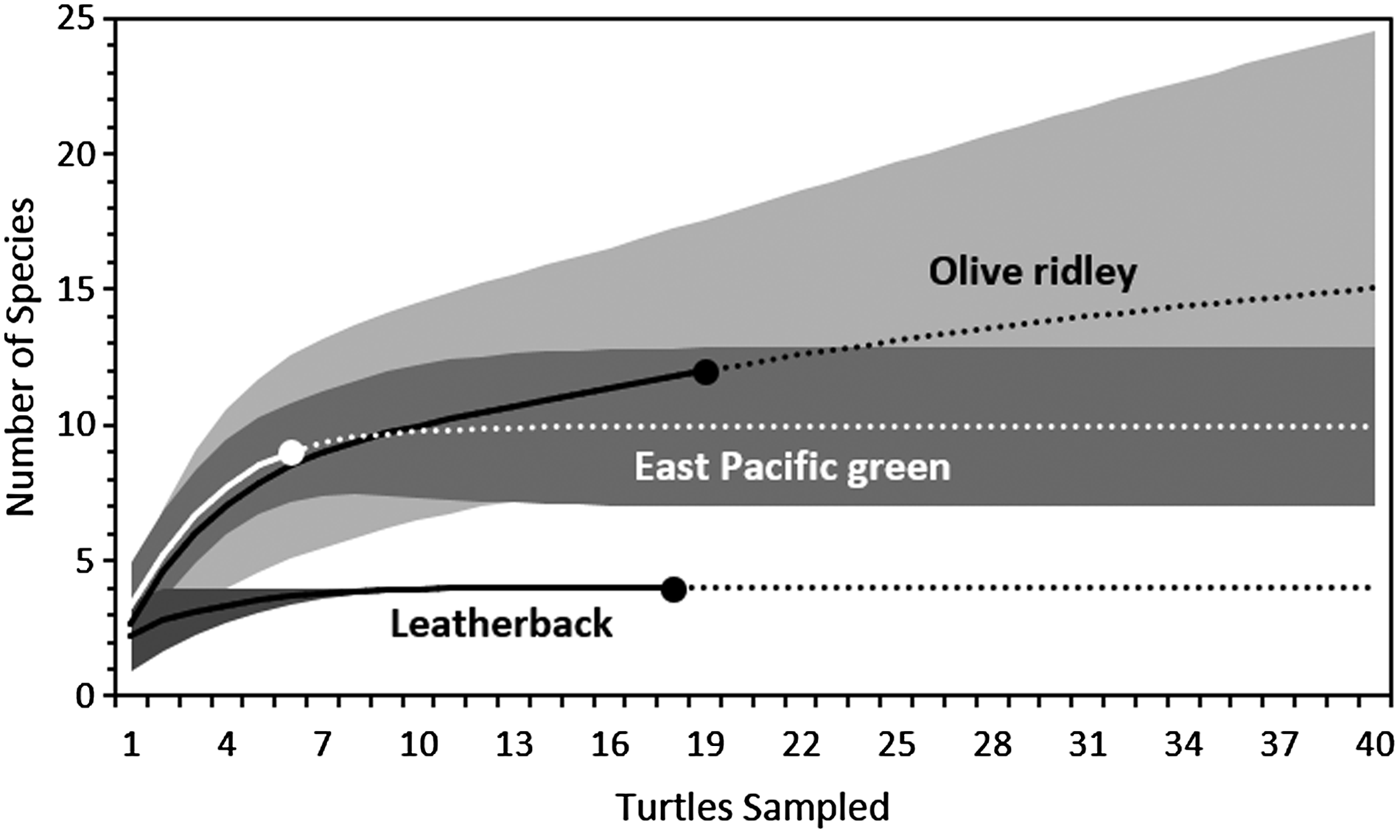
The rarefaction curves indicated that if the sample size was increased to 40 for each host species, only four more taxa would be encountered (three on olive ridley turtles and one on green turtles; Figure 2). This suggests that we recorded the majority of epibiont taxa found in nesting sea turtles in PNMB. Consequently, the differences in sample size between the sea turtle species should be suitable for statistical comparison and should not have a pronounced effect on the species diversity indices.
