Introduction
West Indies, as other islands, are considered as natural laboratories for understanding species formation, colonization, diversification and extinction (MacArthur & Wilson, Reference MacArthur and Wilson1963; Woods, Reference Woods and Woods1989; Ricklefs & Bermingham, Reference Ricklefs and Bermingham2008). They are composed of groups of islands with different origins, ages and geography. The Greater Antilles are old, large and have possibly conserved interactions with the surrounding continents, i.e. North and Central America, since their origin. Consequently, the history of their fauna is complex, with possible repeated episodes of colonization. The Lesser Antilles originated more recently (less than 20 Myr ago), partly as a consequence of the active volcanic activity yet going on. They were never connected to each other, neither to the continent. Thus, as oceanic islands, it is generally assumed that they have been colonized by over-water dispersal (Darlington, Reference Darlington1957) prior to any possibility of vicariance. The entomo-fauna of these islands is fairly poor, for the number of both species and individuals per species. This may be a consequence of their recent colonization associated with difficulties of adaptation to endemic flora. Cyclocephala tridentata Fabricius 1801 (Coleoptera: Scarabaeidae: Dynastinae) is among the rare species which apparently can pullulate. This root-eater insect found an abundant source of food with sugar cane cultures, intensely developed since the 18th century. It belongs to the largest genus of Dynastinae, mainly distributed in Central and South America (Endrödi, Reference Endrödi1985). Four other species of Cyclocephala occur in Guadeloupe, including a strictly endemic one, C. insulicola Arrow 1937, morphologically very close to C. tridentata, but limited to highlands of the rain forest of Basse-Terre, while C. tridentata occurs all around, in the rest of Guadeloupe and other islands. Because of its limited distribution area, C. insulicola was considered as a possible vicariant of C. tridentata (Chalumeau, Reference Chalumeau1983). Two continental species, C. maffafa and C. melanocephala, occur as a sub-species (C. maffafa grandis Burmeister 1847 and C. melanocephala rubiginosa Burmeister 1847). The last species, C. immaculata Olivier, 1789, is limited to the north of Grande-Terre.
C. tridentata is also known from Dominica and Martinique. Its possible presence in South America needs evaluation (Endrödi, Reference Endrödi1985). Populations from Dominica were considered as a sub-species, C. t. dominicensis, while those of Guadeloupe and Martinique remained in the same sub-species C. t. tridentata (Chalumeau, Reference Chalumeau1983). A first cytogenetic investigation in Guadeloupe revealed some surprising data (Dutrillaux et al., Reference Dutrillaux, Xie and Dutrillaux2007). The karyotypes of C. insulicola and C. m. grandis, composed of sub-metacentrics, were very close to each other and fairly comparable to those of other Dynastinae species. At contrast, the karyotype of C. t. tridentata, mainly composed of acrocentrics and highly polymorphic, appeared very different from others. It was postulated that it was highly derived from an ancestral karyotype close to that of other species.
