INTRODUCTION
Parasites in the genus Sarcocystis have a 2-host life cycle with mainly herbivores or omnivores as intermediate hosts and carnivores as definitive hosts (Dubey et al. Reference Dubey, Speer and Fayer1989). A single herbivore may serve as the intermediate host for several Sarcocystis species. In cervids, several Sarcocystis species are commonly found in the same host (Odening, Reference Odening1998). Six Sarcocystis species have been described from reindeer based on both sarcocyst morphology and molecular data (Gjerde, Reference Gjerde1986; Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007). Seven Sarcocystis species have been described from moose, 3 based on cyst morphology and molecular data, 1 by cyst morphology only, and 2 by DNA sequencing only (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008). Altogether, 5 species have been described from roe deer, 2 based on both cyst morphology and molecular data, and 3 by morphology only (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2009).
Three morphologically different sarcocyst types have previously been reported from European red deer (Cervus elaphus hippelaphus) or North American elk (Cervus elaphus canadensis). (1) Sarcocysts without discernible protrusions when viewed in the fresh state by light microscopy (LM), but with delicate band-like protrusions when observed by transmission electron microscopy (TEM). This species was named Sarcocystis cervicanis by Hernández-Rodríguez et al. (Reference Hernández-Rodríguez, Martínez-Gómez and Navarrete1981a, Reference Hernández-Rodríguez, Martínez-Gómez, Navarrete and Acosta-Garciab) and Sarcocystis wapiti by Speer and Dubey (Reference Dubey1982), who actually failed to recognize the protrusions. This species was referred to as an unnamed Sarcocystis sp. by Entzeroth et al. (Reference Entzeroth, Neméseri and Scholtyseck1983), and as Sarcocystis cf. grueneri by Wesemeier and Sedlaczek (Reference Sedlaczek and Wesemeier1995a). (2) Sarcocysts with delicate hair-like protrusions, representing in part a species that was named Sarcocystis sybillensis by Dubey et al. (Reference Dubey, Jolley and Thorne1983), but particularly the species referred to as Sarcocystis cf. capreolicanis by Wesemeier and Sedlaczek (Reference Sedlaczek and Wesemeier1995a) and by Kutkienė (Reference Kutkienė2003). (3) Sarcocysts with erect, finger-like protrusions tentatively identified as Sarcocystis cf. hofmanni by Wesemeier and Sedlaczek (Reference Sedlaczek and Wesemeier1995a) and by Kutkienė (Reference Kutkienė2003), and as a Sarcocystis sp. similar to S. hofmanni by Stolte et al. (Reference Stolte, Bockhardt and Odening1996). These species descriptions have been based on studies using either LM of fresh cysts, TEM or scanning electron microscopy (SEM), or a combination of these methods. No molecular data have previously been published from Sarcocystis species in red deer or elk.
Morphological studies of sarcocysts from different cervid hosts have shown that they harbour Sarocystis species with highly similar cyst morphology. Molecular studies of Sarcocystis DNA isolated from sarcocysts in moose, reindeer and roe deer have verified that some Sarcocystis species with similar cyst morphology in these 3 hosts, i.e., Sarcocystis alces/Sarcocystis tarandivulpes/Sarcocystis gracilis; Sarcocystis alceslatrans/Sarcocystis rangi; and Sarcocystis hardangeri/Sarcocystis ovalis/Sarcocystis oviformis, are indeed separate species, but closely related genetically (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2009).
Sarcocystis species have been assumed to be intermediate host specific (Dubey et al. Reference Dubey, Speer and Fayer1989), but a few molecular studies indicate that the same Sarcocystis species may infect 2 closely related intermediate hosts, i.e., cattle and water buffalo (Yang et al. Reference Yang, Zuo, Yao, Chen, Yang and Zhang2001; Li et al. Reference Li, Yang, Zuo, Attwood, Chen and Zhang2002; Chen et al. Reference Chen, Zuo and Hu2003). Sarcocystis neurona has also been shown to use several intermediate hosts (Mullaney et al. Reference Mullaney, Murphy, Kiupel, Bell, Rossano and Mansfield2005). Thus, only molecular methods can truly determine whether sarcocysts with similar morphology in different hosts belong to the same species or to separate species.
The slowly evolving small subunit (ssu) rRNA gene is commonly used for species identification and is well suited for studying the phylogenetic relationships of Sarcocystis species due to a relatively large amount of variability along this gene between members of the genus Sarcocystis (Morrison et al. Reference Morrison, Bornstein, Thebo, Wernery, Kinne and Mattsson2004). Studies of Sarcocystis species in moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008), reindeer (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007) and roe deer (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2009) have shown that molecular data based on the ssu rRNA gene provide useful complementary information to morphological data, in order to differentiate between species with similar cyst morphology and for correct species identification of cysts with an atypical appearance. Most Sarcocystis species in moose, reindeer and roe deer have shown little or no intra-species variation along the ssu rRNA gene. However, several variants of the ssu rRNA gene appear to occur in Sarcocystis scandinavica of moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008), and indications of several variants were also recognized as double peaks on DNA sequencing chromatograms, or slightly different DNA sequences of different isolates of Sarcocystis rangiferi and S. tarandi of reindeer (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007).
The aims of this study were (1) to identify and characterize, by morphological and molecular methods, Sarcocystis species occurring in Norwegian red deer, (2) to determine whether Sarcocystis species in red deer were genetically different from those previously found in reindeer, roe deer and moose and (3) to investigate the phylogenetic relationships between Sarcocystis species in red deer and those from other hosts.
