Introduction
False brome grass, Brachypodium distachyon (L.) Beauv, has been proposed as a bridge between rice (Oryza sativa L.) and temperate cereal crops for genomics research (Draper et al., Reference Draper, Mur, Jenkins, Ghosh-Biswas, Bablak, Hasterok and Routledge2001). B. distachyon has a number of advantages as a model species such as annual growth habit, self-fertility, short generation time, small genome (~355 Mb) (Bennett and Leitch, Reference Bennett and Leitch2005) and five pairs of readily identifiable chromosomes. Some biological features in B. distachyon that are not found in rice but are important for temperate cereal crops include freezing tolerance, vernalization requirement and host resistance to specific pathogens or insect pests (Draper et al., Reference Draper, Mur, Jenkins, Ghosh-Biswas, Bablak, Hasterok and Routledge2001). The genomic infrastructure of this species has continued to develop (Hasterok et al., Reference Hasterok, Draper and Jenkins2004, Reference Hasterok, Marasek, Donnison, Armstead, Thomas, King, Wolny, Idziak, Draper and Jenkins2006; Vogel et al., Reference Vogel, Garvin, Leong and Hayden2006a, Reference Vogel, Gu, Twigg, Lazo, Chingcuanco, Hayden, Donze, Vivian, Stamova and Coleman-Derrb; Huo et al., Reference Huo, Gu, Lazo, Vogel, Coleman-Derr, Luo, Thilmony, Garvin and Anderson2006, Reference Huo, Lazo, Vogel, You, Ma, Hayden, Coleman-Derr, Hill, Dvorak, Anderson, Luo and Gu2008). Whole genome sequencing of B. distachyon is underway (http://www.brachypodium.org/). Nevertheless, much basic information about this species is still lacking. For example, no linkage map of B. distachyon is available. In addition, although B. distachyon accessions have varying vernalization requirements and different reactions to several plant pathogens (Draper et al., Reference Draper, Mur, Jenkins, Ghosh-Biswas, Bablak, Hasterok and Routledge2001; Routledge et al., Reference Routledge, Shelley, Smith, Talbot, Draper and Mur2004; Allwood et al., Reference Allwood, Ellis, Heald, Goodacre and Mur2006), genetic variation in response to other biotic or abiotic stresses has not been reported. Such information could help facilitate utilization of B. distachyon resources in crop breeding.
The greenbug, Schizaphis graminum Rondani, and Russian wheat aphid, Diuraphis noxia Mordvilko (RWA hereinafter), are the two most important aphid pests of wheat in the southern Great Plains of the USA. They are especially notorious for periodic changes of prevailing biotypes in the field, rendering currently deployed host resistance genes useless. Over 30 greenbug biotypes (Burd and Porter, Reference Burd and Porter2006) and five RWA biotypes (RWA1–RWA5) (Burd et al., Reference Burd, Porter, Puterka, Haley and Peairs2006) have been identified. Greenbug biotypes E and I and RWA biotypes RWA1 and RWA2 currently dominate wheat fields (Burd and Porter, Reference Burd and Porter2006; Puterka et al., Reference Puterka, Burd, Porter, Shufran, Baker, Bowling and Patrick2007). Although a number of host resistance genes (R genes) against the two aphids have been identified in wheat, none have been cloned. Map-based cloning of genes in wheat is still a formidable task because of its very large genome size (~16,000 Mb). Our long-term goal is to elucidate the molecular mechanisms of host resistance against aphid pests in cereal crops. Because wheat is phylogenetically closer to B. distachyon than to rice (Gaut, Reference Gaut2002), we are exploring the potential of B. distachyon-greenbug as a model system for plant–aphid interactions in the grass genome. In this study, we examined the reaction of B. distachyon diploid accessions to infestation by four different greenbug and RWA biotypes. Through database mining of B. distachyon expressed sequence tag (EST) and genomic DNA sequences, we developed microsatellite or simple sequence repeat (SSR) markers and used them to evaluate genetic diversity among six B. distachyon accessions.