DNA-based techniques have proven useful in many areas of entomological research, particularly in the study of taxonomic and phylogenetic relationships (reviewed in Caterino et al., Reference Caterino, Cho and Sperling2000) and population genetics (Behura, Reference Behura2006). Several gene regions have been investigated but the two regions most often targeted for sequencing in insect systematics are mitochondrial DNA (mtDNA) and nuclear ribosomal DNA (rDNA) (Simon et al., Reference Simon, Frati, Beckenbach, Crespi, Liu and Flook1994; Caterino et al., Reference Caterino, Cho and Sperling2000; Greenstone, Reference Greenstone2006; Stouthamer, Reference Stouthamer, Bigler, Babendreier and Kuhlmann2006). The genes most commonly used include cytochrome oxidase I and II (COI, COII) for mtDNA, and the 16S and 12S subunits of rDNA. Generally, the COI gene is also widely used in DNA barcoding that involves the sequencing of a particular fragment of DNA as a way of identifying an organism. Comparison of the COI sequence from an unidentified specimen to DNA sequence databases of identified and characterized species may allow the identification and phylogenetic classification of the specimen. As universal primers are used in the initial PCR assay, barcoding can identify new or previously unknown species, cryptic species or strains. However, as a single character (i.e. one portion of a gene) is used for the identification of a specimen, this technique may have limited phylogenetic resolution unless combined with other genetic, morphological and/or ecological information (Moritz & Cicero, Reference Moritz and Cicero2004; Dasmahapatra & Mallet, Reference Dasmahapatra and Mallet2006). Therefore, DNA taxonomy is a current controversy (e.g. Hebert et al., Reference Hebert, Cywinska, Ball and deWaard2002; Lipscomb et al., Reference Lipscomb, Platnick and Wheeler2003; Tautz et al., Reference Tautz, Arctander, Minelli, Thomas and Vogler2003; Will & Rubinoff, Reference Will and Rubinoff2004). Regardless of differing theoretical or practical views about the larger ‘barcoding’ programme, a portion of variable DNA sequence can aid diagnosis minimally. Circumstances might include the identification of a limited set of well-known taxa or groups in which traditional diagnostic techniques are exceptionally difficult. Current examples have focused primarily on the identification of species when specimens are especially similar or indistinguishable using traditional types of data.
To understand phylogenetic and taxonomic relationships between Cyclocephala species in Lesser West Indies and the origins of island populations, a dual chromosomal and molecular study was developed in specimens from Guadeloupe, Dominica and Martinique, which indicate a northward colonization of these islands.
Material and methods
All animals were captured by light traps in the three islands. Captures were performed at several locations of Basse-Terre of Guadeloupe, near: Petit-Bourg (16°12′N, 61°39′W), Deshayes (16°19′N, 61°45′W), Sofaïa (16°18′N, 61°43′W) and in the west versant of the rain forest (16°11′N, 61°45′W) in March or December 2006, 2007 and 2009. All insects from Dominica were caught at Trafalgar Falls (15°19′N, 61°22′W) in December 2009. Those from Martinique were caught at Le Prêcheur (14°48′N, 61°13′W), Sainte-Anne (14°26′N, 60°53′W) and Ajoupa-Bouillon (14°50′N, 61°08′W) in November 2009.
The insects were kept alive in earth and fed with apple during one to ten days, until they could be dissected in the INRA laboratory from Guadeloupe (Domaine Duclos) or MNHN laboratory in Paris. Dividing cells for cytogenetic studies were obtained from mid gut or testis (Angus, Reference Angus1982, Reference Angus1988) and treated according to our usual methods (Dutrillaux et al., Reference Dutrillaux, Moulin and Dutrillaux2006, Reference Dutrillaux, Pluot-Sigwalt and Dutrillaux2010).
Sequencing analysis of the mitochondrial COI gene
Mitochondrial DNA was isolated from 152 individuals: 124 specimens Cyclocephala tridentata (87 from Guadeloupe, 26 from Dominica and 11 from Martinique), 17 Cyclocephala melanocephala rubiginosa (eight from Guadeloupe and nine from Martinique), four Cyclocephala insulicola from Guadeloupe and five Cyclocephala maffafa. A 525 bp segment at the 3′ end of the COI gene was amplified using the primers C1J 1718 (5′-GGAGGATTTGGAGGTTGATTAGTTCC-3′) and C1N 2191 (5′-CCCGGTAAAATTAAAATATAAACTTC-3′) (Simon et al., Reference Simon, Frati, Beckenbach, Crespi, Liu and Flook1994). PCR reactions (50 μl) contained 200–500 ng DNA, 10× Taq buffer, 2 mM MgCl2, 0.2 mM of each dNTP, 50 pmoles of each primer and 1 U Taq polymerase (Invitrogen, Carlsbad, CA, USA). The cycling conditions consisted of an initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 40 s and extension at 72°C for 1 min, with a final extension at 72°C for 10 min. All amplified samples were screened for polymorphism using the Single-Strand Conformation Polymorphism (SSCP) method. This method allows the detection of single base polymorphisms in short DNA stretches due to mobility differences of single-stranded DNA fragments during electrophoresis in polyacrylamide gels (Orita et al., Reference Orita, Iwahana, Kanazawa, Hayashi and Sekiya1989). Preliminary SSCP tests were performed with samples known to carry different sequences. Samples that showed the same SSCP pattern were grouped, and representative samples from each profile were purified using QIAquick PCR purification kit (QIAGEN Cat. No. 28106, Valencia, CA, USA) and were sequenced directly and bi-directionally by Macrogen Inc. Nucleotide sequences were aligned using ClustalX (Larkin et al., Reference Larkin, Blackshields, Brown, Chenna, McGettigan, McWilliam, Valentin, Wallace, Wilm, Lopez, Thompson, Gibson and Higgins2007).
