Introduction
Species distribution models have been used to predict the distributions of a wide range of taxa (e.g. Guisan and Thuiller, Reference Guisan and Thuiller2005; Araújo and Guisan, Reference Araújo and Guisan2006), and are increasingly employed for crop wild relatives (Guarino, Reference Guarino, Guarino, Ramanatha Rao and Reid1995; Afonin and Greene, Reference Afonin, Greene, Greene and Guarino1999; Greene et al., Reference Greene, Hart and Afonin1999; Jarvis et al., Reference Jarvis, Williams, Williams, Guarino, Caballero and Mottram2005; Maxted et al., Reference Maxted, Dulloo, Ford-Lloyd, Iriondo and Jarvis2008; Parra-Quijano et al., Reference Parra-Quijano, Iriondo and Torres2012). The Crop Wild Relatives and Climate Change project of the Global Crop Diversity Trust and Millennium Seed Bank Partnership, Royal Botanic Garden, Kew (http://www.cwrdiversity.org) uses a gap analysis methodology developed by Ramírez-Villegas et al. (Reference Ramírez-Villegas, Khoury, Jarvis, Debouck and Guarino2010), which incorporates the use of the maximum entropy model ‘Maxent’ (Phillips et al., Reference Phillips, Anderson and Schapire2006) to support collection planning for crop wild relatives. The input data for Maxent include the geographic origins of both genebank and herbarium specimens, and statistics representing the current climate, i.e. a set of temperature and precipitation parameters. The Maxent output distribution maps intend to give an indication of locations where the species may be present.
Collectors of plant genetic resources (PGR) are interested in material with new genetic diversity, preferably from species that can be crossed with the cultivated taxa for crop improvement. Such PGR can typically be collected from regions that have not been sampled previously. The gap analysis method for crop gene pools (Ramírez-Villegas et al., Reference Ramírez-Villegas, Khoury, Jarvis, Debouck and Guarino2010) assesses the priority with which a certain crop wild relative should be collected using the sampling representativeness score (SRS), estimating gross representation in genebanks; the geographic representativeness score (GRS), estimating the comprehensiveness of genebank collections regarding the geographic range of the species; and the environmental representativeness score (ERS), estimating comprehensiveness based on a principal component analysis (PCA) of the full environmental range of the modelled species. As such, it is possible to get an overview of populations that are under-represented in genebanks and which may contain novel genetic diversity. The GRS and ERS depend upon Maxent distribution models. In this method, it is implicitly assumed that the herbarium data provide the full coverage of the distribution area of the species. In practice, herbarium data are incomplete and sampling bias can result in the systematic exclusion of certain regions of the species distribution from the presence data. As a result, the distribution deduced with an associative species distribution model such as Maxent is not necessarily the complete distribution of the species. Maxent improves its models by excluding part of the presence data from the training sample to subsequently use it as a test sample. However, this test sample is selected randomly, so it does not systematically exclude a specific area, which mimicks the detection of an undiscovered region of the species distribution.
Here we use the distribution of the wild relatives of lettuce (Lactuca spp.) as a case study to investigate how the predicted distribution of Lactuca saligna depends on the input occurrence data. In addition, we compare the Maxent distribution predictions against the expedition data of the endemic Lactuca georgica and the cosmopolitan Lactuca serriola. The results are utilized to discuss the applicability of Maxent to support PGR collecting missions.
Materials and methods
Study material
The wild relatives of lettuce were chosen as the focus of the case study due to high available knowledge and data of this crop, and the coincidence with a Lactuca collecting mission in the Trans-Caucasus organized by the Centre for Genetic Resources, The Netherlands (CGN) in 2013. Moreover, the wild relatives of lettuce represent a wide variety of both niche and distribution sizes, ranging from pan-temperate distributions for L. serriola (D'Andrea et al., Reference D'Andrea, Broennimann, Kozlowski, Guisan, Morin, Keller-Senften and Felber2009; Alexander, Reference Alexander2013) and L. saligna, to the narrow endemism of L. georgica in the Caucasus region (Zohary, Reference Zohary1991), and thus permit an evaluation of modelling methods for a variety of species types.