Materials and methods
Plant materials
Seeds of four diploid accessions, BD2 (PI185133), BD3 (PI185134), BD18 (PI245730) and BD29 (PI639818) were acquired from the USDA Small Grain Collection at Pullman, WA (http://www.ars-grin.gov/). Five inbred lines, Bd1-1 (PI170218), Bd2-3, Bd3-1, Bd18-1 and Bd21 (PI254867), which had undergone five to six generations of selfing, were kindly provided by David Garvin (USDA-ARS, St Paul, MN). BD2, BD3 and BD18 were the respective parental lines of Bd2-3, Bd3-1 and Bd18-1. Three species in the grass genome: Aegilops tauschii (Coss.) Schmal. (accession AL8/78), rice (O. sativa L. cv. Nipponbare) and perennial ryegrass (Lolium perenne L. cv. Palmer) were also employed in phylogenetic analysis. Seeds of the ryegrass cultivar ‘Palmer’ were provided by Lloyd Nelson (Texas AgriLife Research at Overton, TX), and the Ae. tauschii AL8/78 seeds were supplied by Mingcheng Luo (University of California, Davis, CA).
Aphid infestation procedure
Infestation procedure followed Weng and Lazar (Reference Weng and Lazar2000). To study reactions of the six B. distachyon lines to aphid infestations, greenbug biotypes C (GBC) and E (GBE) and RWA biotypes RWA1 and RWA2 were used. GBC and GBE colonies were reared in an isolated room at Texas AgriLife Research, Bushland, TX. RWA1 and RWA2 were kindly provided by J. P. Michaud (Kansas State University, Hays, KS) and Gary Puterka (USDA-ARS, Stillwater, OK), respectively. Wheat cultivars or germplasm lines ‘TAM 107’, ‘TAM 110’, ‘Halt’ and ‘94M370’ were used as resistant controls. Wheat cultivar ‘TAM 105’ was the susceptible control in all tests. Seeds of each line were germinated in 10.2 cm square plastic pots (Hummert International, Earth City, MO) filled with a commercial potting mix (Sunshine Mix LC1, BWI Companies, Dallas, TX). Seedlings were watered regularly. At the three-leaf stage, each pot was thinned to four seedlings per pot. In tests with each aphid biotype, all pots were arranged randomly in the growth chamber with five replicates per genotype. Feeding responses were recorded after 14 d of infestation for the greenbug and 3 weeks for the RWA when the susceptible controls were killed. The screening test for each biotype was repeated at least twice. Because the feeding response in all tested samples could be easily and clearly rated as either R (resistant) or S (susceptible), no statistical analysis of aphid screening data was applied.
Database mining and SSR marker development
Database mining and SSR marker development followed Weng et al. (Reference Weng, Azhaguvel, Michels and Rudd2007). The B. distachyon EST and bacterial artificial chromosome (BAC) end genomic DNA sequences were downloaded from NCBI (National Center for Biotechnology Information, Bethesda, MD; http://www.ncbi.nlm.nih.gov). In total, 20,449 B. distachyon ESTs (12,798 kb) and 2,185 genomic DNA sequences (~1,222 kb) were examined. Briefly, the DNA sequences were scanned for simple perfect SSRs using the SSR Primer Discovery Program (http://hornbill.cspp.latrobe.edu.au/cgi-binpub/), in which the minimum length of SSRs was set as 10 bp, and only SSRs with repeat units of 2–5 bp in the motif were reported. The SSRs with mono- and hexanucleotide repeats were not counted and were not included in the final statistics. For a 10 bp SSR, one occurrence may comprise a repeat of five dinucleotides, four trinucleotides, three tetranucleotides or two pentanucleotides. The SSR motifs include both strands of the DNA sequence. CGG, for example, also includes GCC and the reverse complements GGC and CCG. The total lengths of each repeat type were used to estimate abundance of SSRs in the genome. The output was downloaded into Microsoft Excel for further analysis.