For all haplotypes, base composition, nucleotide variation, polymorphic and parsimony informative sites were assessed using MEGA version 4.0 (Tamura et al., Reference Tamura, Dudley, Nei and Kumar2007). Phylogenetic associations among lineages were assessed with PAUP* 4.0 beta 10 version (Swofford, Reference Swofford1998). To determine the appropriate model of sequence evolution and statistically compare successively nested more parameter-rich models for this data set, the program MODELTEST Version 3.6 (Posada & Crandall, Reference Posada and Crandall1998) was used. With a statistical significance of P=0.01 the HKY85 model (Hasegawa et al., Reference Hasegawa, Kishino and Yano1985), with gamma correction, obtained the best likelihood score and was thus selected for the neighbor-joining (NJ) analysis. Parsimony and ML trees were constructed under the heuristic search option with 100 random-taxon-addition replicates and tree bisection–reconnection branch swapping, using PAUP*. Node support was assessed on the basis of 1000 bootstrap replicates.
A Bayesian analysis was also performed with mrbayes version 3.1 (Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001), under the HKY85 model of sequence evolution. Depending on the data set, random starting trees run for 2×106 to 8×106 generations were used, sampled every 100 generations. Burn-in frequency was set to the first 25% of the sampled trees. For all trees, the sequences of Cyclocephala mafaffa grandis were used as outgroup. The software Network 3.1.0.1 (Fluxus Technology Ltd, downloaded from http://www.fluxus-technology.com/sharenet.htm) was used to construct a median joining (MJ) network based on the restriction site data.
Results
Chromosomes
Cyclocephala tridentata (CT)
Guadeloupe (CTT)
Following our study of ten males (Dutrillaux et al., Reference Dutrillaux, Xie and Dutrillaux2007), 36 additional specimens were karyotyped. All had 2n=20 chromosomes, with a number of acrocentrics ranging from 8 to 14 (fig. 1a). All large non-acrocentrics had a very unusual conformation, with long heterochromatic centromeric regions recalling HSRs (homogeneously staining regions) carrier of amplified DNA. When sub-metacentrics were present in a karyotype, their homo- or heterozygote status could be identified by bivalent analyses at pachynema (fig. 1a). We could estimate that up to six chromosome pairs can be involved in this polymorphism. The karyotype of a sterile specimen, without spermiogenesis, captured in the rain forest at an altitude of about 450 m was obviously hybrid between C. t. tridentata and C. insulicola (fig. 1b), with seven acrocentric/sub-metacentric heterozygote pairs, none of them carrying HSR-like structures, as in C. insulicola karyotype (not shown). Considering their homo- or heterozygote status, 18 different karyotype formulae could be established in addition to that of the hybrid.