MATERIALS AND METHODS
Materials
Red deer
Muscle tissue samples from the heart (n=37), oesophagus (n=37), and diaphragm (n=34) of a total of 37 adult red deer were obtained from animals killed during the regular hunting season in September–October 2006 (17 animals), 2007 (14 animals), and 2008 (6 animals). The animals originated from Kvam (2 deer) and Etne (12 deer) municipalities in Hordaland County and from Vikebygd municipality (23 deer) in Rogaland County, in Western Norway. Etne and Vikebygd are neighbouring municipalities on the south side of the mouth of the Hardanger fjord, whereas Kvam municipality is situated on the north-western side of this fjord. The samples were frozen shortly after collection and stored at −20°C for 4–6 months (year 2006 and 2008), or 17–18 months (year 2007) before examination immediately upon thawing.
Reindeer
Sarcocysts were isolated in 2006 from fresh samples of the oesophagus and diaphragm from 2 semi-domesticated reindeer. The samples originated from animals which had been slaughtered at a field abattoir in Nordland County in Northern Norway. The sarcocysts were kept frozen at −20°C until molecular examination in 2008.
Light microscopy and cyst isolation
The muscle tissue was examined under a stereomicroscope and individual sarcocysts were excised and examined under a light microscope as previously described (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007). Photographs were taken with a Leica DC480 digital camera. A few cysts were treated with ultrasound as previously described to remove remnants of the host cell and thus better revealed their surface structure (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008).
Scanning electron microscopy
Cysts with different surface morphology, as determined by LM, were fixed in 3% glutaraldehyde in cacodylate buffer (pH 7·4) following excision from the oesophagus or diaphragm. The cysts were dried according to one of the following procedures. (1) Several fixed cysts were enclosed in small permeable nylon bags (Nytal®), before being dehydrated in an ascending ethanol series, comprising 30–100% ethanol, and critical-point dried in an E3100 Critical Point Dryer (Bio-Rad, UK), with carbon dioxide as transitional fluid. (2) Cysts were fixed and kept in 1·5 ml Eppendorf tubes, while being dehydrated in an ascending ethanol series, comprising 30–100% ethanol, and air-dried from 100% hexamethyldisilazane (HMDS) after replacement of 100% ethanol and different ethanol-HMDS mixtures with 100% HMDS in several steps. Finally, a small volume of 100% HMDS, containing the cysts, was allowed to evaporate at room temperature for 3–4 h. Dried cysts were mounted with carbon tabs on aluminium specimen holders and coated with a 30 nm layer of platinum using a SC510 Sputter Coater (Bio-Rad, UK) and then examined and photographed with a Philips XL30 ESEM scanning electron microscope (Philips, The Netherlands).
DNA isolation and PCR amplification
After cyst isolation and preliminary species identification by LM, individual cysts were placed in 1·5 ml Eppendorf tubes containing 20 μl of distilled water and kept frozen at −20°C until DNA was isolated from randomly selected cysts of each type. In addition, DNA was isolated from 2 cysts of different species that had been examined by SEM to confirm their identity by molecular methods. After SEM examination, those 2 cysts were retrieved from their aluminium holders and placed in 20 μl of distilled water in separate Eppendorf tubes. Total DNA was extracted from the sarcocysts according to previously described methods (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007).
The complete ssu rRNA gene was amplified as previously described (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007). To identify some of the isolates, only the second half of the gene was sequenced, using the primer combination S3/Primer B.
The first internal transcribed spacer region (ITS-1) of S. rangiferi and S. tarandi from reindeer and 2 morphologically similar species from red deer was PCR amplified according to the following protocol. Each reaction contained 4 μl aliquots of the DNA solution, 25 μl of HotStarTaq Master Mix (Qiagen GmbH, Germany), 8 μg bovine serum albumin, 20 pmol of each primer 18S14F and ITS1-FR (Rosenthal et al. Reference Rosenthal, Dunams and Pritt2008), and RNase-free water to make a final volume of 50 μl. PCR cycling conditions were: Initial Hot Start at 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 43·2°C for 45 s, 72°C for 75 s, and a final extension at 72°C for 10 min. A negative control was included in each reaction.
Cloning
Part of the ssu rRNA gene and the entire ITS-1 region of 2 isolates each of S. rangiferi and S. tarandi from reindeer and 2 similar species from red deer were cloned. Purified PCR products from the amplification of Sarcocystis DNA with primer combination Primer 1L/Primer 3H were used as templates in the cloning reactions for the ssu rRNA gene. Purified PCR products from the amplification of Sarcocystis DNA with primer combination 18S14F/ITS1-FR were used as templates in the cloning reactions for the ITS-1 region. TOPO TA Cloning® Kit for Sequencing with One Shot® TOP10 Chemically Competent E. coli (Invitrogen, Scotland) was used to clone the DNA fragments according to the manufacturer's instructions. Plasmid DNA was purified using QIAprep Spin Miniprep Kit (Qiagen GmbH, Germany) according to the manufacturer's instructions. Five clones of each of the 2 isolates of each of the 2 species from both hosts were sequenced at both the ssu rRNA gene (40 sequences) and the ITS-1 region (40 sequences).
DNA sequencing and sequence analysis
Quantity and purity of the PCR products and plasmid DNA were evaluated on a NanoDrop® ND-1000 Spectrophotometer (NanoDrop Technologies, USA). The amplified DNA fragments were sent to Eurofins MWG Operon, Germany, for sequencing on both strands using the same forward and reverse primers as for the PCR and cloning reactions. The application Contig Express in Vector NTI® Advance 10 software (Invitrogen, Scotland) was used to assemble sequences from each of the isolates and clones into complete ssu rRNA gene and ITS-1 sequences.
Sequence identity of different isolates or clones of the same Sarcocystis species was determined on EMBOSS (the European Molecular Biology Open Software Suite) (http://www.ebi.ac.uk/emboss/align/) using the default options. EMBOSS was also used to determine sequence variability between Sarcocystis species sequenced in this study and Sarcocystis species from reindeer, moose and roe deer sequenced in previous studies.