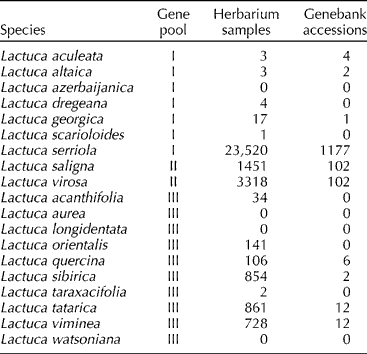
We collected information about known Lactuca populations from both herbarium and genebank databases (Table 1). The International Lactuca Database, Eurisco and the National Plant Germplasm System were used as the sources of information about genebank accessions. For herbarium samples, we consulted the Global Biodiversity Information Facility as well as obtained occurrence records directly from herbaria and researchers (see online supplementary Table S1). We used the inventory by Van Treuren et al. (Reference Van Treuren, Coquin and Lohwasser2012) to select the species known to belong to the gene pool of cultivated lettuce (Lactuca sativa) (Table 1) and to check for synonyms (see online supplementary Table S2) and reassignments from other genera. Van Treuren et al. (Reference Van Treuren, Coquin and Lohwasser2012) conducted a survey of the International Plant Names Index, which revealed a total number of 538 Lactuca spp., of which 357 referred to synonyms and basionyms, whereas for another 51, taxonomic status and their belonging to the genus Lactuca was questionable. Of the remaining 130 species, 20 are generally considered to be part of the lettuce gene pool (Table 1).
Table 1 Occurrence data of the species from the primary (I), secondary (II) and tertiary (III) gene pools of Lactuca sativa in herbaria and genebanks

The data were cleaned; records without and with only coarse geographic information were removed. In addition, we removed the duplicate species-specific locations. For the remaining locations, we used DIVA-GIS (vs 7.5, www.diva-gis.org) (Hijmans et al., Reference Hijmans, Guarino and Mathur2012) to cross-check the match between longitude/latitude combination and the stated country.
Species distribution modelling
Current climatic conditions were downloaded from http://www.worldclim.org at a scale of 2.5 arcminutes, including 19 bioclimatic variables (Table 2). Maxent (Phillips et al., Reference Phillips, Anderson and Schapire2006) uses a presence-only dataset as inputs and background points as pseudo-absences. The required 10,000 background points were selected randomly in each of the continents with species-specific occurrence data to ameliorate sample bias (VanDerWal et al., Reference VanDerWal, Shoo, Graham and Williams2009). Populations that were located outside the grid of climate cells, e.g. in the sea, were removed from the dataset.
Table 2 Nineteen bioclimatic variables used as input for the model, downloaded at a scale of 2.5 arcminutes from http://www.worldclim.org

Robustness of predicted distributions
L. saligna was chosen to assess the robustness of the Maxent projections in the marginal areas of the distribution range, since this species has a pan-temperate distribution with sufficient data points to exclude specific regions. Together with L. serriola and L. virosa, L. saligna serves as an important source of novel diversity for exploitation in the development of novel lettuce varieties by breeding companies (Lebeda et al., 2009).
Occurrence locations of L. saligna included North America, Australia and Europe, and from there extending into the eastern Mediterranean and Caucasus (see online supplementary Fig. S1). To assess the robustness of the projections, we excluded the L. saligna occurrence samples in, respectively, the Greek region (GRC, 88 occurrences), the Israeli region (ISR, 32 occurrences) and the Eurasian region (EUR, 684 occurrences) (see online supplementary Fig. S1). The Eurasian region was excluded to serve as a benchmark for the other predictions. The Greek and Israeli regions were very distinct marginal areas within the Eurasian distribution, from a geographic point of view (see online supplementary Fig. S1). Maxent was run for each of the three new datasets and the output was compared with the model based on the original, complete dataset. We used a tenfold division of the input data, each fold replicating the model using the consecutive parts as the test sample, while the remaining 90% of the input data were used as the training sample. For visual comparison, we used the Maxent projections, based on this tenfold cross-validation for each dataset. In addition, we correlated the Maxent estimated probabilities of occurrence of each 2.5-arcminute cell in each of the excluded regions that resulted from the models with the reduced and the original dataset. This indicates how the estimated probability of occurrence in a single cell changes when the occurrence data are excluded from the dataset on which the probability model was based.
Relationship with expedition data
A Lactuca expedition to the Trans-Caucasus was organized by CGN in 2013, which resulted in 94 unique collection locations. We compared the presence and absence locations of L. georgica and L. serriola with the model predictions for these locations. The model predictions were made without including the data of the sites visited during the expedition. So, while the modelling was done after the completion of the expedition, we mimicked the data availability prior to the collecting expedition, as if the modelling was done in its preparation. L. georgica and L. serriola were chosen because these two species were collected in a fair number of populations (32 and 55, respectively). In addition, they represent two opposites of the endemism spectrum, with L. georgica being endemic to the Trans-Caucasus, while L. serriola having a pan-temperate distribution, with very many data points (Table 1). To investigate the effect of zooming in on the target area, a new L. serriola model was made using only the 133 known occurrence samples from the expedition region, again without including the data from the newly sampled populations. The predicted probability of occurrence was determined for the 2.5-arcminute grid cell in which a population was found. All probabilities were classified in categories ranging from 0.1 to 1.0, and the number of presence and absence locations were summed per class.