Fig. 1. (a) C-banded karyotype of C. tridentata from Guadeloupe with three large sub-metacentrics (N°1, 1 and 3) carrier of amplified heterochromatin (dark). Homozygosity for pair 1 and heterozygosity for pairs 3 and 8 is demonstrated by bivalents staining at pachytene stage of meiosis I (large elements on the right). C, centromere locations on bivalents. (b) C-banded karyotype of a hybrid C. tridentata×C. insulicola. The large acrocentrics (right) are from C. tridentata. The large sub-metacentrics (left) are from C. insulicola and do not carry amplified heterochromatin. (c) C-banded karyotype of C. tridentata dominicensis carrier of four large acrocentrics. The homo- and heterozygote status has been determined by pachynema analysis, as shown in (a). (d) C-banded karyotype of C. tridentata from Martinique. The small telomeric C-bands are variable, but all specimens had only sub-metacentric chromosomes. (e) C-banded karyotype of C. melanocephala rubiginosa from Martinique. One pair of small chromosomes (N° 7 or 8) and the X are acrocentric. Chromosome 1 is more metacentric than in other species. Bar=10 μm.
Dominica (CTD)
The 11 karyotyped specimens had 2n=20 chromosomes. The number of acrocentrics ranged from 3 to 6. None of the large sub-metacentrics had the HSR-like structures found in specimens from Guadeloupe (fig. 1c). Meiotic analyses indicated that three pairs were involved in this polymorphism, probably by pericentric inversions.
Martinique (CTM)
The nine specimens studied also had 2n=20 chromosomes. All were sub-metacentric. Most of them had a discrete C-banding at telomeric regions, not detected in specimens from Guadeloupe and Dominica (fig. 1d). No variations of euchromatin were observed among the specimens from the three different investigated locations of the island.
Cyclocephala melanocephala rubiginosa (CMR)
Fairly similar to that of CTM, its karyotype differed by pair N°1, more metacentric and pairs N°8 and X, acrocentric (fig. 1e). Only specimens from Martinique (CMRM) were karyotyped.
Cyclocephala mafaffa grandis (CMG)
Described in Dutrillaux et al. (Reference Dutrillaux, Xie and Dutrillaux2007), its karyotype differs from that of CTM by the morphology of its chromosome X, acrocentric as in CMR, and the lack of telomeric heterochromatin.
Cyclocephala insulicola (CIN)
Also described in Dutrillaux et al. (Reference Dutrillaux, Xie and Dutrillaux2007), its karyotype, is very similar to that of CTM, but does not exhibit telomeric heterochromatin.
Nucleotide analysis and phylogenetic reconstruction
Sequences obtained, primers excluded, corresponded to a 525 bp segment. Haplotypes were evaluated and compared to published sequences in Genbank. All corresponded to new sequences and thus were submitted to GenBank. Their accession numbers are: JF309340, JF309341, JF309342, JF309344, JF309345, JF309355 and JF309356 for C. t. tridentata from Guadeloupe, JF309360, JF309361, JF309358 and JF309363 for C. tridentata from Martinique, JF309343, JF309354 and JF309349 for C. insulicola, JF309350 and JF309359 for C. mafaffa grandis, JF309351, JF309352, JF309353 and JF309357 for C. t. dominicensis and finally JF309364, JF309362, JF309348, JF309347 and JF309346 for C. melanocephala rubiginosa.
Nuclear mitochondrial pseudogenes (numts) seem to commonly occur in most eukaryotic species studied so far (Richly & Leister, Reference Richly and Leister2004; Cameron et al., Reference Cameron, Sullivan, Song, Miller and Whiting2009). However, the absence of insertions, deletions or in-frame stop codons within the sequences studied indicated that they correspond to functional mitochondrial COI gene fragments and are not derived from numts. The high percentage (59.3% on average) observed for the A+T content, is a common feature of animal mitochondrial genes (Brown, Reference Brown and Macintyre1985). Out of the 525 sites, 141 were variable while 132 of them were informative for parsimony. Among the 141 variable sites, a pattern typically observed in segments under strong functional constraints was revealed given that 78.7% are third codon positions, while first and second codon positions are much more conserved (19.9% and 1.4%, respectively). A number of insect molecular studies using COI are in accordance with these findings (Cognato & Sperling, Reference Cognato and Sperling2000; Villalba et al., Reference Villalba, Lobo, Martín-Piera and Zardoya2002). Similarly, of the 141 variable sites, only ten result in amino acid substitution. Sequence divergence within groups and species ranged from 0.2% for C. t. dominicensis (CTD) to 1.2% for C. melanocephala rubiginosa (CMR). The lowest sequence divergence (3.7%) was observed between C. t. tridentata (CTT) and C. t. dominicensis (CTD) and the highest (18.2%) between C. t. tridentata (CTT) and C. melanocephala rubiginosa (CMR). With some minor differences, the median joining (MJ) network (fig. 2) and the phylogenetic trees based on the Bayesian analysis (fig. 3), as well as on pairwise haplotype divergence (NJ) (tree not shown, available on request), showed similar topologies. The main branches exhibited high bootstrap values (NJ) and high posterior probabilities (Bayesian analysis). In all analyses, specimens of C. tridentata from the three islands as well as the three other species were clustered in clearly separated groups without clade overlaps. Using C. mafaffa grandis as outgroup, both trees showed that, within C. tridentata species, the group of C. tridentata from Martinique had a more basal (ancestral) position followed by the groups from Domenica and Guadeloupe (fig. 3).