DNA sequence alignments
Ssu rRNA gene
Complete ssu rRNA gene sequences from the Sarcocystis species sequenced in this study were used in the phylogenetic analyses along with ssu rRNA gene sequences of Sarcocystis species from other intermediate hosts, retrieved from GenBank (Table 1). A representative sample of the subfamily Sarcocystinae was generated by including Sarcocystis species with different intermediate and definitive hosts. Based on a guide tree using the neighbour-joining method generated by the application AlignX in Vector NTI® Advance 10 software (Invitrogen, Scotland), species within the subfamily Toxoplasmatinae with the shortest evolutionary distance to the subfamily Sarcocystinae were chosen as outgroup-species to root the tree. The following species were selected: Isospora orlovi, Besnoitia jellisoni, Neospora caninum and Toxoplasma gondii. All available ssu rRNA gene sequences of Sarcocystis species from cervids were included in the analyses. However, for taxa represented by more than 1 sequence with sequence variation at less than 10 nucleotide positions, the sequences were merged into a consensus sequence using the IUPAC codes for nucleotide positions with more than 1 possible character state. All available clones of S. scandinavica from moose, and a Sarcocystis sp. isolated from sika deer, were included in the analyses.
Table 1. Taxon name of species used in the phylogenetic analysis, their known intermediate host(s), and GenBank Accession number(s) of their ssu rRNA gene sequence(s)
(The Sarcocystis species from red deer sequenced in the present study are in boldface.)

To infer the phylogeny of the different Sarcocystis species from red deer, all ssu rRNA gene sequences were aligned, based on the secondary structure of the rRNA molecules according to previously described methods (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008). The final alignment contained 82 taxa, based on 101 ssu rRNA gene sequences, and 1900 aligned nucleotide positions.
A separate analysis was also run including only clones of S. tarandi and S. rangiferi from reindeer and the S. tarandi-like and S. rangiferi-like species from red deer, as well as the sequences of Sarcocystis sp. type D from moose and a Sarcocystis sp. from Japanese sika deer. Sarcocystis buffalonis and S. hominis were chosen as outgroup-species in this analysis.
ITS-1
Complete ITS-1 sequences of all clones were aligned using AlignX in Vector NTI. Three clones of Sarcocystis cruzi (GenBank Accession numbers EF622174, EF622175 and EF622176) were selected as outgroup taxa for the phylogenetic analyses. The final alignment contained 43 taxa and 778 aligned nucleotide positions.
Phylogenetic analysis
MrModeltest (Posada and Crandall, Reference Posada and Crandall1998) was used to select the model of sequence-evolution with the best fit to the data set, and the GTR+I+G model was chosen for the ssu rRNA analyses and GTR+G for the ITS-1 analyses.
GARLI (Zwickl, Reference Zwickl2006) was used to run maximum-likelihood analyses of all the aligned ssu rRNA gene and ITS-1 sequences. Bootstrap support was calculated from 100 repetitions and the other parameters were set to default.
Phylogenetic relationships among the aligned sequences were also examined with Bayesian methods using MrBayes 3.1.2 (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003), with the default values. In order to determine when the chains had converged, the output tree files were analysed using the program AWTY (Wilgenbusch J.C., Warren D.L., Swofford D.L. 2004. AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. http://ceb.csit.fsu.edu/awty.).
RESULTS
Prevalence of Sarcocystis in red deer
Sarcocysts were found in all 37 adult red deer examined (100%). Thirty four red deer were positive for Sarcocystis in the diaphragm, 34 red deer were infected in the oesophagus, and 7 animals had sarcocysts in the heart.
Sarcocyst types and ssu rRNA gene sequences
Two types of sarcocysts were seen by gross examination of the oesophagus and diaphragm: about 1–2 mm long, elongated and cigar-shaped cysts and about 1 mm long and ovoid cysts. These tissues also contained small slender cysts that were difficult to detect without the aid of a stereomicroscope. No cysts were seen grossly in cardiac muscle, but a few cysts were recognized with some difficulty using a stereomicroscope.
Sarcocysts with 4 different types of surface morphology (types 1–4) were seen by LM of samples from the oesophagus and diaphragm: (1) slender spindle-shaped cysts with thin hair-like protrusions, which could only be seen after removal of the surrounding host cell, (2) slender spindle-shaped cysts with upright, finger-like protrusions, (3) thick, cigar-shaped cysts with upright, finger-like protrusions and (4) oval cysts encapsulated by fibrous material, which after removal of this layer displayed slanting, tongue-like protrusions. Cysts of types 1 and 2 were mostly microscopic and indistinguishable in situ, whereas cysts of types 3 and 4 were macroscopically visible. Eleven animals were infected with only a single cyst type. Eight deer were infected with 2 cyst types. Twelve deer were infected with 3 cyst types and 6 animals had a mixed infection with 4 cyst types.
In cardiac muscle, only a few small (<0·5 mm), sac-like cysts were detected. These cysts either had hair-like protrusions like type 1 cysts, or upright, finger-like protrusions similar to those of type 2 and 3 cysts.
The same 4 cyst types were also seen by SEM, whereas 5 different ssu rRNA gene sequence types, representing 5 different Sarcocystis species, were obtained after DNA sequencing and cloning. All 5 species will be further described in the following sections and 1 new species will be named.
Description of Sarcocystis hjorti n. sp
General remarks
Cysts of S. hjorti (Type 1 by LM and SEM) were found in the diaphragm and/or oesophagus of 35 (95%) red deer examined. Six (16%) animals also had a cardiac infection. Sarcocysts in the same animal varied considerably in size. The cysts in the diaphragm and oesophagus were more numerous and larger than those in cardiac muscle.