Over 6,000 EST-SSRs were identified from database mining. Only SSRs with a minimum length of 20 bp (for EST-SSRs) or 16 bp (for genomic SSRs) were selected for phylogenetic analysis. SSRs in this subset were subjected to a Basic Local Alignment and Search Tool (BLAST) (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990) search against the NCBI B. distachyon database to remove primers with redundant sequences. This resulted in 160 EST- and 21 genomic DNA-derived SSRs in this study. Detail information on all 181 SSRs is provided in Supplementary Table 1, available online only at http://journals.cambridge.org
PCR amplification
Genomic DNAs were extracted from young leaves by the CTAB method (Murray and Thompson, Reference Murray and Thompson1980). Each PCR contained 20 ng template DNA, 0.5 μM each of two primers and 1 × PCR master mix (Promega Inc., Madison, WI) in a total volume of 10.0 μl, which was performed in a PTC-200 thermocycler (Bio-Rad, Hercules, CA). A single touch-down PCR program was used (Weng et al., Reference Weng, Li, Devkota and Rudd2005).
For most SSR markers, the variation in the size of the PCR products allowed the patterns to be resolved unambiguously in 4% high-resolution agarose gels stained with ethidium bromide. For SSR markers generating DNA fragments of very similar size from different DNA templates, PCR products were size-fractioned in non-denaturing polyacrylamide gels and banding patterns were visualized by SYBR Gold staining (Molecular Probes, Eugene, OR). For quality control, markers detecting null alleles in PCR were repeated at least once to rule out the possibility of the failure of PCR amplification. For PCR products amplified by genomic SSRs, only fragments similar to the expected sizes were scored for data analysis.
Data analysis
Data from 160 EST-SSRs and 21 genomic SSRs were analysed. PCR products from each SSR marker (locus) were scored and the polymorphic information content (PIC) value (Botstein et al., Reference Botstein, White, Skolnick and Davis1980) was estimated to assess the informativeness of each marker. PIC was calculated using the formula PIC = 1 − ∑(P i)2, where P i represents the frequency of the ith allele. Null and non-polymorphic alleles were not included in computing PIC.
Two methods were used to evaluate SSR-based genetic diversity. In the first method, similarity estimates of microsatellite alleles were obtained using the ‘City Block’ approach. Haplotypes were clustered with hierarchical clustering using the Cluster 3.0 program (http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/software.htm). In the second method, the marker data were analysed with the software package PHYLIP 3.66 (Felsenstein, Reference Felsenstein1989; http://evolution.genetics.washington.edu/phylip.html), in which 1000 bootstrapped datasets were created for construction of pairwise distance matrices (Nei and Li, Reference Nei and Li1979). The resulting datasets were subjected to neighbour-joining cluster analysis (Saitou and Nei, Reference Saitou and Nei1987), and a consensus tree was constructed with rice, Ae. tauschii and perennial ryegrass as out-groups.
Results
Aphid feeding responses of six B. distachyon accessions
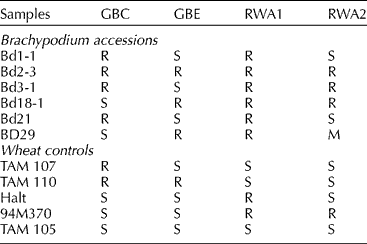
The infestation test results with the RWA and the greenbug on six diploid B. distachyon accessions and the wheat controls are summarized in Table 1. On susceptible TAM 105 seedlings, RWA2 feeding caused leaf chlorosis and leaf rolling (Burd et al., Reference Burd, Burton and Webster1993). Symptom development in seedlings of B. distachyon lines Bd1-1 and Bd21 upon RWA2 aphid feeding was similar to the susceptible wheat controls. Thus, lines Bd1-1 and Bd21 were scored as susceptible to RWA2. Similar to the resistant wheat controls, no symptoms were observed in Bd2-3, Bd3-1 and Bd18-1 in response to RWA2 feeding. Thus, Bd2-3, Bd3-1 and Bd18-1 were scored as resistant to RWA2. All the B. distachyon lines appeared to be resistant to RWA1.