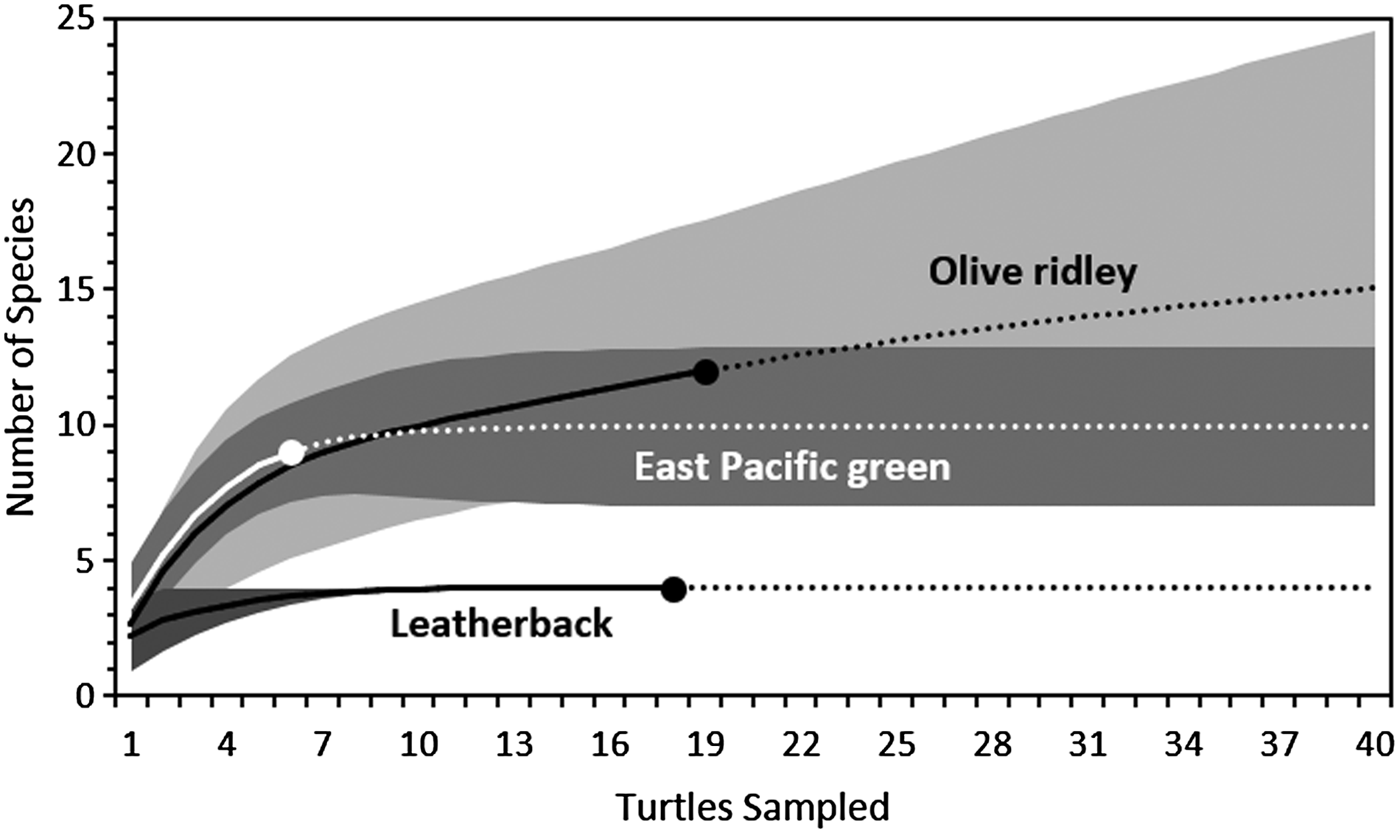
Fig. 2. Species-richness rarefaction curves for the epibionts per host host species found on leatherback (Dermochelys coriacea), olive ridley (Lepidochelys olivacea) and green (Chelonia mydas) turtles nesting on Parque Nacional Marino Las Baulas, Costa Rica. Solid lines represent the modelled rate of increase for species richness with increasing sample size, while dotted lines represent extrapolated data to predict species richness up to 40 samples. Dark grey, grey and light grey areas represent the 95% confidence interval for the rarefaction curves for leatherback, green and olive ridley turtles respectively. For clarity, the lower limits of the confidence interval for olive ridley turtles is hidden behind that of the green turtles.