Morphological features of sarcocysts (type 1)
Light microscopy of sarcocysts
Cysts in the oesophagus and diaphragm were slender and spindle-shaped, measuring about 1·0–3·1×0·08–0·2 mm. They had thin, flexible, hair-like protrusions, which were about 10–12 μm long and only visible after removal of host cell material (Fig. 1a–c). The much smaller cysts in the heart were sack-like and 0·25–0·35×0·1 mm in size (Fig. 1d). Some of the cysts in cardiac muscle seemed to have a smooth surface without protrusions, but this was most likely due to a thin layer of host-cell material, covering the delicate hair-like protrusions (Fig. 1e), since such cysts were found to belong to this species by molecular methods.

Fig. 1. Sarcocystis hjorti n. sp. in red deer. Light microscopic appearance of fresh cysts. (a) Portion from the middle region of a partly disrupted cyst revealing a thin wall with delicate hair-like protrusions (P). (b) Pointed tip of slender cyst with hair-like protrusions (P). (c) Larger magnification of portion of cyst in (b) showing densely packed protrusions (P). (d) Small cyst from cardiac muscle still partly enclosed by its host cell, but displaying a thin, smooth wall (arrows) where its surface is exposed. (e) Surface of another small cyst from cardiac muscle with barely visible hair-like protrusions (P). Scale bars =10 μm in (a), (b), (c) and (e), 100 μm in (d).
Scanning electron microscopy
The cyst surface had numerous flexible, slender, hair-like protrusions, about 0·4 μm wide at their base and narrowing towards their tip (Fig. 2a–e). The length of the protrusions was difficult to ascertain due to their winding course, but was at least 10 μm. Most of the protrusions were fairly flattened, particularly at their bases (Fig. 2d and e). The protrusions were regularly distributed in rows across the surface, with a fairly uniform distance between their bases in a given cyst.

Fig. 2. Sarcocystis hjorti n. sp. in red deer. Scanning electron micrographs of cysts. (a) Nearly complete, but broken, about 1·3 mm long cyst, partly enclosed by its host cell (Hc). (b and c) Different portions of cyst in (a), showing cyst surface covered with densely packed hair-like protrusions. (d and e) Higher magnification of surface of another cyst, showing details of the protrusions (P) protruding from the cyst surface (S). The protrusions are fairly flattened at their bases, possibly due to SEM processing. Scale bars =100 μm in (a), 10 μm in (b) and (c), 2 μm in (d) and (e).
Molecular characteristics
Four complete and 3 partial ssu rRNA gene sequences were obtained from 5 different isolates, including 2 isolates from cardiac muscle. The complete or partial gene sequences obtained from all isolates were identical. Two nucleotide positions had double peaks on the chromatogram in all isolates. The complete ssu rRNA gene sequence has been deposited in GenBank with Accession number GQ250990.
The ssu rRNA sequence of S. hjorti was identical with that of the unnamed Sarcocystis sp. Type E from moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008) i.e., GenBank Accession number EU282017. The sequence from moose also had 2 undetermined nucleotides and one of them was located in the same position as the undetermined nucleotide in the sequences from red deer.
Host
Type host
Red deer (Cervus elaphus) and moose (Alces alces) are known intermediate hosts for S. hjorti. The definitive host(s) have not been identified, but are presumably canines (see later about phylogenetic position).
Type specimens
Type material, consisting of sarcocysts excised from muscular tissues from red deer, and photographs from LM and SEM examinations of the sarcocyst wall, have been deposited at the National History Museum, Oslo, Norway, collection number NHMO-Prot00006.
Locality
Sarcocysts of S. hjorti were found in red deer from Hordaland and Rogaland Counties in South-Western Norway in the present study, and in a single moose from Eastern Norway (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008).
Etymology
The species name is derived from the Scandinavian common word for red deer and also partly comprising the Norwegian word for species of the family Cervidae.
Description of Sarcocystis tarandi
General remarks
Type 2 sarcocysts were found in the diaphragm and/or oesophagus of 16 (43%) red deer examined, all of which originated from Etne and Vikebygd municipalities. Fourteen of these animals were also infected with S. rangiferi. Three animals (8%) had cysts with similar surface morphology in the heart. The cysts were small and slender, but generally larger and thicker in the oesophagus than in the diaphragm. Thin, finger-like protrusions gave the cyst wall a thick and striated appearance, which also could be seen in fresh cysts enclosed by their host cell. Type 2 cysts were morphologically indistinguishable from those of S. tarandi in reindeer (Gjerde, Reference Gjerde1984a). Molecular data and phylogenetic analyses strongly suggested that Type 2 cysts in red deer and S. tarandi cysts in reindeer should be considered to be the same species.
Morphological features of sarcocysts
Light microscopy of sarcocysts
The cysts in the diaphragm and oesophagus were slender and spindle-shaped with tapering pointed ends (Fig. 3a and b), measuring 1·0–2·3×0·07–0·15 mm. The cyst surface had densely packed, upright, thin, finger-like protrusions, which were 7–8 μm long and 1·5–2 μm wide (Fig. 3b). The protrusions appeared to be round to polygonal, and to be distributed in rows in a hexagonal pattern across the surface (Fig. 3c). A few cysts, 0·2–0·3×0·06–0·13 mm in size, with similar, about 5 μm long protrusions, were isolated from cardiac muscle (Fig. 3d and e), but it could not be determined from their appearance whether they belonged to S. tarandi or S. rangiferi (type 3). However, 1 such cyst (Fig. 3d) was determined with certainty to be a cyst of S. tarandi after sequencing part of the ssu rRNA gene.

Fig. 3. Sarcocystis tarandi in red deer. Light microscopic appearance of fresh cysts. (a) A typical slender spindle-shaped cyst, fairly long (2·3 mm). The thick wall is seen as a lighter zone at the periphery. (b) Tip of another cyst with densely packed upright protrusions (P), which give the cyst wall a thick, striated appearance. (c) Surface view of a cyst with densely packed protrusions (P). (d) Small cyst from cardiac muscle, with a thick striated wall, subsequently confirmed to be S. tarandi by molecular methods. (e) Another small cyst from cardiac muscle, possibly of S. tarandi, but not confirmed by molecular methods. Scale bars=100 μm in (a), 10 μm in (b), 5 μm in (c), 50 μm in (c) and (d).