Table 1 Responses of six Brachypodium distachyon accessions and the wheat controls to infestation of greenbug biotypes C (GBC) and E (GBE), and Russian wheat aphid biotypes RWA1 and RWA2a
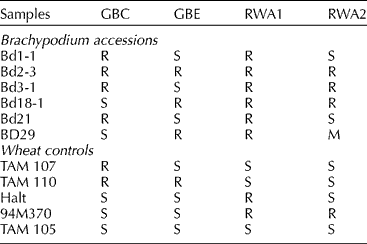
a R, resistant; S, susceptible; M, intermediate resistance.
The B. distachyon responses to greenbug feeding were similar to those seen in wheat, resembling leaf senescence, which developed much faster in susceptible genotypes than in resistant ones (Weng et al., Reference Weng, Lazar, Michels and Rudd2004). Lines Bd1-1, Bd2-3, Bd3-1 and Bd21 gave resistant response to GBC. Lines Bd18-1 and BD29 gave susceptible response to GBC. Differential response was also observed in response to GBE feeding. Bd2-3, Bd18-1 and BD29 gave resistant response and Bd1-1, Bd3-1 and Bd21 gave susceptible response.
Because the initial aphid infestation was large (on average more than 50 per seedling), the symptoms caused by greenbug or RWA feeding developed quickly in both wheat and B. distachyon plants. Usually, susceptible B. distachyon and wheat seedlings were killed by greenbug feeding within 10 d after infestation, and 2–3 weeks in the case of RWA. Resistant B. distachyon plants with no vernalization requirement (Bd3-1 and Bd21) were able to grow to maturity, while the aphid populations on them continued to decline.
Overall, except for Bd1-1 and Bd21, which had similar responses, all other B. distachyon accessions gave differential responses to the infestation by GBC, GBE and RWA2 (Table 1). Feeding response in all lines could be clearly rated as either ‘R’ or ‘S’ except in BD29, which showed symptoms intermediate between typical resistant and susceptible lines upon RWA2 infestation. Symptoms appeared similar between the inbred lines Bd2-3, Bd3-1, Bd18-1 and their respective parental lines BD2, BD3 and BD18 indicating no effect of inbreeding on aphid feeding response in these genotypes. The distinct responses of different accessions to the four biotypes were consistent in repeated experiments.
Microsatellite abundance and distribution in B. distachyon EST sequences
A total of 12,798 kb of the expressed portion (ESTs) of the B. distachyon genome was scanned for microsatellite abundance, and the results are summarized in Table 2. Of 20,499 ESTs screened, 6,977 (34.03%) SSR-containing ESTs with bi-, tri-, tetra- or pentanucleotide SSR motifs were identified. A significant number of ESTs (19.26%) harboured more than one SSR. Overall, approximately 0.80% of the 12,798 kb EST sequences contained SSRs motifs with on average one SSR in every 1.85 kb EST sequence. Because the mono- and hexanucleotide SSR repeats were not counted in this study, comparing the SSR abundance of B. distachyon with other grass species was difficult. If only the di-, tri-, tetra- and pentanucleotide SSR motifs were considered, the abundance of SSRs in EST sequences of B. distachyon (Table 2) was very similar to that found in several cereal crop species such as rice, wheat and barley (La Rota et al., Reference La Rota, Kantety, Yu and Sorrells2005).
Table 2 Abundance and motif distribution of SSRs in the Brachypodium distachyon genome

a Di, dinucleotide repeats; tri, trinucleotide repeats; tetra, tetranucleotide repeats; penta, pentanucleotide repeats.
Analysis of the distribution of the four repeat types revealed that trinucleotide repeats predominated the coding regions of the B. distachyon genome, accounting for 75.7% of all SSRs identified (Table 2). The other three repeat types had similar frequencies (dinucleotide 7.3%, tetranucleotide 9.8% and pentanucleotide 7.2%) in the EST sequences analysed.
