Introduction
Angiostrongylus cantonensis is the main aetiological agent of eosinophilic meningoencephalitis in humans (Graeff-Teixeira et al., Reference Graeff-Teixeira, Da Silva and Yoshimura2009). Its life cycle includes mollusks as intermediate host and rodents as definitive hosts, with the adult worms living inside the pulmonary arteries (Alicata, Reference Alicata1965). Humans can become infected through the ingestion of undercooked mollusks, crustaceans or vegetables contaminated with third-stage larvae (L3). In human hosts, the third-stage larvae (L3) do not complete the cycle and are trapped in the central nervous system.
Cerebral angiostrongyliasis is an emerging global public health concern, with outbreaks documented across the world, including in Southeast Asia, Pacific Islands, parts of South and Central America and the Caribbean (Wang et al., Reference Wang, Lai, Zhu, Chen and Lun2008; Martin-Alonso et al., Reference Martin-Alonso, Foronda, Quispe-Ricalde, Feliu and Valladares2011; do Espírito-Santo et al., Reference do Espírito-Santo, Pinto, da Mota and Gryschek2013; Burns et al., Reference Burns, Bicknese, Qvarnstrom, DeLeon-Carnes, Drew, Gardiner and Rideout2014). To help elucidate the mechanisms and effects of cerebral angiostrongyliasis in humans, both whole genome sequencing (Morassutti et al., Reference Morassutti, Perelygin, De Carvalho, Lemos, Pinto, Frace, Wilkins, Graeff-Teixeira and Da Silva2013; Yong et al., Reference Yong, Eamsobhana, Lim, Razali, Aziz, Rosli, Poole-Johnson and Anwar2015a) and a complete mitochondrial genome (Yong et al., Reference Yong, Song, Eamsobhana, Goh and Lim2015b; Červená et al., Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019) comparative transcriptomic analysis between the fifth-stage larvae and A. cantonensis adult lineages from China have been performed in previous studies (Yong et al., Reference Yong, Eamsobhana, Lim, Razali, Aziz, Rosli, Poole-Johnson and Anwar2015a).
In the current study, we used de novo transcriptome assembly, complete functional annotation and metabolic reconstruction from adult worms of A. cantonensis isolated from Porto Alegre (30°01′40″ south; 51°13′43″ west), in the southernmost Brazilian state, Rio Grande do Sul, with the aim of identifying the expressed and active genes in adult worms to improve our understanding of this disease and its main aetiological agent. Our findings represent a valuable source of novel molecular targets for the diagnosis and treatment of cerebral angiostrongyliasis. The annotation results are available in the database AngiostrongylusDB at: http://angiostrongylus.lad.pucrs.br/admin/welcome.
Materials and methods
Isolation of worms and RNA extraction
Angiostrongylus cantonensis female worms were recovered (42 days post-infection) from the pulmonary arteries and right-heart cavities of experimentally infected Wistar Rattus norvegicus, washed in saline solution, and stored at ‒80°C in RNAlater (Qiagen, Inc., Valencia, CA) until use. Approximately 30 mg of worms were homogenized in 600 μL of lysis buffer RA1 using a T8 homogenizer (IKA WORKS, Inc., Wilmington, NC, USA). Total RNA was isolated using the NucleoSpin RNA II kit (Machery-Nagel, Inc., Bethlehem, PA, USA), according to the manufacturer's protocol.
De novo assembly, prediction of coding regions and functional annotation
The A. cantonensis adult worm transcriptome was sequenced at Macrogem using Illumina HiSeq2500, obtaining 2 × 150 paired end reads in the fastq format. The Illumina adapter sequences were removed from the reads, and low-quality reads towards the 3′ and 5′ ends of the reads were trimmed. Then, the reads were scanned with a 4 base-wide sliding window, and leading or trailing bases with an average Phred quality score lower than 20 were removed using Trimmomatic (Bolger et al., Reference Bolger, Lohse and Usadel2014). Reads shorter than 50 bp were also discarded. Transcriptome de novo assembly was performed using Trinity 2.0.6 (Grabherr et al., Reference Grabherr, Haas, Yassour, Levin, Thompson, Amit, Adiconis, Fan, Raychowdhury, Zeng, Chen, Mauceli, Hacohen, Gnirke, Rhind, di, Birren, Nusbaum, Lindblad-Toh, Friedman and Regev2011) with default k-mer. Gene open reading frames (ORFs) or protein coding regions within the transcripts were predicted in TransDecoder (https://github.com/TransDecoder/TransDecoder/wiki) using the Ab-initio model, while homology searches were performed using the blastp results against all the proteins of the Swiss-Prot databases, PFAM functional domains (El-Gebali et al., Reference El-Gebali, Mistry, Bateman, Eddy, Luciani, Potter, Qureshi, Richardson, Salazar, Smart, Sonnhammer, Hirsh, Paladin, Piovesan, Tosatto and Finn2019) and THMM 2.0c (Transmembrane helice Markov model) software packages. Only predicted ORFs that were at least 100 amino acids long were retained. Subsequently, non-redundant (NR) sequences were obtained by comparing against all sequences in Cluster Database at High Identity with Tolerance (CD-HIT; http://weizhongli-lab.org/cd-hit/) using a cut-off of 100% identity (parameters: -c 1.0, -aL 1.0, -AL 1.0). The genes corresponding to these ORFs were recovered using the Perl script and functionally annotated using BLASTx (BLAST+ v2.2) (Camacho et al., Reference Camacho, Coulouris, Avagyan, Ma, Papadopoulos, Bealer and Madden2009) against NR (NCBI, non-redundant database) with an E-value cut-off of 1.0 × 10−6 and with InterProScan5 (Jones et al., Reference Jones, Binns, Chang, Fraser, Li, McAnulla, McWilliam, Maslen, Mitchell, Nuka, Pesseat, Quinn, Sangrador-Vegas, Scheremetjew, Yong, Lopez and Hunter2014). All annotations were performed and recovered using Blast2GO (Conesa et al., Reference Conesa, Götz, García-Gómez, Terol, Talón and Robles2005). Genes annotated with gene ontology (GO) term Oviposition were recovered using a complete list of GO annotation level terms assigned with Blast2GO with bash programming language scripts. The ORFs were annotated with BLASTp against TrEMBL/UniprotKB with an E-value cut-off of 1.0 × 10−6 and annotation recovery with Blast2GO.