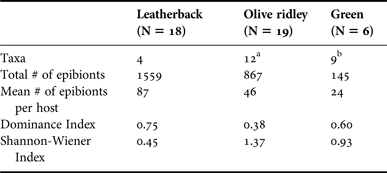
Leatherback turtles only hosted four epibiont taxa, yet they hosted a mean of 86 epibionts per host (Table 2). Furthermore, the low Shannon–Wiener Index (0.45) and high Dominance Index (0.75) of epibiont communities on leatherback turtles indicated that these communities were dominated by a single taxon. This taxon was the barnacle Platylepas coriacea, which constituted 85.1% of all the epibionts sampled from leatherback turtles. Contrastingly, Platylepas coriacea was not observed on either olive ridley or green turtles.
Table 2. Measures of overall epibiont abundance, dominance, and species richness per host species of leatherback (Dermochelys coriacea), olive ridley (Lepidochelys olivacea), and green (Chelonia mydas) turtles nesting on Parque Nacional Marino Las Baulas, Costa Rica.

a Does not include the unidentified lepadomophs.
b Does not include the unidentified Platylepas sp. or Caecum sp.
Olive ridley turtles hosted 12 epibiont taxa, which is more than either leatherback or green turtles, and hosted a mean of 46 epibionts per host. Furthermore, the high Shannon–Wiener Index (1.37) and low Dominance Index (0.38) of the epibiont communities of olive ridley turtles suggests that their epibionts were present in more equal abundances than the epibionts on either leatherback or green turtles (Table 2). Two barnacle species Platylepas decorata and Stomatolepas elegans, an unidentified limpet, a muricid snail, and the leech Ozobranchus branchiatus were exclusively found on olive ridley turtles.
Green turtles hosted nine epibiont taxa but only hosted a mean of 24 epibionts per host, less than either leatherback or olive ridley turtles. Furthermore, the epibiont communities of green turtles were of intermediate diversity when compared with those of olive ridley and leatherback turtles as illustrated by a Shannon–Wiener Index value of 0.93. They also had an intermediate dominance value of 0.60, which probably reflects the presence of the barnacle Chelonibia testudinaria on every individual sampled. Caecum sp., Hyachelia tortugae, Megabalanus coccopoma and the tanaid were exclusively found on green turtles.
Only seven of the 18 epibiont taxa were recorded on more than one host turtle species. Conchoderma virgatum was found on both leatherback and olive ridley turtles, while filamentous algae, Balaenophilus manatorum, Chelonibia testudinaria, Conchoderma virgatum, Platylepas hexastylos and Podocerus chelonophilus were found on olive ridley and green turtles. Only a single taxon, Lepas spp., was found on all three sea turtle species.
Taxonomic considerations
The epibiont taxa that could not be identified to species level or quantified readily are listed below. (1) It is likely that both Lepas hilli and Lepas anatifera were present in the leatherback samples, yet we refer to them collectively as Lepas spp. due to the difficulty of differentiating between these species (Lazo-Wasem et al., Reference Lazo-Wasem, Pinou, Peña de Niz and Feuerstein2011). (2) Many of the turtles had patches of filamentous algae either attached directly to the carapace or to other epibionts. As algae can be near microscopic, easily overlooked, and difficult to quantify, we recorded which individuals had ‘clumps’ of algae attached to the carapace of the basibiont, but did not record the presence of algae when it was only small strands that were attached to individual epibionts. (3) Two blind shells, micromolluscs of the genus Caecum, were found in a single sample, yet these are probably not true epibionts. Blind shells are gastropods that commonly feed on the microflora of sand and they could have been present in the sand that the turtle had thrown on its carapace during nesting. Thus, they were not included in Table 3 nor in any measures of species richness or diversity. (4) A single olive ridley individual was found with 20 stalked barnacles that were only identifiable as lepadomorphs as their capitula (shell and body) were broken off and only the stalks remained. (5) Two green turtles were found with a total of 10 barnacles that could only be identified as Platylepas sp. due to the difficulty surrounding the taxonomy of Platylepas decorata (Frick & Zardus, Reference Frick and Zardus2010).