Results
Quality of the models
The four Maxent models for L. saligna (the full dataset and those excluding, respectively, GRC, ISR and EUR) and the model for L. georgica were classified as valid models (see online supplementary Table S3) according to the gap analysis protocol (Ramírez-Villegas et al., Reference Ramírez-Villegas, Khoury, Jarvis, Debouck and Guarino2010), for which the average test area under the curve (AUC) should be larger than 0.7, the standard deviation of the test AUC smaller than 0.15 and the proportion of the potential distribution area with a standard deviation of the estimated probability of occurrence >0.15 should be smaller than 10%. The L. serriola model including all occurrence data was not considered valid, as a result of the low average test AUC of 0.65. However, when the analysis was limited to the expedition region, this resulted in a valid model for L. serriola.
Robustness of model predictions for Lactuca saligna
For L. saligna, excluding the occurrence data points in the GRC resulted in small changes in the estimated probabilities of occurrence in this region. The patterns in probability distributions were very similar between the two models (Fig. 1, maps) with a good correlation between the two models (Fig. 1, scatter plot) with a small decrease in estimated probabilities from the model where the occurrences in the GRC were omitted when compared with the model that included all occurrences.

Fig. 1 Projected probabilities of occurrence for Lactuca saligna in the Greek region (GRC) when including and excluding the occurrence data of this region on a scatter plot showing the changes in projected probabilities for the GRC when omitting the occurrence data.
Excluding the occurrence data points in the ISR led to large changes in the estimated probabilities of the occurrence of L. saligna in this region (Fig. 2). The similarity in the patterns of probability distributions was recognizable (Fig. 2, maps), but the decrease in estimated probabilities from the model in which the local occurrence data were omitted was substantial (Fig. 2, scatter plot).

Fig. 2 Projected probabilities of occurrence for Lactuca saligna in the Israeli region (ISR) when including and excluding the occurrence data in this region on a scatter plot showing the changes in projected probabilities for the ISR when omitting the occurrence data.
When all the occurrences in the EUR were excluded, the probabilities of the occurrence of L. saligna changed drastically (Fig. 3, scatter plot), showing increases and decreases depending on the location. The pattern of the potential distribution changed from an emphasis on Western Europe to a most probable occurrence in the Middle East and Central Asia (Fig. 3, maps). The maximum estimated probability of occurrence increased from 0.75 to 0.97. However, the margins of the potential distribution of L. saligna were very similar between the two models.

Fig. 3 Projected probabilities of occurrence for Lactuca saligna in the Eurasian region (EUR) when including and excluding the occurrence data in this region on a scatter plot showing the changes in projected probabilities for the EUR when omitting the occurrence data.
Model predictions in relation to expedition data
Figure 4 shows the presence and absence of both L. georgica and L. serriola in 94 unique locations sampled during the CGN Lactuca expedition in 2013 in relation to the Maxent estimated probabilities of occurrence in the matching 2.5-arcminute grid cells. The model performed quite well for L. georgica, as at locations with low estimated probabilities of occurrence, only absence of the species was observed, while at the locations with high estimated probabilities of occurrence, observed presence was considerably higher than absence of the species. In contrast, the L. serriola projection showed very little differentiation across the expedition region, restricting the estimated probability of occurrence to only a few classes in the middle of the potential range. The majority of locations fell within the 0.4–0.5 probability class, at which more or less equal numbers of absences and presences were observed. The new L. serriola model, excluding all occurrence data but the ones in the expedition region, showed much more differentiation. However, the estimated probability of occurrence appeared to be a poor predictor for the presence and absence of the species.

Fig. 4 Number of locations in the region under study where Lactuca georgica and Lactuca serriola were present or absent plotted against the estimated probabilities of occurrence at these locations, based on the model including all occurrence data (L. georgica and L. serriola) and only occurrence data in the region under study (L. serriola Caucasus (CAU)).