Transcriptome quality evaluation
The Trinity Assembly statistics, such as Nx statistics (e.g. the contig N50 value), total trinity genes, total of trinity transcripts and other transcripts, were obtained with the Perl script ‘TrinityStats.pl’ of the Trinity toolkit. The completeness and contiguity of the A. cantonensis transcriptome was assessed by comparing their assembly transcripts to benchmarking sets of universal single-copy (BUSCO) of the Eukaryota and Nematoda in BUSCO v3 (Simão et al., Reference Simão, Waterhouse, Ioannidis, Kriventseva and Zdobnov2015), based on the content of near-universal single-copy orthologues selected from the OrthoDB v9 database. The full-length transcript or near full-length transcript was identified using BLASTX against the predicted proteome of Dictyocaulus viviparus (GCA_000816705.1), Ancylostoma ceylanicum (GCA_000688135.1), Ancylostoma duodenale (GCA_000816745.1) and Caenorhabditis elegans (GCF_000002985.6) obtained from WormBase (https://parasite.wormbase.org/index.html), with an E-value cut-off of 1 × 10−20 and parameters -max_target_seqs 1. The resulting BLASTX table (outfmt6) was processed with the Perl script ‘analyze_blastPlus_topHit_coverage.pl’ from the Trinity toolkit. For each Blast hit in the target protein database, the best matching Trinity transcript was identified and selected. The percentage of Blast hit lengths covered by the Trinity transcript was identified. Global alignment between the encoding proteins was performed using Needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/).
Transcript abundance estimation
To estimate transcript abundance, sequenced pair-end reads were re-aligned to the assembled transcripts for quantification by using RSEM software (RNA-Seq by Expectation Maximization) using the Trinity script (align_and_estimate_abundance.pl) in the Trinity toolkit. TPM values with cut-off ⩾100 were considered highly expressed, while 3 ⩽ TPM < 100 were considered moderately expressed, according to previous studies (Shadeo et al., Reference Shadeo, Chari, Vatcher, Campbell, Lonergan, Matisic, van Niekerk, Ehlen, Miller, Follen, Lam and MacAulay2007; Piras et al., Reference Piras, Chiow and Selvarajoo2018; Duan et al., Reference Duan, Flock, Jue, Zhang, Jones, Al Seesi, Mandoiu, Pillai, Hoffman, O'Neill, Zinn, Govoni, Reed, Jiang, Jiang and Tian2019).
Annotation and reconstruction of metabolic pathways
The annotation of signal transduction pathways from the transcriptome of A. cantonensis was performed through KEGG Automatic Annotation Server (KAAS) (Moriya et al., Reference Moriya, Itoh, Okuda, Yoshizawa and Kanehisa2007), which provides functional annotation of genes by BLAST comparisons against the manually curated KEGG GENES database (Kanehisa et al., Reference Kanehisa, Sato, Kawashima, Furumichi and Tanabe2016). To accomplish this, the proteomes in the representative set for eukaryotes were used as a reference. The set contained the following organisms: hsa: Homo sapiens, mmu: Mus musculus, rno: Rattus norvegicus, dre: Danio rerio, dme: Drosophila melanogaster, ath: Arabidopsis thaliana, sce: Saccharomyces cerevisiae, ago: Ashbya gossypii, cal: Candida albicans, spo: Schizosaccharomyces pombe, ecu: Encephalitozoon cuniculi, pfa: Plasmodium falciparum 3D7, cho: Cryptosporidium hominis, ehi: Entamoeba histolytica, eco: Escherichia coli K-12 MG1655, nme: Neisseria meningitidis MC58, hpy: Helicobacter pylori 26695, bsu: Bacillus subtilis subsp. subtilis 168, lla: Lactococcus lactis subsp. lactis Il1403, mge: Mycoplasma genitalium G37, mtu: Mycobacterium tuberculosis H37Rv, syn: Synechocystis sp. PCC 6803, aae: Aquifex aeolicus, mja: Methanocaldococcus jannaschii, ape: Aeropyrum pernix jointly with available proteomes of nematodes: cel: Caenorhabditis elegans, cbr: Caenorhabditis briggsae, nai: Necator americanus, bmy: Brugia malayi, loa: Loa, tsp: Trichinella spiralis and trematode: smm: Schistosoma mansoni. The results contained a list of KO assignments (KEGG Orthology) and functional annotations for the A. cantonensis transcripts that were processed with the Perl script. Complete and almost complete modules were assigned with Reconstruct Module of KEGG (https://www.genome.jp/kegg/tool/map_module.html), while metabolic pathway figures were obtained with KEGG Mapper (https://www.genome.jp/kegg/tool/map_pathway2.html). The complete modules were tighter functional units for pathways, and protein complexes often corresponded to sub-pathways in the KEGG pathway maps. Each module was identified by M and was manually defined as a combination of K numbers (representing individual genes in each genome) revealing whether a module was complete, and a gene set was present. The list of A. cantonensis genes with KOs assigned (Supplementary Table S6) with KASS was used for module reconstruction.
Database of A. cantonensis: AngiostrongylusDB
The annotation of the A. cantonensis transcriptome is available as refined data established in the relational structure of MySQL Database Manager System 14 (https://dev.mysql.com/), and can be accessed using the web interface AngiostrongylusDB, http://angiostrongylus.lad.pucrs.br/admin/welcome, with access to the control profiles for queries, curation of the data or inclusion of new annotations using the programming language Ruby on Rails 4.2.6. This allows for the easy maintenance and adaptation of research requirements in the database and provides a framework that facilitates navigation between the application's functionalities. The web interface AngiostrongylusDB provides the following annotation datasets: (i) functional annotation retrieved from BLASTx and BLASTp against non-redundant protein sequence database (NCBI); (ii) annotation of KASS; (iii) UniProt Knowledge base (UniProtKB) and (iv) search facilitator corresponding to filters for these annotation listings. These listings contain links to the refined details of each list item according to the sequence identifier and links for the editing or curation of these data, depending on the user's profile.