Table 3. Results of the SIMPER procedure to identify the relative contribution of each epibiont species to any dissimilarity between the epibiont communities of leatherback (Dermochelys coriacea), olive ridley (Lepidochelys olivacea), and green (Chelonia mydas) nesting on Parque Nacional Marino Las Baulas, Costa Rica.

Statistical comparisons
The ANOSIM identified statistically significant differences in the epibiont community structures of leatherback and olive ridley turtles (P < 0.001) as well as leatherback and green turtles (P < 0.001); however, no statistically significant difference was found in epibiont community structure of olive ridley and green turtles (P = 0.496). The SIMPER analysis showed that the dissimilarity between leatherback and olive ridley turtles was primarily due to differences in the frequency of Platylepas coraicea (21.73%; Table 3). Conversely, the largest difference between green turtles and both leatherback and olive ridley turtles (21.04 and 14.05% respectively) was due to differences in the frequency of Chelonibia testudinaria (Table 3).
The results from the HCA were consistent with the results of the ANOSIM. The epibiont communities of leatherback turtles were less similar to olive ridley and green turtles (distance = 209, bootstrap probability of 100%) than either olive ridley or green turtles were to each other (distance = 162, bootstrap probability of 27%) (Figure 3). Furthermore, the epibiont communities of olive ridley and green turtles at PNMB were more similar to conspecific populations nesting at Ostional and Jalisco than to sympatrically nesting, non-conspecifics.

Fig. 3. Hierarchical cluster analysis using Ward's method for the epibiont community structures of leatherback (Dermochelys coriacea), olive ridley (Lepidochelys olivacea) and green (Chelonia mydas) turtles nesting on Parque Nacional Marino Las Baulas, Costa Rica as well as olive ridley and green turtles nesting on Jalisco, Mexico. Each model was bootstrapped through 20,000 random permutations to determine the bootstrap probability for each node (values displayed above each node).
DISCUSSION
Interest in sea turtle epibiosis has grown in recent years, yet the majority of this research has been descriptive. To move the field forward, we applied a hypothesis-driven approach to investigate how the habitat preferences of sea turtles may affect their epibiont communities.
Based on the assertion that a necessary prerequisite for epibiosis is a geographic and ecological overlap between the host and the epibiont (Frick & Pfaller, Reference Frick, Pfaller, Wyneken, Lohmann and Musick2013), we proposed two alternative hypotheses to explain the epibiont community structure of nesting sea turtles. If epibiont communities of nesting sea turtles are primarily determined by the location of the hosts’ nesting area, we predicted that the epibiont communities of sympatrically nesting sea turtles should be similar. Alternatively, if epibiont communities of nesting sea turtles are primarily determined by the location of the hosts’ foraging habitats, then we predicted that the epibiont communities of sympatrically nesting turtles should be different and instead influenced by the location of the foraging areas.. For leatherback, olive ridley and green turtles nesting in PNMB, we found the leatherback turtles in PNMB have epibiont communities that are statistically distinct from those of the sympatrically nesting olive ridley and green turtles. Furthermore, the epibiont communities of olive ridley and green turtles in PNMB were more similar to conspecifics from other nesting beaches: Ostional (50 km away) and Jalisco (2300 km away), than to each other.
The similarity between the epibiont communities of distantly nesting conspecifics suggests that these individuals may share behaviours or biotic traits that are important for determining epibiont community structure. As sea turtles can migrate thousands of km between nesting and foraging areas, even distantly nesting conspecifics can still share foraging habitats (Bailey et al., Reference Bailey, Benson, Shillinger, Bograd, Dutton, Eckert, Morreale, Paladino, Eguchi, Foley, Block, Piedra, Hitipeuw, Tapilatu and Spotila2012; Hart et al., Reference Hart, Blanco, Coyne, Delgado-Trejo, Godley, Jones, Resendiz, Seminoff, Witt and Nichols2015). Thus, as olive ridley turtles share both oceanic foraging habitats with leatherback turtles and coastal foraging habitats with green turtles, it would be expected for them to have intermediary epibiont communities between leatherback and green turtles. Yet the epibiont communities of olive ridley turtles were far more similar to green turtles than leatherback turtles. As the prevalence of coastal or oceanic foraging in olive ridley turtles appears largely dependent on the prevailing oceanographic conditions (Plotkin, Reference Plotkin2010), this could suggest that over the duration of this study the prevailing oceanographic conditions favoured coastal foraging in olive ridley turtles. Indeed, the study period overlapped with an El Niño event, which has been previously linked to increased coastal foraging in olive ridley turtles (Plotkin, Reference Plotkin2010).