Fig. 2. Median-joining (MJ) network showing the mutational relationships among 25 Cyclocephala haplotypes detected in a sample of 152 individuals. Each coloured scheme represents a haplotype, and each group is represented by a different scheme. Each haplotype is identified by its corresponding number, as in the text. Vertical bars on connecting lines between schemes represent a single mutational change, while numbers in bold indicate the total of mutational changes separating the different groups. Red schemes indicate individuals from Guadeloupe, green from Dominica and yellow from Martinique. For the three letter codes, see the Results section. CIN/CTT3 corresponds to the hybrid (fig. 1b). Its haplotype indicates that the mother was CIN.

Fig. 3. Phylogenetic tree resulting from the Bayesian analysis, clustering the 25 different haplotypes from Cyclocephala groups. The topology of the clusters was similar for the NJ tree. Numbers above branches of the major clusters correspond to posterior probabilities from the Bayesian analysis. Because different specimens showed the same haplotype, haplotypes in the tree have been chosen randomly. For the three letter codes, see the Results section. Each haplotype is identified by its corresponding number, as in text. CIN/CTT3 corresponds to the hybrid (fig. 1b). Its haplotype indicates that the mother was CIN. Trees were routed with sequences of Cyclocephala mafaffa grandis.
To estimate the time of separation between groups and species, two different base substitution rates were applied: a divergence rate of 2.3% per million year (Myr), which has been obtained for mitochondrial genes in various organisms (e.g. Brower, Reference Brower1994; Carisio et al., Reference Carisio, Cervella, Palestrini, DelPero and Rolando2004), and a divergence rate of 3.54% Myr for the COI gene based on ‘known’ dates (Papadopoulou et al., Reference Papadopoulou, Anastasiou and Vogler2010). Based on the above molecular clocks, separation times that might have occurred between groups and species ranged from 1.0 table 1).
Table 1. Pairwise estimates of nucleotide divergence (×102) among Cyclocephala species and groups.
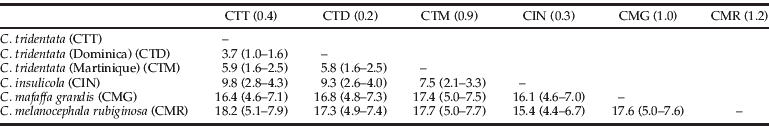
Numbers in the first line, next to the three letter codes indicate nucleotide divergence within taxa. Numbers in parentheses, next to pairwise nucleotide divergences, indicate estimates of the time of separation between groups and species in million years (Myr).