Results
De novo assembly of transcriptome and completeness assessment
The transcriptome of A. cantonensis (Supplementary Fig. 1) generated 58 418 364 paired reads (A2_1 and A2_2, respectively) with 93.58% of these reads having a Phred score >Q30 (per base sequence quality). De novo assembly generated 82 779 transcripts, with 53 467 genes with an N50 length of 1870, a median contig length of 535, an average contig length of 1034.77 and a total of assembled contigs of 85 657 588 bp (Supplementary Table S1). The completeness of Transcriptome Assessment by the Benchmarking Universal Single-Copy Orthologues (BUSCO v3) indicated that 87.1% (N = 852) of the 978 core Metazoan genes were identified as complete, fragmented 1.3% (N = 14), while 11.6% (N = 113) of the genes were missing from the assembly (Supplementary Table S1). Regarding the eukaryotic core genes, 98.3% (N = 298) of the genes were identified as complete, 1.0% (N = 3) as fragmented and 0.7% (N = 2) as missing. The quantity of the full-length or nearly full-length transcripts, determined by the number of transcripts that were reconstructed to full-length (100% alignment) or near full-length (>70% alignment) by alignment to proteome of nematodes, is shown in Table 1. We identified a total of 14 502 full-length or near full-length (100–70%) using as reference the proteomes of A. ceylanicum, D. viviparus, A. duodenale and C. elegans (Table 1 and Supplementary Table S1). Our transcriptome has 22 292 NR proteins codified, more than the reference genome of A. cantonensis deposited at the NCBI Genome database (GCA_009735665.1, Xu et al., Reference Xu, Xu, Sun, Xu, Zeng, Shan, Yuan, He, He, Yang, Luo, Wei, Wu, Liu, Xu, Dong, Song, Zhang, Yu, Wang, Zhang, Fang, Gao, Lv and Wu2019) what has 10 314 proteins and a genome size of 293 Mb.
Table 1. Full-length transcript reconstruction comparative analysis of the transcriptome of Angiostrongylus cantonensis adult worms

The cumulative number of proteins of A. ceylanicum, D viviparous, A. duodenale and C. elegans recovery in BLASTx that aligned by at least one transcript in the assembly of the A. cantonensis transcriptome at 80–100% of coverage. Transcripts identified in A. cantonensis were annotated as full-length transcripts if they matched a protein in the reference proteome database with an E-value threshold of 1 × 10−20. Pct_cov_Hit, percentage of coverage of top matching hits of reference proteome aligned across more than X% (80–100) with the A. cantonensis transcript. PC, protein counts of target reference proteome aligned by at least one transcript of A. cantonensis. PCS, protein counts sum of reference proteins from reference proteome aligned at X% (80–100) coverage by at least one A. cantonensis transcript. Assembly ID of A. ceylanicum: GCA_000688135.1, D. viviparous: GCA_000816705.1, A. duodenale: GCA_000816745.1, C. elegans: GCF_000002985.6.
Functional annotation of the transcriptome
From the 82 779 transcripts, 50 207 coding regions were predicted using TransDecoder with an ab initio model and homology for known sequences and functional domains. Subsequently, 22 292 NR-predicted transcripts were obtained with CD-HIT using 100% identity with all other default parameters unchanged. The distribution of the most significant hits (Top-Hits) in the BLASTx results against the NR database (NCBI) is shown in Fig. 1 and Supplementary Table S1. From the 22 292 proteins, nematode parasites accounted for 94.33% of all hits, most of them (7175 proteins, 32.10%) with high similarity to A. ceylanicum, 5596 (26.82%) to D. viviparus, 4706 (21.05%) to Haemonchus contortus, 1292 (5.78%) to N. americanus, 891 (3.98%) with A. duodenale, 652 (2.91%) to Oesophagostomum dentatum and 288 (1.28%) to A. cantonensis, 84 (0.37%) to C. elegans, 397 (1.77%) to other organisms, and the remaining 870 (3.89%) did not show similarity to any other organisms in the NR database. Taken together, all 22 292 sequences were annotated using BLASTx, InterProscan and GO (Supplementary Table S2). The datasets of the annotation of A. cantonensis and the fasta files were stored in a database created in MySQL and can be accessed at https://angiostrongylus.lad.pucrs.br/admin.

Fig. 1. Species distribution of predicted homologues in the transcriptome of Angiostrongylus cantonensis adult worms. Homologues were predicted using a BLASTX search against the Nr NCBI database at an E-value cut-off of 1.0 × 10−6. The top eight species with the most homologues are shown.
GO annotation [biological processes, molecular functions (MF) and cellular components] at level 2 were predicted for the 22 292 total mRNA sequences of adults of A. cantonensis (Fig. 2 and Supplementary Table S2). The top GO term assignments for all mapped genes in the biological processes (BP) category included: cellular process (N = 10 281), metabolic process (N = 9545 proteins), developmental process (N = 6824), multicellular organism process (N = 6799), biological regulation (N = 6106), regulation of biological process (N = 5324), localization (N = 4933), reproduction (N = 4303), cellular component organization or biogenesis (N = 3422), response to stimulus (N = 3278), locomotion (N = 2831), reproductive process (N = 2639), signalling (N = 2234), positive regulation of biological process (N = 1617), negative regulation of biological process (N = 1405), multi-organism process (1190), growth (N = 948), behaviour (N = 829), cell proliferation (N = 292) and biological adhesion (N = 253) (Fig. 2A). In the MF category, the GO terms assignment include: binding (N = 9064), catalytic activity (N = 7567), transport activity (N = 1260), regulator (N = 544), molecular transducer activity (N = 499), transcription regulator activity (N = 440), structural molecule activity (N = 421), translation regulator activity (N = 230), antioxidant activity (N = 82), molecular carrier activity (N = 30), cargo receptor activity (N = 18), small molecule sensor activity (N = 4) and protein tag (N = 2) (Fig. 2B). For the cellular component category, the GO terms assigned include: cell (N = 8613), cell part (N = 8542), organelle (N = 6009), membrane (N = 4394), protein containing complex (N = 3429), membrane part (N = 3313), organelle part (N = 3184), membrane enclosed lumen (N = 922), supramolecular complex (N = 651), extracellular region (N = 466), extracellular region part (372), cell junction (N = 347), synapse (N = 283), synapse part (N = 173), virion part (N = 53), virion (N = 53), nucleoid (N = 13), other organism (N = 4) and other organism part (N = 4) (Fig. 2C). The GO annotation (biological processes, MF and cellular components) at level 9 was also predicted for all sequences (Supplementary Table S2). We found 1053 GO terms, where the top five most abundant GO terms were: alpha-amino acid metabolic process (GO:1901605) with 1218 transcript sequences; nucleic acid-templated transcription (GO:0097659) with 1173 transcript sequences; hermaphrodite genitalia development (GO:0040035) with 1173 transcript sequences; regulation of RNA biosynthetic process (GO:2001141) with 984 transcript sequences and regulation of vulval development (GO:0040028) with 447 transcript sequences. Within the predicted proteins from ORFs, we identified 3012 signatures present in the PFAM, 578 in SMART and 618 in the SUPERFAMILY database. The top 20 most abundant signatures identified by these software are shown in Fig. 3A–3C, respectively.

Fig. 2. Functional categorization of transcripts of A. cantonensis based on GO annotations terms at level 2. Functional annotation was performed using Blast2GO. The transcriptome was functionally mapped to GO terms and annotated using an E-value-hit-filter of 1.0 × 106 with all other parameters as default.