If the hosts’ foraging areas are strongly influencing epibiont communities, then we also predicted that turtle species with more diverse foraging behaviour should have more diverse epibiont communities. Fitting these predictions, leatherback turtles, which occupy large, yet relatively uniform, oceanic foraging habitats, had the lowest epibiont diversity. Olive ridley turtles, which occupy a range of coastal and oceanic habitats, had the highest epibiont diversity. Finally, green turtles, which occupy a range of small, yet relatively complex, coastal foraging habitats, had similarly high epibiont diversity.
While it might be assumed that sympatrically nesting sea turtles would have similar epibiont communities if they are exposed to similar communities of potential epibionts in the habitats near the nesting beach, this might does always occur. Different sea turtle species may occupy widely different habitats during the inter-nesting interval (the time between consecutive nesting events in a single nesting season). For example, inter-nesting leatherback turtles tracked from PNMB tended to occupy waters up to 100 km from the nesting beach (Shillinger et al., Reference Shillinger, Swithenbank, Bograd, Bailey, Castelton, Wallace, Spotila, Paladino, Piedra and Block2010), whereas olive ridley and green turtles from nearby beaches rarely moved more than 25 and 5 km from the nesting beach respectively (Plotkin et al., Reference Plotkin, Byles, Rostal and Owens1995; Blanco et al., Reference Blanco, Morreale, Seminoff, Paladino, Piedra and Spotila2013).
The greater similarity between the inter-nesting habitats of olive ridley and green turtles than leatherback turtles could therefore explain the observed differences between epibiont communities. If this is the case, then it may also be expected that leatherback turtles, which have the largest and potentially most diverse inter-nesting habitats, would also host the greatest diversity of epibionts. Yet this was not true and leatherback turtles actually have the least diverse epibiont communities (leatherbacks: four taxa; olive ridleys: 12 taxa; greens: nine taxa). Furthermore, we discovered that the epibiont communities of conspecific individuals from distant nesting beaches were more similar to each other than sympatrically nesting non-conspecifics. It is highly unlikely that these distantly nesting conspecifics have more overlap in their nesting habitats than even sympatrically nesting non-conspecifics. Thus, we conclude that the location of inter-nesting habitats is not the primary factor influencing the diversity and constituency of epibiont communities.
Another potential explanation for the difference between host species could be that leatherback turtles provide unique challenges to epibiont colonization when compared with olive ridley or green turtles. For example, leatherback turtles can dive to depths exceeding 1000 m (Houghton et al., Reference Houghton, Doyle, Davenport, Wilson and Hays2008), while olive ridley and green turtles rarely descend beyond 200 m (Hays et al., Reference Hays, Åkesson, Broderick, Glen, Godley, Luschi, Martin, Metcalfe and Papi2001; Polovina et al., Reference Polovina, Howell, Parker and Balazs2002). Deep diving leatherback turtles experience rapid changes in pressure and temperature, which may be inimical for some epibiont taxa. Consequently, only epibionts that were able to withstand such temperature changes would be able to survive on a leatherback turtle host. Nevertheless, many shallow water invertebrates can survive rapid changes in pressure and temperature far exceeding those that may be experienced by a diving leatherback turtle (Mestre et al., Reference Mestre, Thatje and Tyler2009; Robinson et al., Reference Robinson, Thatje and Osseforth2009; Oliphant et al., Reference Oliphant, Thatje, Brown, Morini, Ravaux and Shillito2011). Thus, differences in diving behaviour between these three sea turtle species does not entirely explain their differences in epibiont communities.