Discussion
Ancestral karyotype and chromosome evolution
Although always composed of 20 chromosomes, the karyotypes of Cyclocephala species from Lesser Antilles exhibit a large range of variations. Autosomes may be exclusively sub-metacentric, as in CMG, CIN and CTM, or principally acrocentric, as in CTT. The X chromosome may be also either sub-metacentric, as in CTT, CTD, CTM and CIN or acrocentric as in CMG and CMR. No chromosomal data exist for other Cyclocephalini, but some other Dynastinae were karyotyped. Most often, autosomes are sub-metacentric, and the X chromosome is acrocentric (euchromatin on long arm only) and carries the Nucleolus Organizer Region (NOR) on its short arm (more or less heterochromatin rich) (Dutrillaux et al., Reference Dutrillaux, Xie and Dutrillaux2007; Vidal et al., Reference Vidal, Giacomozzi and Riva1977; Vidal & Giacomozzi, Reference Vidal and Giacomozzi1978; Vitturi et al., Reference Vitturi, Colomba, Volpe, Lannino and Zunino2003; personal data). The X is also recurrently acrocentric in Melolonthinae, Rutelinae, and Cetoniinae (Dutrillaux et al., Reference Dutrillaux, Mercier, Xie and Dutrillaux2008 and unpublished data; Moura et al., Reference Moura, Souza, Melo and Lira-Neto2003), suggesting that the acrocentric morphology may be ancestral for Dynastinae. For this chromosome as for autosomes, inversions appear to have frequently occurred during evolution. One of them originated the sub-metacentric X of CTT, CTD, CTM and CIN, transposing the NOR at the extremity of the long arm. It occurred after the divergence of CMR and CMG.
The precise morphology of the ancestral sub-metacentric autosomes remains impossible to reconstruct. However, the fairly distal location of the centromere of chromosome 1, observed in some of the Cyclocephala species only, may be of recent origin. Still more recent appear to be the accumulations of acrocentrics, observed in CTD and CTT. These data make possible to propose a scheme of chromosome evolution (fig. 4). C. melanocephala ancestors would have separated first, followed by those of C. mafaffa. These two species are still present in the American continent. C. insulicola and C. tridentata from Martinique ancestors would have diverged, after the inversion of the X, without acquisition of any other structural rearrangement. Then, the accumulation of inversions, involving chromosomes 1 to 6, isolated C. t. dominicensis, and finally C. t. tridentata. This scheme fits very well with the reconstruction obtained by COI gene sequence analysis.

Fig. 4. Phylogenetic tree based on chromosome rearrangements having occurred since the divergence from the presumed ancestral karyotype of Cyclocephalini (left). For the three letter codes, see the Results section. ○, pericentric inversions; ▪, heterochromatin (H) addition. Numbers under symbols indicate the chromosomes involved. Chromosome polymorphism in CTT and CTD is not indicated. On the right, summary of the tree reconstructed by COI gene analysis.
Genome evolution and biogeography
Two species, C. melanocephala and C. mafaffa, occur in the American continent. The first one is also present, as a sub-species, in the three investigated islands, whereas the second one is present in Guadeloupe only. In the chromosomal scheme of fig. 4, they diverged shortly after the origin of the tree, but are clearly separated by inversions. They are also well separated by a high number of gene mutations (fig. 2), with a high genetic distance (17.6%). C. mafaffa is genetically homogeneous, as is C. melanocephala from Guadeloupe. At contrast, C. melanocephala from Martinique is heterogeneous. This may indicate the unique colonization of Guadeloupe vs repeated colonization of Martinique. Both chromosomal and mitochondrial DNA schemes indicate the monophyly of C. insulicola and C. tridentata, with C. tridentata from Martinique and C. insulicola which remain close to each other and to the origin. C. t. dominicensis and C. t. tridentata are clearly derived, both genetically and chromosomally, with C. t. dominicensis in intermediate position between C. t. tridentata and C. insulicola/C. tridentata from Martinique. Thus, the colonization by C. tridentata followed a northward direction, at least from Martinique to Guadeloupe, probably by over-water dispersal. According to Endrödi (Reference Endrödi1985), the species also occurs in Suriname and Colombia. Thus, it may originate from South America.