Fig. 3. Top 20 most abundant signatures identified in PFAM (A), SMART (B) and SUPERFAMILY databases (C). The most abundant functional domains identified in the coding predicted proteins of A. cantonensis are shown.
Estimation of transcript abundance in adult A. cantonensis worms
The FPKM and TPM (>400) values for the 20 top highly expressed genes are given in Table 2 and Supplementary Table S3. Among the highly expressed genes were those coding for: hypothetical protein DICVIV_07050, an unnamed protein product, rRNA promoter binding protein, von Willebrand factor type D domain protein, nematode fatty acid retinoid binding protein, ribosomal protein L40, collagen alpha-1(I) chain-like, core histone H2A/H2B/H3/H4, major sperm protein (MSP), Hsp20/alpha crystallin family, putative cuticle protein, intracellular globin, 14-3-3 protein, aspartyl protease inhibitor, Ubiquitin, glyceraldehyde-3-phosphate dehydrogenase, type I, parasitic stage specific protein 1, chondroitin proteoglycan 3, putative DNA-binding response regulator CreB, putative DNA-binding response regulator CreB and Transthyretin-like family protein (Table 2). Among the top 20 transcripts we found signal peptides for parasitic stage specific protein 1, von Willebrand factor type D domain protein and nematode fatty acid retinoid binding protein (Supplementary Table S4). Altogether 1694 predicted proteins were identified with signal peptide (Supplementary Table S4).
Table 2. Highly expressed genes in female adult worms from A. cantonensis, southern Brazil isolate from Porto Alegre, after transcriptome analysis

TPM, transcripts per kilobase million; FPKM, fragments per kilobase million.
Annotation, reconstruction of metabolic pathways and functional modules
In total, 4203 coding proteins were assigned a KO (orthologous group) with KASS, annotating 389 metabolic pathways containing at least gene, and 92 complete modules, such as glycolysis module (M00001), citrate cycle, first carbon oxidation (M00010), pyruvate oxidation (M00307), pentose phosphate pathway (M00006), galactose degradation (M00632), glycogen biosynthesis (M00854) and degradation (M00855). The genes necessary for the module of complete fatty acid biosynthesis initiation (M00082), fatty acid biosynthesis, elongation (M00083), fatty acid biosynthesis, elongation, mitochondria (M00085), elongation (M00415), sphingosine degradation (M00100), fatty acid biosynthesis, elongation, endoplasmic reticulum (M00415) and triacylglycerol biosynthesis (M00089). De novo lipid synthesis was found, as well as beta-oxidation for synthesis of palmitoyl-coA (M00086 and M00087). We identified the pathways of phosphatidylcholine (M00090) and phosphatidylethanolamine (M00092), both the largest components of phospholipid of biological membranes. We also identified the module for N-glycan precursor biosynthesis (M00055), N-glycosylation by oligosaccharyltransferase (M00072), N-glycan precursor trimming (M00073), GPI-anchor biosynthesis, core oligosaccharide (M00065), N-glycan metabolism such as glycosaminoglycan biosynthesis and linkage tetra saccharide (M00057). The module M00065 (GPI-anchor biosynthesis, core oligosaccharide), which includes the enzyme PIGA (phosphatidylinositol glycan, class A), catalyses the first step of GPI-anchor synthesis. The transcriptome annotation analysis indicates that A. cantonensis can synthesize de novo purine adenine (M00049) and guanine (M00050), pyrimidines such uridine (M00051) and other ribonucleotides (for more details see module M00052). The modules of metabolic pathways of serine biosynthesis from glycerate-3P (M00020), methionine salvage pathway (M00034) and proline biosynthesis from glutamate are present (M00015). For example, the complete module of glycosaminoglycan biosynthesis was comprised of a heparan sulphate (HS) backbone (M00059) containing all enzymes involved in the synthesis of heparan sulphate or heparin (HP) (Fig. 4). The final product of HS biosynthesis was the form of HS proteoglycan.

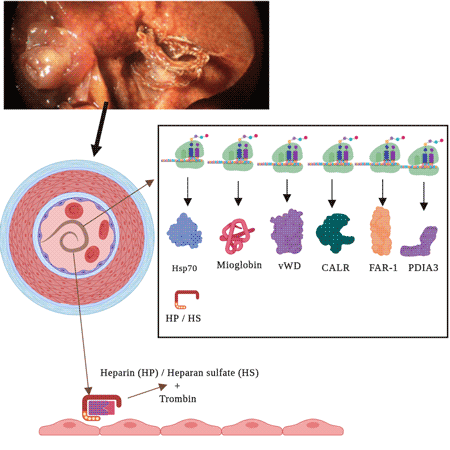
Fig. 4. Pathways of glycosaminoglycan biosynthesis. Biosynthesis of heparan sulphate and heparin backbone. Red rectangles: enzymes annotated in A. cantonensis with KEGG Automatic Annotation Server (KAAS) composing the complete functional module of heparan sulphate and heparin backbone. Other rectangles (white): unidentified enzymes.
Among the incomplete metabolic pathways, genes homologous to gp63 (EC:3.4.24.36, KO: K01404, comp16391_c0, TPM: 4.59), which encodes leishmanolysin of Leishmania sp. promastigotes (Fig. 5) and TcCRT (calreticulin, KO: K08057, comp6270_c0, TPM: 182.93) of Trypanosoma cruzi, were annotated in A. cantonensis (Fig. 6). In this last pathway, ACE (peptidyl-dipeptidase A, EC: 3.4.15.1, KO: K01283, comp30541_c0, TPM: 1.39) was also identified. In addition, the coding sequences for laminin gamma-1 (KO: K05635, LAMC1, ID: comp27259_c0_seq1, TPM: 1.94) and heat shock 70 kDa protein (HSP70) 1/2/6/8 (HSPA1s, KO: K03283, comp19023_c0_seq1, TPM: 362.36), homologous proteins components of the mechanism of infection of Toxoplasma, were also annotated.

Fig. 5. Metabolic pathways of Leishmania spp. Red rectangles: enzymes annotated in A. cantonensis with KEGG Automatic Annotation Server (KAAS) with reciprocal best-hit homologous in Leishmania spp.