Among the 2,185 B. distachyon genomic sequences (1,222 kb) examined, 281 were found to contain 312 SSRs. Because the sample size was too small (0.34% of the 355 Mb B. distachyon genome), the SSR abundance and distribution features of the genomic DNA sequences were not compared with those of the EST sequences. From the 281 genomic SSRs identified, 21 were selected and used in the genetic diversity study.
SSR-based genetic diversity among diploid B. distachyon accessions
From database mining, 181 new B. distachyon SSR markers were developed including 160 EST- and 21 genomic SSRs (see Supplementary Table 1, available online only at http://journals.cambridge.org for details). Of the 160 EST-SSRs, 24 failed to amplify any product in any line, but all 21 genomic SSRs had successful amplification in at least one DNA template tested.
The 136 EST-SSR markers detected in total 289 alleles with an average of 2.13 alleles per marker. In contrast, the 21 genomic SSRs detected an average of 4.0 alleles per locus. The usefulness of these SSR markers was also evaluated by their PIC values, which are listed in supplementary Table 1, available online only at http://journals.cambridge.org. As many as 45.9% of the 157 SSRs were monomorphic among the six accessions, in which 63 were EST-SSRs and nine were genomic SSRs. The average PIC for the 85 polymorphic SSRs was 0.44, ranging from 0.06 (BDeSSR012) to 0.83 (BDgSSR12). In agreement with the low PIC values, the SSR-based polymorphisms among the different B. distachyon accessions were generally low, ranging from 11.7% between Bd2-3 and Bd3-1 to 37.8% between BD29 and Bd21. The average polymorphism for Bd1-1, Bd2-3, Bd3-1, Bd18-1, Bd21 and BD29 across the other five accessions was 31.2, 22.8, 22.2, 23.5, 26.0 and 33.8%, respectively.
The effect of inbreeding on genetic diversity was compared in three pairs of B. distachyon lines. The polymorphism between each of the three inbred lines and their respective parental lines based on 139 EST-SSRs was 4.5, 4.2 and 4.8% for BD2, BD3 and BD18, respectively. Interestingly, except for the marker BDgSSR17 (see Supplementary Table 1, available online only at http://journals.cambridge.org) which detected two polymorphic alleles between Bd3-1 and BD3, no polymorphism was found with the remaining 20 genomic SSRs between all three pairs of lines.
Phylogenetic relationships of B. distachyon with other grass species
Of the 373 alleles detected by 157 SSR markers, 258 (69.2%) were specific to the B. distachyon genome. In total, 59, 40 and 31 alleles were detected in the perennial ryegrass, Ae. tauschii and rice, respectively. It seems that there were no significant differences in cross-species transferability of EST versus genomic SSRs in this study. For example, of the 54 alleles detected in the ryegrass, 39 and 15 were identified by 32 EST (1.2 alleles per SSR) and 10 genomic (1.5 alleles per SSR) markers, respectively. Each EST and genomic SSR was able to detect, on average, 1.2 (28/24) and 1.7 (12/7) alleles, respectively, in the Ae. tauschii genome.
The phylogenetic relationships between the six B. distachyon accessions and three other grass species (rice, Ae. tauschii and perennial ryegrass) were evaluated using the City Block distance matrix-based hierarchical clustering and the genetic distance-based neighbour-joining clustering methods. The phylogenetic tree generated by the City Block approach is shown in Fig. 1, which was the same as that produced with the neighbour-joining method (data not shown). The genetic distance matrix of Nei and Li (Reference Nei and Li1979) based on 1,000 bootstrapped datasets is presented in Table 3, which agreed very well with the phylogenetic relationships of the nine tested samples as revealed by the City Block-based analysis (Fig. 1). For example, from Table 3, the genetic distances of Ae. tauschii to B. distachyon (average over all accessions) and rice were 0.4623 and 0.7303, respectively, suggesting that Ae. tauschii was closer to B. distachyon than to rice.