Scanning electron microscopy
The cyst surface had densely spaced, thin, upright protrusions, about 6 μm long and 1·1–1·4 μm in widest diameter (Fig. 4a–e). The protrusions had a polygonal to oval outline (Fig. 4d and e). However, on many cysts they appeared flattened, probably due to the processing for SEM. The protrusions were aligned in rows across the surface (Fig. 4d and e).

Fig. 4. Sarcocystis tarandi in red deer. Scanning electron micrographs of cysts. (a) Complete, about 0·9 mm long, spindle-shaped cyst completely free from its host cell. (b) Portion of cyst in (a) showing an even periphery due to densely packed cyst wall protrusions (P) of uniform length. (c) Similar view as in (b) from another cyst. (d) Higher magnification showing the finger-like shape and straight posture of the protrusions (P). (e) Detail of protrusions in (d) showing their fairly smooth surface and rounded tip. Scale bars=100 μm in (a), 20 μm in (b) and (c), 2 μm in (d), 1 μm in (e).
Molecular characteristics
Type 2 cysts had ssu rRNA gene sequences which were very similar to those of S. tarandi in reindeer obtained in a previous study (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007). Two sarcocysts (isolates CeSt-I and CeSt-II) from red deer and 2 sarcocysts (isolates RtSt-I and RtSt-II) from reindeer were randomly selected for more comprehensive molecular studies. Initial attempts to sequence the ssu rRNA gene directly after PCR resulted in DNA chromatograms which contained a few double peaks in the first part of the gene sequence. By cloning the PCR products before sequencing, fine single-peak chromatograms were obtained. The complete ssu rRNA gene of 5 clones of each of the 2 isolates from red deer and reindeer was amplified and sequenced, and the entire ITS-1 region of 5 clones of each of the same 4 isolates was amplified and sequenced.
Ssu rRNA gene
The length of the DNA sequences ranged between 1727 and 1734 nucleotides, after some sequences had been truncated to make all sequences start and end at the same nucleotide positions. DNA sequences from red deer differed from each other by 0·1–1·1% and sequences from reindeer differed from each other by 0·1–1·0% (Table 2). DNA sequences from red deer differed from those from reindeer by 0·2–1·2% (Table 2). The sequence variation was mainly due to nucleotide substitutions, but also to 1–3 indels, located particularly along the first half of the ssu rRNA sequences. The ssu rRNA gene sequences of all clones of S. tarandi from red deer and all clones from reindeer have been deposited in GenBank with Accession numbers GQ251011-20 and GQ250967-76.
Table 2. Sequence identities at the small subunit rRNA gene and first internal transcribed spacer region between different clones of Sarcocystis rangiferi (Srf) and Sarcocystis tarandi (St) from red deer (Ce) and reindeer (Rt)
(The roman numbers indicate from which isolate the clones originated.)
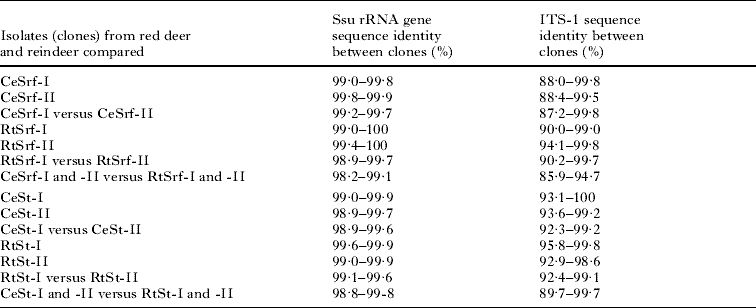
ITS-1
The length of the DNA sequences ranged between 613 and 641 nucleotides, including 134 nucleotides of the ssu rRNA gene and 19 nucleotides of the 5·8 S gene, after all sequences had been adjusted to start and end at the same nucleotide positions. DNA sequences from red deer differed from each other by 0·0–7·7% and sequences from reindeer differed from each other by 0·2–7·6% (Table 2). DNA sequences from red deer differed from those from reindeer by 0·3–10·3% (Table 2). The sequence variation was due to nucleotide substitutions and indels along the entire ITS-1 region, but the large difference in sequence identities between some clones was mainly due to a few indels of up to 15 nucleotides. The complete ITS-1 sequences of all clones of S. tarandi from red deer and all clones from reindeer have been deposited in GenBank with Accession numbers GQ251006-10 and GQ250957-66.
Type specimens
Type material, consisting of sarcocysts excised from muscular tissues from red deer and reindeer, and photographs from LM and SEM examinations of the sarcocyst wall, have been deposited at the National History Museum, Oslo, Norway, collection numbers NHMO-Prot00007 and NHMO-Prot00008.
Description of Sarcocystis rangiferi
General remarks
Type 3 sarcocysts were found in the diaphragm and/or oesophagus of 18 (49%) red deer examined, all of which originated from Etne and Vikebygd municipalities. Some animals were heavily infected with this species in the oesophagus. In animals with a moderate infection, the cysts often had a patchy distribution with several cysts confined to limited areas. The cigar-shaped to ellipsoidal cysts were often macroscopically visible in the diaphragm and oesophagus. The cyst surface was covered with thick, finger-like protrusions, making the cyst wall appear thick and striated, even in cysts still enclosed by their host cell. The host cells did not seem to be enclosed by fibrous material. Otherwise, the cysts were morphologically similar to those of S. rangiferi in reindeer (Gjerde, Reference Gjerde1984a). Molecular data and phylogenetic analyses suggested that Type 3 cysts in red deer and S. rangiferi cysts in reindeer belonged to the same species.