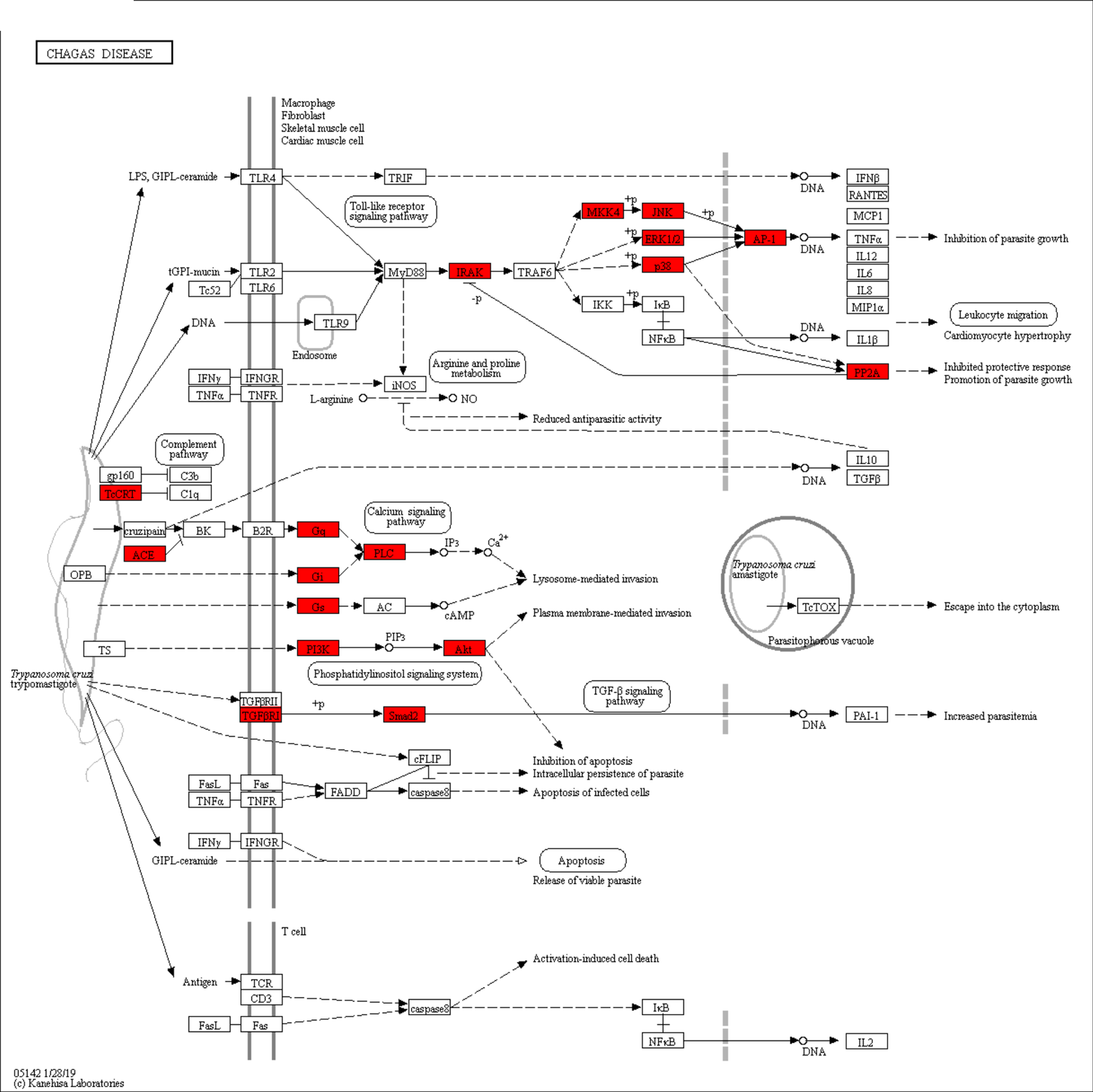
Fig. 6. Metabolic pathways of Trypanosoma cruzi. Red rectangles: enzymes annotated in A. cantonensis with KEGG Automatic Annotation Server (KAAS) with reciprocal best-hit homologous in T. cruzi.
The hemoglobin Hb [hemoglobin subunit alpha (K13822, comp7751_c0_seq1, TPM: 3.70), and its beta subunit (K13823, comp7748_c0_seq1_ORF, TPM: 2.19)] was also annotated (Supplementary Table S6). These sequences were found to have the best hit values (BLASTx, 100% of identity) against the sequences of R. norvegicus (Supplementary Table S6). However, additional sequences of intracellular myoglobin (comp18983_c0_seq1, TPM: 1658.86), homologous for nematode sequences of Syngamus trachea, were also annotated (Supplementary Table S6).
Discussion
In the current study, all NR ORFs in adult A. cantonensis worms from a Brazilian isolate were identified (available at http://angiostrongylus.lad.pucrs.br/admin/welcome). Our assembly had an N50 of 1870, totalling 85 657 Mb and 22 294 NR ORF sequences. We identified and annotated a higher number of predicted proteins than the previously deposited reference genomes of A. cantonensis (Xu et al., Reference Xu, Xu, Sun, Xu, Zeng, Shan, Yuan, He, He, Yang, Luo, Wei, Wu, Liu, Xu, Dong, Song, Zhang, Yu, Wang, Zhang, Fang, Gao, Lv and Wu2019), which could make our current transcriptome analysis a complement for the functional knowledge of the coding proteins present in A. cantonensis. In addition, our reference database makes all ORF sequences annotated available for functional genomics studies. In the reference genome of A. cantonensis (Xu et al., Reference Xu, Xu, Sun, Xu, Zeng, Shan, Yuan, He, He, Yang, Luo, Wei, Wu, Liu, Xu, Dong, Song, Zhang, Yu, Wang, Zhang, Fang, Gao, Lv and Wu2019), the fasta files with annotation are not available (https://parasite.wormbase.org/Angiostrongylus_cantonensis_prjna350391/). Among the sequences with the best BLAST hits within nematodes, only a small number were from A. cantonensis. This is because the A. cantonensis proteome is absent from the NR NCBI database. The species list with the best BLAST hit matched the phylogenetic clade V, in which A. cantonensis is grouped (Kiontke et al., Reference Kiontke, Barrière, Kolotuev, Podbilewicz, Sommer, Fitch and Félix2007; van den Elsen et al., Reference van den Elsen, Holovachov, Karssen, van Megen, Helder, Bongers, Bakker, Holterman and Mooyman2009).
The GO terms are organized hierarchically from the more general level terms to the more specific parent terms (Alexa et al., Reference Alexa, Rahnenführer and Lengauer2006). In our analysis, we found a similar pattern of GO terms as in a previous transcriptome study (Yu et al., Reference Yu, Cao, Long, Tukayo, Feng, Fang and Luo2017), but we have additionally identified the GO terms behaviour, cell population proliferation and biological adherence (Fig. 2). Although the same study by Yu and colleagues aimed to compare L5 and adult worms’ transcriptome in A. cantonensis, the current study focused on the complete functional annotation, ORFs’ identification and reconstruction of metabolic pathways important for the survival of the adult worm within the host.