Fig. 1 Cluster analysis of microsatellite marker haplotypes of six Brachypodium distachyon (BD or Bd) accessions, rice (Oryza sativa cv. Nipponbare), perennial ryegrass (Lolium perenne cv. Palmer) and Aegilops tauschii (accession AL8/78). Haplotype clustering was based on similarity estimates using the ‘City Block’ approach.
Table 3 Genetic distance matrix (Nei and Li, Reference Nei and Li1979) among six B. distachyon accessions, rice (cv. Nipponbare), perennial ryegrass (cv. Palmer) and Ae. tauschii (accession AL8/78)

Discussion
In this study, six B. distachyon accessions were tested for their response to infestation by two cereal aphids: the greenbug (biotypes GBC and GBE) and Russian wheat aphid (biotypes RWA1 and RWA2). Different reactions among the six accessions to aphid feeding of GBC, GBE and RWA2 were observed (Table 1). Resistant and susceptible phenotypes could be easily identified. The feeding response in B. distachyon was similar to that observed in wheat. In addition, because no difference in feeding response was found between each of the three inbred lines (Bd2-3, Bd3-1 and Bd18-1) and their respective parental lines (BD2, BD3 and BD18), host resistance in the B. distachyon accessions to different greenbug or RWA biotypes may be controlled by simply inherited genes. In earlier experiments, we found that Arabidopsis thaliana was not a host of the greenbug (non-host resistance), and the greenbugs infesting rice (cv. Nipponbare) were able to survive but performed very poorly (Weng et al., unpublished data). Considering the closer phylogenetic relationship of wheat with B. distachyon than to rice (Draper et al., Reference Draper, Mur, Jenkins, Ghosh-Biswas, Bablak, Hasterok and Routledge2001; also see discussion below), the greenbug–B. distachyon and RWA–B. distachyon host–insect systems may prove to be useful models to study the molecular mechanisms of R gene-mediated aphid resistance in the grass genome.
One objective of this study was to develop molecular markers in B. distachyon that can be used to evaluate genetic diversity among B. distachyon accessions. We conducted database mining of the B. distachyon EST and genomic sequences for SSRs. It was found that the B. distachyon genome was abundant in microsatellite sequences, and the distribution of microsatellites in the B. distachyon genome was similar to that in wheat and barley (Table 2). Among the four types of SSR motifs examined in the B. distachyon EST sequences, over 75% belong to trinucleotide repeats (Table 2). The predominance of trinucleotide repeats in coding regions seems to be common in cereal genomes such as rice and wheat (La Rota et al., Reference La Rota, Kantety, Yu and Sorrells2005; Peng and Lapitan, Reference Peng and Lapitan2005), which is probably the consequence of negative selection against frame-shift mutations in the coding regions (Metzgar et al., Reference Metzgar, Bytof and Wills2000). By exploring the genomic resources, it is possible to develop a large number of new SSR markers for B. distachyon. This has proven to be a robust and efficient strategy for marker development in the cereal crop genomes (Gupta and Varshney, Reference Gupta and Varshney2000; Theil et al., Reference Thiel, Michalek, Varshney and Graner2003; Nicot et al., Reference Nicot, Chiquet, Gandon, Amilhat, Legeai, Leroy, Bernard and Sourdille2004; Zhang et al., Reference Zhang, Bernard, Leroy, Feuillet and Sourdille2005).
In this study, both EST-SSR and genomic SSR markers were developed and used in evaluating genetic diversity among the six B. distachyon accessions. As compared with EST-SSRs, each genomic SSR marker was able to detect twice as many alleles (on average 4.0 alleles per genomic SSR versus 2.13 alleles per EST-SSR). It has been well documented that EST-SSRs are less polymorphic than genomic SSRs in earlier studies in rice (Cho et al., Reference Cho, Temnykh, Chen, Lipovich, McCouch, Park, Ayers and Cartinhour2000), sugarcane (Cordeiro et al., Reference Cordeiro, Casu, McIntyre, Manners and Henry2001), barley (Thiel et al., Reference Thiel, Michalek, Varshney and Graner2003) and durum wheat (Eujayl et al., Reference Eujayl, Sorrells, Baum, Wolters and Powell2002). Because EST-SSRs detected much fewer alleles, their usefulness in genetic mapping in B. distachyon is limited, and genomic DNA-derived SSRs should be employed. Large-scale development of these markers will be feasible soon as the whole genome sequence of B. distachyon is near completion.