Morphological features of sarcocysts
Light microscopy of sarcocysts
The cysts were cigar-shaped to ellipsoidal with rounded tips, measuring about 0·7–1·7×0·16–0·4 mm (Fig. 5a). The cyst surface had densely packed, upright, fairly thick finger-like protrusions, about 7–8 μm long and about 3 μm wide (Fig. 5b). The protrusions appeared to have a round to polygonal cross-section and to be regularly distributed in rows in a hexagonal pattern across the surface (Fig. 5c).

Fig. 5. Sarcocystis rangiferi in red deer. Light microscopic appearance of fresh cysts. (a) A typical thick, cigar-shaped cyst. The cyst is still enclosed by a thin layer of host cell material, but the thick wall is seen as a light zone at the periphery. (b) Cyst wall with fairly thick, upright finger-like protrusions (P) seen in profile. (c) Surface view of the tip of a partly emptied cyst showing densely packed protrusions (P). Scale bars=100 μm in (a), 5 μm in (b) and (c).
Scanning electron microscopy
The cyst surface had closely spaced upright, prominent protrusions, about 3 μm wide and 7 μm long (Fig. 6a–e). The protrusions had nearly hexagonal bases, as seen on a sonicated cyst where several protrusions had been broken off (Fig. 6b). They were polygonal for the majority of their length, but rounder at their tips. Minute round depressions, about 0·1 μm in diameter, were seen on the surface of the rounded tips (Fig. 6d). The protrusions were regularly arranged in rows across the surface (Fig. 6d and e).

Fig. 6. Sarcocystis rangiferi in red deer. Scanning electron micrographs of cysts. (a) A complete approximately 0·8 mm long, thick cyst, with exposed protrusions (P) and also partly covered by host cell material (Hc). (b) Cyst that has been partly disrupted by ultrasound treatment to remove the host cell. Most of the protrusions (P) are still in place, but some have been broken off near the cyst surface (arrows) revealing their polygonal to hexagonal bases. The break in the cyst reveals inner compartmentalization by septa. (c) Surface view showing densely packed finger-like protrusions. (d) Higher magnification revealing regularly distributed small pits on the rounded tips of the protrusions. Scale bars =100 μm in (a), 30 μm in (b), 10 μm in (c), 1 μm in (d).
Molecular characteristics
Type 3 cysts had ssu rRNA gene sequences which were very similar to those of S. rangiferi in reindeer obtained in a previous study (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007). Two isolates (CeSrf-I and CeSrf-II) from red deer and 2 isolates (RtSrf-I and RtSrf-II) from reindeer were therefore randomly selected for more comprehensive molecular studies. Initial attempts to sequence the ssu rRNA gene directly after PCR, resulted in DNA chromatograms which contained a few double peaks in the first part of the gene sequences. By cloning the PCR products before sequencing, fine single peak chromatograms were obtained. The complete ssu rRNA gene of 5 clones of each of the 2 isolates from red deer and reindeer was amplified and sequenced, and the entire ITS-1 region of 5 clones of each of the same 4 isolates was amplified and sequenced.
Ssu rRNA gene
The length of the DNA sequences ranged between 1734 and 1739 nucleotides, after some sequences had been truncated to make all sequences start and end at the same nucleotide positions. DNA sequences from red deer differed from each other by 0·0–1·0% and sequences from reindeer differed from each other by 0·0–1·1% (Table 2). DNA sequences from red deer differed from those from reindeer by 0·9–1·8% (Table 2). The sequence variation was due to nucleotide substitutions and indels mainly located in the variable regions of the first half of the ssu rRNA sequences. Multiple alignments showed that most of the substitutions and indels were located at the same nucleotide positions in all sequences and that the different clones appeared to form sequence groups of highly similar or identical sequences. The complete ssu rRNA gene sequences of all clones of S. rangiferi from both red deer and reindeer have been deposited in GenBank with Accession numbers GQ251021-30 and GQ250977-86.
ITS-1
The length of the DNA sequences ranged between 587 and 613 nucleotides, including 134 nucleotides of the ssu rRNA gene and 19 nucleotides of the 5·8 S gene, after all sequences had been adjusted to start and end at the same nucleotide positions. DNA sequences from red deer differed from each other by 0·2–12·8% and sequences from reindeer differed from each other by 0·2–10·0% (Table 2). DNA sequences from red deer differed from those from reindeer by 5·3–14·1% (Table 2). The sequence variation was due to single nucleotide substitutions and indels along the entire ITS-1 region. However, multiple alignments showed that in a few regions of the ITS-1 sequence, the clones could be divided into several subgroups, each with highly similar sequences, but clearly different from those of other subgroups. The complete ITS-1 sequences of all clones of S. rangiferi from red deer and all clones from reindeer have been deposited in GenBank with Accession numbers GQ250991-251000 and GQ250947-56.
Type specimens
Type material, consisting of sarcocysts excised from muscular tissues from red deer and reindeer, and photographs from LM and SEM examinations of the sarcocyst wall, have been deposited at the National History Museum, Oslo, Norway, collection numbers NHMO-Prot00009 and NHMO-Prot00010.
Description of Sarcocystis hardangeri and S. ovalis
General remarks
Type 4 sarcocysts were found in the diaphragm and/or oesophagus of 18 (49%) red deer examined. No cysts were found in the heart. The ovoid cysts were macroscopically visible, but tightly enclosed by a fibrous layer, which had to be removed to enable visualization of the tongue-shaped protrusions that ran along the surface. The cysts were morphologically indistinguishable from S. hardangeri in reindeer (Gjerde, Reference Gjerde1984b, Reference Gjerdec), S. ovalis in moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008), and S. oviformis in roe deer (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2009). Molecular examinations revealed that these cysts represented 2 species, S. hardangeri and S. ovalis. The morphological description therefore applies to both of these species in red deer.