The homologue for glyceraldehyde-3-phosphate dehydrogenase (Table 2) was annotated as one of the most expressed mRNAs in A. cantonensis. This enzyme was shown to act as a complement-binding protein, inhibiting the complement-mediated lysis of sensitized erythrocytes (Sahoo et al., Reference Sahoo, Murugavel, Devi, Vedamurthy, Gupta, Singh and Joshi2013), and was present in the parasite secretion product of H. contortus (Vedamurthy et al., Reference Vedamurthy, Sahoo, Devi, Murugavel and Joshi2015), suggesting that it could be a common strategy used by parasites to suppress the host's immune response.
The MSP genes (comp19031_c0 and comp19047_c0; Supplementary Table S3) were found to be highly expressed in adult females of A. cantonensis. Since these genes have not been previously reported in Angiostrongylus species, we investigated the MSP transcripts further and assembled the raw reads of A. cantonensis adult females from another transcriptome study, deposited in the SRA database (SRR3199277) (Yu et al., Reference Yu, Cao, Long, Tukayo, Feng, Fang and Luo2017). As a result, the same ORFs encoding the MSP proteins were found (Supplementary Table S5). MSP genes have been described to encode sperm-specific cytoskeletal proteins, where their expression in nematodes is restricted to sperm (Cottee et al., Reference Cottee, Nisbet, Boag, Larsen and Gasser2004; Strube et al., Reference Strube, Buschbaum and Schnieder2009). The MSP protein of Ascaris males is an essential protein for the development and locomotion of amoeboid sperm (King et al., Reference King, Essig, Roberts and Moerland1994; Kuwabara, Reference Kuwabara2003). In C. elegans, hermaphrodite MSP is secreted to stimulate oocyte maturation and ovulation (Miller, et al., Reference Miller, Nguyen, Lee, Kosinski, Schedl, Caprioli and Greenstein2001; Han et al., Reference Han, Cottee and Miller2010). This study is the first to report on MSP transcripts present in female nematodes. This may be due to MSP mRNA molecules present in recent sperm fertilized females just before collection, since in C. elegans 10% of oocyte mRNAs are of paternal origin from mRNA transferred from sperm to oocytes (Stoeckius et al., Reference Stoeckius, Grün and Rajewsky2014).
An intracellular globin (TPM: 1276.59) highly expressed in A. cantonensis was also annotated. As adult worms inhabit arteries, we hypothesized that the biological role of this protein is associated with the acquisition of blood oxygen and haem group oxygen from the host. The myoglobin-predicted protein identified here presents with 60.1% of identity with the myoglobin sequence (GLB2) of Nippostrongylus brasiliensis. This GLB2 was found to be expressed when the nematode enters the host and on the L4 lung stage as an anaerobic pre-adaptation to the intestine (Bouchery et al., Reference Bouchery, Filbey, Shepherd, Chandler, Patel, Schmidt, Camberis, Peignier, Smith, Johnston, Painter, Pearson, Giacomin, Loukas, Bottazzi, Hotez and LeGros2018). This suggest the metabolic dependence of A. cantonensis on its host, since parasite nematodes lack a complete and functional haem biosynthetic pathway (Rao et al., Reference Rao, Carta, Lesuisse and Hamza2005), and must acquire haem from exogenous sources (Luck et al., Reference Luck, Yuan, Voronin, Slatko, Hamza and Foster2016).
Another transcript that was identified was FAR-1, which encodes an unusual secreted α-helix rich lipid-binding protein found exclusively in nematodes (Rey-Burusco et al., Reference Rey-Burusco, Ibanez-Shimabukuro, Gabrielsen, Franchini, Roe, Griffiths, Zhan, Cooper, Kennedy, Corsico and Smith2015). In agreement with our findings, the transcripts encoding for FARs are expressed in adult females of Aphelenchoides besseyi (Cheng et al., Reference Cheng, Xiang, Xie, Xu, Xie, Zhang and Li2013), A. ceylanicum (Fairfax et al., Reference Fairfax, Vermeire, Harrison, Bungiro, Grant, Husain and Cappello2009), Heterodera avenae and Heterodera filipjevi (Qiao et al., Reference Qiao, Luo, Peng, Luo, Huang, Cui, Li, Kong, Jiang, Chitwood and Peng2016). FAR-1 protein antibodies have been used for the immunization of animals infected with A. ceylanicum (Fairfax et al., Reference Fairfax, Vermeire, Harrison, Bungiro, Grant, Husain and Cappello2009) and B. malayi (Zhan et al., Reference Zhan, Arumugam, Kennedy, Tricoche, Lian, Asojo, Bennuru, Bottazzi, Hotez, Lustigman and Klei2018).
A putative DNA-binding response regulator CreB (TPM: 632.26, 100% of identity with CAR63606.1) was among the top 20 genes that were highly expressed (Table 2) and is specific to A. cantonensis. There were no BLASTx/BLASTp hits found against any other organisms when these sequences were compared against the NR NCBI database and Worm Parasite database. Considering its uniqueness, it has potential for use as a marker in the diagnosis of angiostrongyliasis. Currently, the function of the putative DNA-binding response regulator CreB in A. cantonensis remains unknown. However, corroborated with our findings, the cDNA of this transcript was previously identified in the fourth-stage larvae of A. cantonensis in a previous study (He et al., Reference He, Cheng, Yang, Meng, He, Zheng, Li, Guo, Pan and Zhan2009).
The annotation results of the metabolic pathways indicated that adult A. cantonensis can synthesize and metabolize major organic macromolecules (Supplementary Table S6). Unlike cestodes and schistosomes, which have lost their capacity for the de novo synthesis of lipids and have become entirely dependent on hosts (Barrett, Reference Barrett, Arme and Pappas1983; Tyagi et al., Reference Tyagi, Rosa, Lewis and Mitreva2015b), A. cantonensis has complete modules for the biosynthesis, initiation, elongation of fatty acids, as well as the beta-oxidation of lipids. As a result, A. cantonensis is also able to synthesize several amino acids, namely serine, cysteine, methionine and proline, given that the complete functional modules for these pathways are present (Supplementary Table S6). However, the pathways for the biosynthesis and degradation of histidine, lysine, phenylalanine, threonine, leucine, isoleucine and valine were found to be incomplete, suggesting its metabolic dependence on the host for these amino acids. The parasites Leishmania, Plasmodium and Cryptosporidium lost their ability to synthesize the nine amino acids (Phe, Trp, Ile, Leu, Val, Lys, His, Thr and Met) in nine independent pathways as soon as they evolved the ability to feed on other organisms (Payne and Loomis, Reference Payne and Loomis2006). In a future study, we will use a comparative genomic approach to compare the metabolic dependence of A. cantonensis and Angiostrongylus costaricensis on their hosts.