From the number of alleles that each EST- or genomic SSR marker was able to detect, and the overall low PIC values of these markers (Supplementary Table 1, available online only at http://journals.cambridge.org), it is evident that the genetic diversity among the six B. distachyon diploid accessions tested in this study is generally low. This can also be seen from the SSR-based polymorphism levels among these lines, which varied from 11.7% between Bd2-3 and Bd3-1 to 37.8% between BD29 and Bd21. Calculation of the pairwise genetic distances among these B. distachyon accessions (Table 3) indicated that BD29 was the most diverged from the other five lines. Our observations also showed that BD29 had the longest vernalization requirement (at least 6 weeks) among the six accessions. BD29 was originally collected from Ukraine, whereas the other five lines were from either Iraq or Turkey (USDA Small Grain Collection, http://www.ars-grin.gov/). The high level of polymorphism in BD29 is correlated with its distinct geographical distribution separated over long distances from the other five lines.
The inbred line Bd21 was chosen for whole genome sequencing because of its small stature, no requirement for vernalization and short life cycle. In this study, the SSR-based polymorphism level between BD29 and Bd21 was the highest (37.8%) among all pairs of the six lines. The B. distachyon linkage map under development (Garvin et al., Reference Garvin, Gu, Hasterok, Hazen, Jenkins, Mockler, Mur and Vogel2008) was from a cross between Bd21 and Bd3-1. However, the polymorphism level between these two lines (17.1%) was less than half of that between Bd21 and BD29. Therefore, it seems that either Bd1-1/Bd21 or BD29/Bd21 would be a good choice to develop segregating populations for linkage mapping in B. distachyon. Because both Bd1-1 and Bd21 are susceptible to the biotype E greenbug, which is the prevailing biotype in the field (Burd and Porter, Reference Burd and Porter2006), the cross combination BD29/Bd21 seems to be the most promising for developing a mapping population based on genetic diversity, reaction to greenbug feeding and vernalization responses.
The phylogenetic relationships of B. distachyon, rice, Ae. tauschii and perennial ryegrass were also analysed using the SSR markers developed from the present study. The phylogenetic trees generated from the City Block approach (Fig. 1) and the neighbour-joining method were highly consistent suggesting that the SSR markers used in this study were robust for phylogenetic analysis. From the phylogenetic tree (Fig. 1) and genetic distance matrix (Table 3), it is evident that Ae. tauschii, the D genome donor of common wheat, is closer to B. distachyon than to rice. This result is consistent with conclusions from earlier studies (Shi et al., Reference Shi, Draper and Stace1993; Hsaio et al., Reference Hsaio, Chatterton, Asay and Jensen1994; Catalán et al., Reference Catalán, Shi, Amstrong, Draper and Stace1995; Catalán and Olmstead, Reference Catalán and Olmstead2000; Vogel et al., Reference Vogel, Garvin, Leong and Hayden2006a; Bossolini et al., Reference Bossolini, Wicker, Knobel and Keller2007) and supports the unique position of B. distachyon as a bridge between rice and temperate cereal crops for genomic research. Information from this study should be useful in genetic mapping and map-based cloning of greenbug or RWA resistance genes in B. distachyon.
Acknowledgements
We thank Josh Evans for technical help. The authors are grateful to Drs David Garvin (USDA-ARS, St Paul, MN) for providing B. distachyon seeds, J.P. Michaud (Kansas State University, Hays, KS) and Gary Puterka (USDA-ARS, Stillwater, OK) for providing RWA used in this study. We also thank John Raupp for editing assistance in the preparation of this manuscript. This research was partially supported by the United States Department of Agriculture, National Research Initiative grant (CSREES 2006-35301-16892).