Morphological features of sarcocysts
Light microscopy
Ovoid cysts, measuring about 0·8–1·9×0·3–0·7 mm (Fig. 7a and b). The cysts and their host cell were enclosed by a fibrous layer about 5–7 μm thick (Fig. 7c). No protrusions were seen. After removal of the fibrous layer, the cysts displayed loosely packed, slanting, tongue-shaped protrusions, about 10 μm long and 2 μm wide, on their surface (Fig. 7d–f). The protrusions were aligned in rows and were generally pointing in the same direction across the surface (Fig. 7d).

Fig. 7. Sarcocystis hardangeri and Sarcocystis ovalis in red deer. Light microscopic appearance of fresh cysts. (a) Ovoid cyst subsequently determined to be of S. hardangeri by molecular methods. (b–f) Cysts presumably of S. ovalis (all cysts examined by molecular methods isolated from red deer from this same area belonged to S. ovalis). (b) Complete oval cyst. (c) Tip of cyst enclosed by its host cell and encapsulated by a layer of fibrous material (F). No protrusions are visible. (d) Surface view of a cyst after removal of most of the surrounding material. Regularly aligned, loosely packed, flattened protrusions (P), are running along the cyst surface, pointing in the same direction. (e) Side view of a cyst wall with slanting protrusions (P). (f) Higher magnification of protrusions revealing their flattened tongue-like shape. Scale bars =100 μm in (a) and (b), 20 μm in (c), 10 μm in (d), (e) and (f).
Scanning electron microscopy
The oval cysts had a bulgy surface provided with loosely packed slanting protrusions, running in parallel to the cyst surface, generally in the same direction (Fig. 8a–c). The protrusions were 10–18 μm long (varying between different cysts), about 2 μm wide, and about 0·5 μm thick at their base. The width increased slightly distal to their base, before narrowing again in the distal third to a pointed tip (Fig. 8c). The protrusions were positioned 3–5 μm apart. The cyst surface in between the protrusions had a wrinkled appearance due to numerous minute folds (Fig. 8c).

Fig. 8. Sarcocystis ovalis in red deer. Scanning electron micrographs of cysts. (a) Complete cyst, about 0·6 mm long, with typical oval shape, but with most of the surface still covered by host material (from red deer infected with S. ovalis). (b) Cyst from which most of the host cell (Hc) has been removed, revealing a bulgy surface (S) with loosely packed tongue-shaped protrusions running along the cyst in the same direction. (c) Higher magnification of another area of the same cyst as in (b) showing tongue-like protrusions (P) and a finely wrinkled cyst surface (S) in between them. This cyst was determined to belong to S. ovalis by molecular methods subsequent to the SEM-examination. Scale bars =100 μm in (a), 15 μm in (b), 5 μm in (c).
Molecular characteristics
Three isolates (I–III) were completely sequenced at the ssu rRNA gene. The gene sequence of isolate I, which was obtained from a red deer from Kvam municipality, was identical with that of S. hardangeri in reindeer obtained in a previous study (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007). The sequence of isolate I differed at 45 nucleotide positions from isolate II (97·7% sequence identity), and 46 nucleotide positions from isolate III (97·6% sequence identity). Isolates II and III differed from each other by 3 nucleotide positions (99·8% sequence identity). The sequences of isolates II and III differed by 2–9 nucleotide positions (99·5–99·9% sequence identity) from those of S. ovalis in moose obtained in a previous study (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008), and are therefore considered to belong to this species. Nine additional isolates that were partially sequenced were identical with the corresponding sequences of isolates II and III. In addition, the partial sequence of 1 isolate derived from a cyst that had been examined by SEM, was also identical with that of isolates II and III. All cysts identified as S. ovalis by molecular methods originated from red deer from Etne and Vikebygd municipalities. Complete ssu rRNA gene sequences of S. hardangeri (isolate I) and S. ovalis (isolates II and III) from red deer have been deposited in GenBank with Accession numbers GQ250987-89.
Type specimens
Type material, consisting of sarcocysts of S. hardangeri excised from muscular tissues from reindeer, and photographs from LM and SEM examinations of the sarcocyst wall, has been deposited at the National History Museum, Oslo, Norway, collection number NHMO-Prot00011. Materials from S. ovalis (from moose) have been deposited previously (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008).
Phylogenetic analyses
Ssu rRNA gene
The placement of S. hjorti, S. hardangeri, S. ovalis, S. rangiferi, and S. tarandi was the same in all estimated tree topologies, with similar branch-length estimates and bifurcation support. All 5 species were placed together with other Sarcocystis species that use an even-toed ungulate as their intermediate host and a canine, feline, human or unknown carnivore as their definitive host (Fig. 9). High support was given in the analyses to place S. hjorti with other species that have been shown to use a canine definitive host i.e., S. alceslatrans, Sarcocystis arieticanis, Sarcocystis capracanis, Sarcocystis cruzi, S. grueneri, S. rangi, S. tarandivulpes, and Sarcocystis tenella. Maximum support was given in the analyses to place S. hardangeri, S. ovalis and S. oviformins in a separate clade. S. scandinavica was represented by multiple sequences in the phylogenetic analyses, but since all clones formed a well-supported cluster on the phylogenetic tree, their branches have been collapsed in Fig. 9. With a strong support, the analyses placed the clade comprising isolates of S. tarandi, S. rangiferi, Sarcocystis sp. Type D from moose, and an unnamed species from sika deer, in the same group as other species that use a feline, human or unknown carnivore as definitive host. However, the relationships within this clade could not be determined based on the ssu rRNA gene, as seen from the fairly low bifurcation support values (Fig. 9). The sequences of the S. tarandi-like species from red deer and those of S. tarandi from reindeer formed 1 cluster and showed an interwoven pattern of S. tarandi-sequences from both cervid hosts and also of sequences from the different cloned isolates (Fig. 10). For the S. rangiferi-type sequences, the pattern was more complex. The sequences from reindeer, including 2 previously sequenced isolates, formed a single cluster with sequences from all 4 isolates arranged in an interleaved pattern (Fig. 10). A fairly long branch separated the reindeer sequences from those of red deer. The sequences of isolate 2 from red deer formed a separate cluster, whereas the branching pattern of 3 clones of isolate 1 could not be determined and 1 clone even grouped together with the cluster of sequences of the unnamed Sarcocystis species from sika deer. One sika deer sequence also appeared to be more closely related to those of the S. rangiferi-type sequences of isolate 1 than with the other sika deer sequences. A close relationship of Sarcocystis sp. Type D from moose to S. tarandi and S. rangiferi was also revealed by the analyses.