The complete modules for the biosynthesis and trimming of the N-glycan precursor (M00055) were found to be present in A. cantonensis. The N- and O-glycans are major constituents of glycoproteins and outer layers covered by a carbohydrate-rich glycocalyx or cuticle surface coat. They have attracted the attention of researchers in parasitic nematodes due to their immunogenic and immunomodulatory characteristics (Veríssimo et al., Reference Veríssimo, Graeff-Teixeira, Jones and Morassutti2019). Other worms, such as Schistosoma spp., T. spiralis, B. malayi, Meloidogyne incognita, Bursaphelenchus xylophilus, Pristionchus pacificus and Ascaris sum, lack the ability to synthesize the N-glycan precursor (M00055) (Tyagi et al., Reference Tyagi, Joachim, Ruttkowski, Rosa, Martin, Hallsworth-Pepin, Zhang, Ozersky, Wilson, Ranganathan, Sternberg, Gasser and Mitreva2015a), and may acquire macromolecules from the hosts. For the biosynthesis of GPI-anchor, core oligosaccharide (M00065), which is indispensable for the germline development of the nematode C. elegans (Murata et al., Reference Murata, Nomura, Dejima, Mizuguchi, Kawasaki, Matsuishi-Nakajima, Ito, Gengyo-Ando, Kage-Nakadai, Mitani and Nomura2012), was also found in A. cantonensis. In addition, two PIGA genes involved in the first step of GPI-anchor synthesis (K03857, phosphatidylinositol N-acetylglucosaminyltransferase subunit A, EC 2.4.1.198, ID: comp18316_c0_seq13 and comp18514_c0_seq25, Supplementary Table S2) were also annotated. These PIGA genes are indispensable for the maintenance of the mitotic germline cell number, germline formation and the normal development of oocytes and eggs, with knockout worms displaying 100% lethality (Murata et al., Reference Murata, Nomura, Dejima, Mizuguchi, Kawasaki, Matsuishi-Nakajima, Ito, Gengyo-Ando, Kage-Nakadai, Mitani and Nomura2012). Therefore, this opens a new line of investigation for the treatment of angiostrongyliasis.
Several other candidate targets for the treatment of angiostrongyliasis were found. Chondroitin proteoglycan 3 (CS3, comp19010_c0_seq1: 756.36) was found among the top 20 most highly expressed genes with the highest expression value. Chondroitin proteoglycan 3 is a heterogeneous group of heavily glycosylated proteins with covalently attached glycosaminoglycan (GAG) chains of chondroitin sulphate (CS) (Noborn et al., Reference Noborn, Gomez Toledo, Nasir, Nilsson, Dierker, Kjellén and Larson2018). It was found to have greater expression in the parasitic female stage of Strongyloides ratti compared with other lifecycle stages (Spinner et al., Reference Spinner, Thompson, Emery and Viney2012). Furthermore, it was also reported as essential in the oogenesis or early embryogenesis of C. elegans, given that treated worms were not viable, showed poor gonad formation, and laid fewer eggs (Galvin et al., Reference Galvin, Mizuguchi, Uyama, Kitagawa, Nomura, Dejima, Gengyo-Ando, Mitani and Sugahara2003).
In addition to chondroitin proteoglycan 3, other proteoglycans (HSPGs) may be useful as treatment targets. The HSPGs that can attach to GAGs are chondroitin, dermatan sulphate and heparan sulphate (HS) (Fig. 4A and Supplementary Table S6). At the first time, the complete module for the HS biosynthetic pathways was identified in the transcriptome of A. cantonensis, with the genes showing moderate levels of expression (3 ⩽ TPM < 100) (Supplementary Table S6). The proteoglycans of heparan sulphate exhibit anticoagulant effects (Teien, et al., Reference Teien, Abildgaard and Höök1976) and play important biological roles in development, cell adhesion, cell migrations, cytokine liberation of secretion, growth factors and morphogens (Esko and Selleck, Reference Esko and Selleck2002; Kreuger and Kjellén, Reference Kreuger and Kjellén2012). Among the HS pathway genes, hst-2 (heparan sulphate 2-O-sulphotransferase) an essential enzyme for the HS sulphation of proteoglycans and tyrosine-sulphated protein. The RNAi-mediated inhibition of hst-2 in C. elegans was found to result in abnormal oocyte formation and inhibited egg production (Akiyoshi et al., Reference Akiyoshi, Nomura, Dejima, Murata, Matsuda, Kanaki, Takaki, Mihara, Nagaishi, Furukawa, Ando, Yoshina, Mitani, Togayachi, Suzuki, Shikanai, Narimatsu and Nomura2015).
In humans, the von Willebrand factor (VWF) has two essential functions in haemostasis: (i) mediating the adhesion of platelets to subendothelial connective tissue and (ii) binding to blood clotting factor VIII. Patients who lack VWF have shown defects both in blood clotting and in the formation of platelet plugs at vascular injury sites (Sadler, Reference Sadler2008). The protein domains of von Willebrand/integrin A have been proposed to play a biological role in nematodes as a cell adhesion factor (Whittaker and Hynes, Reference Whittaker and Hynes2002). The functional Von Willebrand factor type D domain (vWF-D) is present in different forms of vitellogenin (Vg), the precursor of egg-yolk proteins in both vertebrates and invertebrates (Akasaka et al., Reference Akasaka, Kato, Kitajima and Sawada2013; Sun et al., Reference Sun, Hu, Liu, Gao and Zhang2013). The fact that vWF-D transcript isoforms (comp6474_c0_seq1, comp6476_c0_seq1 and comp6473_c1_seq1) were found to be highly expressed in the adult worms of Angiostrongylus gave rise the hypothesis of a possible survival strategy, since worms live inside arteries and are in close contact with the vascular endothelium. The predicted proteins of these transcript isoforms contain signal peptides, lipoprotein N-terminal, and vWF-D domains, as predicted in SMART (Supplementary Table S4). We hypothesized that HS and vWF-D modulate coagulation and prevent thrombosis in the intravascular habitat of A. cantonensis.