Discussion
Species distribution modelling
Species distribution models have been used for a number of decades and for many purposes (see the review by Elith and Leathwick, Reference Elith and Leathwick2009), such as the consequences of climate change on species distributions and the assessment of the distribution of an invasive species. With the growing availability of digital records from natural history museums, herbaria and genebanks coupled with the demand for mapped predictions, the incentive to put this source of presence-only data to use has been increasing (Elith and Leathwick, Reference Elith and Leathwick2009). Many different modelling techniques exist, and the discussion on how to best model presence-only data continues. It is now common to compare presence data with background or pseudo-absence data, by e.g. using regression methods such as generalized linear models, generalized additive models or multivariate adaptive regression splines, but also GARP (Genetic Algorithm for Rule Set Production, Stockwell and Peters, Reference Stockwell and Peters1999), ENFA (Ecological Niche Factor Analysis, Hirzel et al., Reference Hirzel, Hausser, Chessel and Perrin2002) and Maxent (Phillips et al., Reference Phillips, Anderson and Schapire2006), in chronological order. Elith et al. (Reference Elith, Graham, Anderson, Dudík, Ferrier, Guisan, Hijmans, Huettmann, Leathwick, Lehmann, Li, Lohmann, Loiselle, Manion, Moritz, Nakamura, Nakazawa, Overton, Townsend Peterson, Phillips, Richardson, Scachetti-Pereira, Schapire, Soberón, Williams, Wisz and Zimmermann2006) reviewed and compared these methods. More recently, a platform was developed to combine different techniques for ensemble forecasting of species distributions (Araújo and New, Reference Araújo and New2007), called BIOMOD (R-package, http://R-Forge.R-project.org) (Thuiller et al., Reference Thuiller, Lafourcade, Engler and Araújo2009). This software is able to fit and compare different model classes in an attempt to reach more robust forecasts by treating the methodological uncertainties in different models. The latest version of BIOMOD, biomod2, also includes Maxent as one of the techniques.
In this study, we chose to use Maxent as the modelling technique to align our results with the gap analysis method (Ramírez-Villegas et al., Reference Ramírez-Villegas, Khoury, Jarvis, Debouck and Guarino2010), which is used as input for many planned collecting expeditions in the Crop Wild Relatives and Climate Change project. With this project in mind, we aimed to provide information about the robustness and accuracy of Maxent with regard to such collecting expeditions. Apart from these considerations, the accessibility and relative ease of use of the Maxent software compared with the others mentioned are valuable assets for application by non-experts in the planning of collecting expeditions. However, for increasing the strength of the predictions, using and comparing different modelling techniques as is done in BIOMOD is probably a valuable contribution. Compared with Maxent, this does require increased need for processing power, technical knowledge and time, which might limit the usability of BIOMOD for the non-expert.
Robustness of Lactuca saligna models
From the comparison of the different L. saligna models, we conclude that the Greek region is not climatically distinct within the known L. saligna distribution area (Fig. 1). In contrast, the Israeli region appeared to be very distinct (Fig. 2). Comparing the response curves of the Maxent predictions with the different environmental variables (see online supplementary Fig. S2), we found the Israeli region to differ from other areas within the distribution range, especially regarding the mean annual temperature (BIO1), the maximum temperature of the warmest month (BIO5), the variation in the precipitation over the seasons (BIO15), and the total precipitation of the warmest quarter (BIO18). Figure 2 indicates that the model is not capable of predicting this latter region as a potential distribution area when data from this region are excluded, while the unique climatic conditions may indicate the presence of potentially interesting diversity. For the Eurasian region of investigation, although the estimated local probabilities differ substantially between the two models (Fig. 3), the distribution borders are very similar. The changes in distribution pattern indicate that the south-eastern region, where the probabilities in occurrence increase when the Eurasian occurrences are excluded, is climatically more similar to the other regions in the world where L. saligna is sampled. From this it follows that, in addition to the Israeli region, Western Europe may be considered a relatively exceptional climatic region in the L. saligna distribution area.
From this analysis, we conclude that the outputs of Maxent are generally robust, yet on a local scale, on which a collecting mission is typically planned, the estimated probabilities of occurrence can differ to a larger extent. This depends on whether populations in the specific region have been sampled before (and the data used as input for the model), and on the climatic relatedness to other sampled regions within the distribution. It is important to note that all species distribution models project suitable habitat by climate association, and thus that none can predict potential distribution in regions that are climatically unique compared with regions where the species has been sampled. Particularly for the purpose investigated here, in search of unique PGR, this is an important limitation. However, excluding occurrence data from the perceived range margins of the species may result in important information about local adaptation to distinct climatic conditions. A PCA of the climatic data from the occurrence locations can provide the same information and may be a good starting point for such an analysis. There are other possibilities to obtain information about local adaptation, e.g. using SNP (single nucleotide polymorphism) data to confirm that a population is genetically different from similar populations, or checking phenotypic characteristics. Such methods tend to be used at a later stage, while the method that we suggest here can be done with currently available information about population locations to get an indication about where such useful locally adapted populations may be present.