The relationships between C. tridentata and C. insulicola, indicated by both gene and chromosomal evolution, were totally unexpected. The two species are morphologically not much different; and, in Guadeloupe, the distribution of C. t. tridentata surrounds that of C. insulicola, which is limited to the forest of altitude from Basse-Terre. The latter was thus logically considered as a vicariant form of the former. Obviously, neither mitochondrial DNA nor chromosomes favor this interpretation. The only possible origins of C. insulicola are either C. tridentata from Martinique or, more likely, a common ancestor from South America. Thus, the colonization of Guadeloupe by C. insulicola is independent from that done by C. tridentata and came either directly from the continent or via Martinique. Mitochondrial data indicate an ancient origin and thus favor a continental origin. C. insulicola appears to be a relict species, only maintained in forest of altitude. We noticed that C. t. tridentata was present in 2008–2009 at a place where only C. insulicola was found ten years before. One specimen, morphologically considered as a sterile male (gonad atrophy) of C. insulicola, was found to be hybrid between C. insulicola and C. t. tridentata by both mitochondrial DNA and karyotype analyses (fig. 2). Thus, the population of C. insulicola may be in regression and that of C. t. tridentata in progression towards highlands. In Dominica, where there is no C. insulicola, C. tridentata occurs in forests at various altitudes. The origin of the Lesser Antilles is double: one ancient (around 50 Myr) corresponds to carbonate sediments, and another, more recent (less than 5 Myr), to volcanic formations. These datings are indeed compatible with the molecular clock giving species or group separations of 1.0 to 7.9 Myr.
Genome polymorphism
Data on mitochondrial DNA indicate that all the species from Guadeloupe and Dominica have low variations, while those from Martinique (C. melanocephala and C. tridentata) appear more heterogeneous. This may be interpreted as the consequence of the old colonization of Martinique, compared to Dominica and Guadeloupe.
Interestingly, no chromosomal variation was observed in species from Martinique. This well-established karyotype indicates these species are far from periods of active chromosomal speciation processes, which is not in contradiction with the DNA heterogeneity. The situation is very different for C. tridentata in Dominica and Guadeloupe. Considering the variations of the acrocentric number and the bivalent synapsis of meiotic cells, we estimated that a minimum of four and 18 different karyotypic formulae existed in Dominica and Guadeloupe, respectively. Such a high chromosomal polymorphism seems to be quite exceptional in Coleoptera. The status of Dominica, with three to six acrocentrics is clearly intermediate between that of Martinique, without acrocentric, and that of Guadeloupe, with 9 to 13 acrocentrics. This fits with DNA data and supports the idea that the chromosome polymorphism has been maintained in Dominica, after migrations to Guadeloupe, and in Guadeloupe, since the beginning of its colonization. Once again, mitochondrial DNA and chromosome data are not contradictory, but quite complementary.
During the last decade, the COI gene was used to accurately place individuals into the correct higher taxonomic levels but also into the correct species. In a survey, aiming to examine the extent of sequence diversity at COI in the major animal phyla, Hebert et al. (Reference Hebert, Cywinska, Ball and deWaard2002) analyzed more than 13,000 congeneric pairs. They found that in 11% of the 891 species of Coleoptera examined, COI sequence divergence ranged from 3.0 to 8.0%. Therefore, based on the COI sequence divergence of the present study, it is worth wondering on the taxonomic status of C. tridentata from the three islands. Sequence divergences between C. t. tridentata - C. t. dominicensis and C. tridentata (Martinique) were 3.7 and 5.9%, respectively, and between C. t. dominicensis and C. tridentata (Martinique) 5.8%. Thus, their geographical, chromosomal and genetic separation suggests they may constitute three true species. A reassessment of their comparative morphology is in progress to propose their new taxonomic status.
Acknowledgements
Molecular analyses were financed by the Postgraduate Courses ‘Biotechnology-quality Assessment in Nutrition and the Environment’ and ‘Applications of Molecular Biology-Genetics. Diagnostic Biomarkers’ of the Department of Biochemistry and Biotechnology, University of Thessaly, Greece. We thank Claudie Pavis, who facilitated our access to INRA laboratories in Guadeloupe and the two anonymous reviewers for their constructive comments.