Furthermore, a leishmanolysin homologue (TPM: 4.59) with moderate expression was also annotated in A. cantonensis. Leishmanolysin (gp63) was first identified in Leishmania promastigotes, where it facilitates migration through the host extracellular layer, as well as it affects the AK, MAP and IRAK-1 kinase signalling pathways (McGwire et al., Reference McGwire, O'Connell, Chang and Engman2002). This protein plays a crucial role in migration through the extracellular matrix and basement membrane proteins. This may facilitate the access of parasites to the blood or lymph circulation, enabling dissemination to distant sites (Ghosh et al., Reference Ghosh, Bandyopadhyay, Kole and Das1999). The gp63 homologue in schistosomes has been expanded to at least 12 putative family members compared to a single orthologue in humans, fruit fly and C. elegans (Brindley et al., Reference Brindley, Mitreva, Ghedin and Lustigman2009; Zhou et al., Reference Zhou, Zheng, Chen, Zhang, Wang, Guo, Huang, Zhang, Huang, Jin, Dou, Hasegawa, Wang, Zhang, Zhou, Tao, Cao, Li, Vinar, Brejova, Brown, Li, Miller, Blair, Zhong, Chen, Liu, Hu, Wang, Zhang, Song, Chen, Xu, Xu, Ju, Huang, Brindley, McManus, Feng, Han, Lu, Ren, Wang, Gu, Kang, Chen, Chen, Chen, Wang, Yan, Wang, Lv, Jin, Wang, Pu, Zhang, Zhang, Hu, Zhu, Wang, Yu, Wang, Yang, Ning, Beriman, Wei, Ruan, Zhao and Wang2009). Functional studies on schistosome cercaria have found that this protein may facilitate tissue invasion (Curwen et al., Reference Curwen, Ashton, Sundaralingam and Wilson2006). The homologue to the metalloprotease leishmanolysin in Entamoeba histolytica, MSP-1 (EhMSP-1), was shown to be a surface metalloprotease involved in the regulation of amoebic adherence and cell motility (Teixeira et al., Reference Teixeira, Sateriale, Bessoff and Huston2012).
The coding sequences for peptidyl-dipeptidase A (angiotensin-converting enzyme, peptidyl-dipeptidase A, EC 3.4.15.1) were identified in A. cantonensis with low levels of expression. Caenorhabditis elegans peptidyl-dipeptidase A lacks crucial active site residues and cannot function as a typical ACE. However, its coding gene is essential for larval development and adult morphogenesis, and is expressed in hypodermal cells, the developing vulva, and ray papillae of the male tail (Brooks et al., Reference Brooks, Appleford, Murray and Isaac2003).
Finally, we annotated the coding sequence transcripts for HSP70 in A. cantonensis, the homologue for HSP70 of Toxoplasma gondii (TgHSp70), which is also known as TgHSp70 cyclophilin 18 (TgCyp18). TgHSp70 is highly antigenic, and mice immunized with rTgHSP70 or rTgHSP70 were found to exhibit a significantly reduced number of cysts in the brains and reduced tissue damage (Czarnewski et al., Reference Czarnewski, Araújo, Oliveira, Mineo and Silva2017). The secreted T. gondii TgHSp70 is involved in the triggering of cysteine–cysteine chemokine receptor 5 (CCR5) in dendritic cells and macrophages, and induces interleukin-12 (Ibrahim et al., Reference Ibrahim, Bannai, Xuan and Nishikawa2009). In H. contortus, HSP70 is expressed in all larval stages and adult forms, with the highest expression found in eggs (Zhang et al., Reference Zhang, Zhou, Yang, Chen, Yan and Du2013). It is one of the most abundant proteins in S. mansoni egg secretions (Cass et al., Reference Cass, Johnson, Califf, Xu, Hernandez, Stadecker, Yates and Williams2007), and was found in the somatic extracts of the larval and adult stages of T. spiralis. Mice vaccinated with T. spiralis HSP70 exhibit strong humoral immune responses and significant reduction in the number of parasites (Wang et al., Reference Wang, Zhu, Yang, Yang, Gu, Wei, Hao, Boireau and Cui2009). Moreover, HSP70 is a potent immunogen in several parasitic infections, such as B. malayi (Selkirk et al., Reference Selkirk, Denham, Partono and Maizels1989), Onchocerca volvulus (Rothstein et al., Reference Rothstein, Higashi, Yates and Rajan1989), Wuchereria bancrofti (Ravi et al., 2004), Echinostoma caproni (Higón et al., Reference Higón, Monteagudo, Fried, Esteban, Toledo and Marcilla2008) and Litomosoides sigmodontis (Hartmann et al., Reference Hartmann, Singh, Rathaur, Brenz, Liebau, Fleischer and Breloer2014). However, further investigation is needed in order to evaluate A. cantonensis Hsp70 as a potential vaccine candidate.
In the current study, several highly expressed genes involved in essential pathways for A. cantonensis survival and immune system modulation inside of blood vessels of R. norvegicus, some of these mechanisms appear conserved during evolution, because they have already been described in other parasites. Here, the full-length transcripts were also identified, which may contribute to the development of vaccine candidates and aid functional genomics studies in elucidating their biological roles. Overall, this study provides a comprehensive source of open data for use in future research on Angiostrongylus nematodes.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182021000469
Data
The raw FASTQ files analysed during the current study are publicly available and have been deposited in BioProject under accession number PRJNA549533. The FASTA files of all ORFs and functional annotations generated during this study are publicly available at http://angiostrongylus.lad.pucrs.br/admin/welcome.
Acknowledgements
The authors would like to thank the Laboratório de Alto Desempenho (LAD) of the PUCRS Polytechnic School and the High-Performance Computing Technician Rafael Lorenzo Bellé for all technical support and Dr Malcolm Jones for the fruitful discussions about A. cantonensis biology. The graphical abstract was created with BioRender.com.
Author contributions
L. M. P. was responsible for conceptualization and design/analysis of bioinformatics, interpretation of data, wrote and revision the article. M. P. G. J. carried out web database construction, took part in writing of the article and revision. B. D. B. participated in part of bioinformatic analysis and revision of the article. A. R. R. and A. B. Z. participated in part of bioinformatic analysis. C. C. T. was responsible for the conceptualization and design of the project, search for financial support, participated in the writing and revision of the manuscript. A. L. M. was responsible for the conceptualization and design of the project, responsible for all experimental laboratory analyses, search for financial support, participated in the writing and revision of the article.
Financial support
This study was supported by the Conselho Nacional de Pesquisa e Desenvolvimento Tecnológico do Brasil [Grant numbers CNPq 401904/2013-0 (2013) and CNPq PQ1D 307005/2014-3 (2014)] and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior do Brasil (Edital 32, 2010).
Conflict of interest
The authors declare there are no conflicts of interest.