A more likely alternative explanation for the unique epibiont communities of leatherback turtles compared with olive ridley and green turtles is differences in the surface properties of the skin and carapace. The skin of leatherback turtles, unlike the ‘hard-shelled’ olive ridley and green turtles, is smooth and not armoured by scutes. In addition, the carapaces of leatherback turtles are covered in skin, while those of olive ridley and green turtles are composed of exposed keratin and are likely to be far more inert. The surface properties (e.g. chemical composition, texture or wettability) of leatherback substratum may therefore be more different from olive ridley and green turtles than the latter two are to each other. Leatherback skin may provide a challenging substrate for many epibionts, which would explain the lack of diversity of epibionts that were found on leatherback turtles in this study.
Lastly, it is possible that population trends may also be influencing epibiont communities. Leatherback turtles have declined by over 98% in the Eastern Pacific Ocean in the past 25 years (Spotila et al., Reference Spotila, Reina, Steyermark, Plotkin and Paladino2000; Santidrián Tomillo et al., Reference Santidrián Tomillo, Vélez, Reina, Piedra, Paladino and Spotila2008; N.J. Robinson, unpublished data), while olive ridley and green turtle populations appear far more stable (Fonseca et al., Reference Fonseca, Murillo, Guadamúz, Spínola and Valverde2009; Delgado Trejo & Díaz, Reference Delgado Trejo, Diaz, Seminoff and Wallace2012). The rapid decline in the population size of leatherback turtles could have also led to a disassociation between these turtles and their epibionts. Indeed, there may be a minimum population density of hosts that are required to support a viable population of epibionts. This could explain why Stomatolepus elegans is commonly found on leatherback turtles in the Atlantic (Carriol & Vader, Reference Carriol and Vader2002; Frick et al., Reference Frick, Zardus and Lazo-Wasem2010), where leatherback turtle populations are stable (IUCN, 2014), but is notably absent on leatherback turtles in this study in the East Pacific. If this is the case, however, then it is surprising that olive ridley turtles in the East Pacific, which also host Stomatolepus elegans, do not also provide a reservoir of epibionts that may also be transferred to leatherback turtles.
In this study, we provided an example of how basic hypotheses about sea turtle epibiosis can be developed and tested statistically. Demonstrating the insights that can be gained from these methods, we recommend that future studies adopt a similar hypothesis-driven approach to investigate the geographic and ecological factors influencing the community structure of sea turtle epibionts. As research continues on this subject, we may uncover valuable insights into the ecology of these epibionts that will facilitate the use of epibionts as indicators of the hosts’ migratory behaviour (see Pfaller et al., Reference Pfaller, Alfaro-Shigueto, Balazs, Ishihara, Kopitsky, Mangel, Peckham, Bolten and Bjorndal2014), reveal the spread of potentially epibiont-borne diseases (see Greenblatt et al., Reference Greenblatt, Work, Balazs, Sutton, Casey and Casey2004, Lazo-Wasem et al., Reference Lazo-Wasem, Pinou, Peña de Niz, Salgado and Schenker2007), and unravel the co-evolutionary relationships between sea turtles and their epibionts (see Pinou et al., Reference Pinou, Lazo-Wasem, Dion and Zardus2013).
ACKNOWLEDGEMENTS
Epibiont collection was conducted under MINAET permits (ACT-OR-DR-143-14 and ACT-OR-DR-030-15). This research was performed in accordance with the Purdue University Animal Care and Use Committee. Samples were exported from Costa Rica under a SINAC permit (DGVS-181-2015). We would like to thank the park rangers at Parque Nacional Marino Las Baulas for their tireless work to protect the nesting sea turtles and their habitats. The staff at the Goldring-Gund Marine Biology Station, Earthwatch Institute volunteers and WCSU Hep Club student volunteers provided assistance in the field. Pilar Santidrián Tomillo and Randall Ureña provided assistance with permitting. Lourdes Rojas provided assistance with identifying taxa. Joe Pfaller, Brett O. Butler, and two anonymous reviewers provided feedback that greatly improved this manuscript.
FINANCIAL SUPPORT
Funding was provided by the Goldring Family Foundation, Earthwatch Institute, The Betz Chair Endowment of Drexel University, The Schrey Chair Endowment of Indiana University-Purdue University Fort Wayne, and The Leatherback Trust. Additional funding was also provided by a WCSU AAUP grant to T. Pinou.