L. saligna is native to Eurasia and North Africa (Lebeda et al., Reference Lebeda, Dolezalová, Feráková and Astley2004a, Reference Lebeda, Dolezalová and Astleyb; GRIN, 2014), and when omitting the Eurasian occurrence locations, we have essentially predicted the native distribution of the species from its non-native distribution. While this is not a logical procedure for collection planning, it gives indication of robustness of the predictions and the possibility to predict species' presence in a region where the species has not been sampled. Omitting many data points from the native area of the species then makes an interesting benchmark study with which to compare the other results from the Greek and Israeli regions. Interestingly, this procedure could be used to indicate the potential origin of populations in the non-native distribution area (Alexander, Reference Alexander2013).
Relationship with expedition data
L. georgica is an endemic (Zohary, Reference Zohary1991) species that lives in an equilibrium environment where natural competition determines the distribution of species. L. serriola is a ruderal species, thus living in disturbed environments (Grime, Reference Grime1977), with a cosmopolitan distribution (D'Andrea et al., Reference D'Andrea, Broennimann, Kozlowski, Guisan, Morin, Keller-Senften and Felber2009; Alexander, Reference Alexander2013). The widespread distribution of the latter and the very many data points that are included in the initial model (Table 1) probably account for the rather undifferentiated model projections observed for L. serriola. It is interesting to note that, although not very informative, this model does provide a good prediction of its occurrence, with a similar number of presence and absence locations across the 50% probability region. When we limited the model to the expedition region, the number of probability classes increased substantially. However, actual presence and absence locations appeared to correlate poorly with the corresponding probability of occurrence according to the modelling (Fig. 4). The L. serriola absence locations are mostly L. georgica presence locations, representing fairly undisturbed habitats. In addition, the ruderal nature of L. serriola, combined with its global distribution, explains its relative insensitivity to climatic conditions. Thus, the Maxent model for this global, ruderal species would not have been informative or otherwise useful for the Trans-Caucasus expedition, not even when the projections would have been restricted to the region of interest. This is in line with the gap analysis protocol as suggested by Ramírez-Villegas et al. (Reference Ramírez-Villegas, Khoury, Jarvis, Debouck and Guarino2010), who excluded weedy species from the analysis. Here it needs to be noted that the collection of widespread and ruderal species such as L. serriola does not require any modelling support, since such species can be easily located. In contrast, expected and actual presence data correlated well for the endemic L. georgica, living in pristine habitat. In fact, a population was sampled in a region where the local experts had not expected it, while the model predicted a high probability of occurrence at this location. In the case that the projection for L. georgica would have been available prior to the collecting mission, the expedition route would have been slightly adjusted to explore a nearby region where also high probabilities of occurrence were estimated. Thus, Maxent distribution models may be useful to support collecting missions and based on our findings, this may particularly apply to endemic species growing in relatively undisturbed habitats.
Concluding remarks
Based on our results, we recommend organizers of collecting missions to run Maxent or similar Species distribution models for their species of interest prior to the expedition in complement to expert knowledge on species distributions. Given sufficient input data, particular faith may be given to the model results for endemic species among the range of relevant crop wild relatives. The resulting maps should be combined with the knowledge of local authorities to identify potential new populations of these species. In addition, excluding occurrence data from the perceived range margins of the species may result in important information about local adaptation to distinct climatic conditions.
To increase access to the methodology, avoiding the necessity of installing and operating the software, a web-based version of Maxent, including the worldclim.org climatic dataset and standard model settings, would greatly facilitate the application of species distribution modelling in the preparation phase of collecting missions, and would be particularly useful for plant genetic resources conservation efforts with limited resources.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S1479262114000847
Acknowledgements
This study was part of the Fundamental Research Programme on Sustainable Agriculture (KB-12-005.03-003) funded by the Dutch Ministry of Economic Affairs. M. C. was additionally funded by the Open Programme of the Netherlands Organisation of Scientific Research (NWO) Data and methods were contributed by the Adapting Agriculture to Climate Change Project, which is managed by the Global Crop Diversity Trust with the Millennium Seed Bank Partnership of the Royal Botanic Gardens, Kew and supported by the Government of Norway. For further information see the project website: http://www.cwrdiversity.org/
The authors acknowledge the contribution of the breeding companies Agrisemen BV, Enza Zaden Research and Development BV, Monsanto Holland BV, Nunhems Netherlands BV and Rijk Zwaan Breeding BV to the Lactuca expedition in 2013.