Fig. 9. Inferred Bayesian phylogenetic tree for the Sarcocystidae, based on ssu rRNA gene sequences, and scaled according to branch lengths. The numbers in the figure show posterior probability support value. Accession numbers of all species used in the analysis are shown in Table 1. Sarcocystis scandinavica was represented by several sequences, but always formed a cluster on the phylogenetic tree, and hence its branches were collapsed. The range of branch lengths within the species-cluster of S. scandinavica is depicted by a dashed line. The phylogenetic relationships between S. rangiferi, S. tarandi, Sarcocystis sp. Type D of moose, and an unnamed Sarcocystis sp. from sika deer are shown in Fig. 10.

Fig. 10. Inferred Bayesian phylogenetic tree for Sarcocystis rangiferi and S. tarandi of red deer (red dots) and reindeer (blue dots), Sarcocystis sp. Type D of moose (green dot), and an unnamed Sarcocystis sp. from sika deer (yellow dots), based on small subunit rRNA gene sequences, and scaled according to branch lengths. The numbers behind the species name indicate from which of the two isolates, from either red deer or reindeer, the sequenced clone originated.
ITS-1
The ITS-1 sequences of the S. tarandi-like species from red deer and those of S. tarandi from reindeer clustered together and were interleaved on the phylogenetic tree (Fig. 11). In contrast, the ITS-1 sequences of the S. rangiferi-like species from red deer and those of S. rangiferi from reindeer formed several clusters, with the isolates of each cluster being derived from only 1 host species. However, all S. rangiferi-type sequences were clearly separated from the S. tarandi-cluster.

Fig. 11. Inferred Bayesian phylogenetic tree for Sarcocystis rangiferi and S. tarandi of red deer (red dots) and reindeer (blue dots), based on sequences of the first internal transcribed spacer, and scaled according to branch lengths. The numbers behind the species name indicate from which of the two isolates, from either red deer or reindeer, the sequenced clone originated.
DISCUSSION
In this study, we found 5 different Sarcocystis species in Norwegian red deer. We assigned the name S. hjorti for the new species, which had been identified as Sarcocystis sp. Type E in moose by molecular methods previously (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008). However, this is the first time that the species has been described as a distinct species morphologically and that this particular ssu rRNA gene sequence has been linked to a certain sarcocyst type. The other 4 species had similar sarcocyst morphology and ITS-1 sequences and/or ssu rRNA sequences to Sarcocystis species previously described from reindeer and moose i.e., Sarcocystis hardangeri, Sarcocystis ovalis, Sarcocystis tarandi and Sarcocystis rangiferi. We have described all 5 species in detail by LM, SEM, ssu rRNA gene sequence characteristics and phylogenetic analyses. In addition the ITS-1 region of S. tarandi and S. rangiferi from reindeer and red deer was sequenced and analysed to investigate whether this DNA sequence would resolve species relationships better than the more slowly evolving ssu rRNA gene. We will discuss each species separately and compare them with similar species previously reported from red deer and elk, as well as from other cervid hosts.
S. hjorti
S. hjorti was the most prevalent species found in the muscle tissues examined from red deer. By light microscopy fresh cysts of S. hjorti were initially thought to represent 2 different Sarcocystis species, since no protrusions were visible on some of the small cysts found in the heart. These small cysts were initially suspected to represent a species with cysts similar to those of S. cervicanis in red deer (Hernández-Rodríguez et al. Reference Hernández-Rodríguez, Martínez-Gómez, Navarrete and Acosta-Garcia1981b), S. wapiti in wapiti (Speer and Dubey, Reference Dubey1982) and S. grueneri in reindeer (Gjerde, Reference Gjerde1986). However, molecular analyses showed that the small cysts belonged to S. hjorti. Thus, the thin hair-like protrusions were most likely hidden under remnants of host cell material as the enclosing host cell was difficult to remove from those small cysts in the heart. The delicate hairs were more easily seen on larger cysts in the oesophagus and diaphragm.
Sarcocysts with similar hair-like protrusions to those of S. hjorti have previously been reported from red deer and elk. In Germany, Wesemeier and Sedlaczek (Reference Sedlaczek and Wesemeier1995a) found such cysts both in a free-ranging red deer and in a red deer and an elk that had been born and raised in a zoo. They referred to this species as S. cf. capreolicanis because of its resemblance to S. capreolicanis in roe deer (Erber et al. Reference Erber, Boch and Barth1978). Kutkienė (Reference Kutkienė2003) also referred to the hairy cysts she found in red deer from Lithuania as S. cf. capreolicanis. It remains to be determined by molecular methods whether this species is identical with S. hjorti in red deer in the present study.
Neither Wesemeier and Sedlaczek (Reference Wesemeier and Sedlaczek1995a) nor Kutkienė (Reference Kutkienė2003) mentioned a paper by Dubey et al. (Reference Dubey, Jolley and Thorne1983), describing the new species S. sybillensis from elk in USA. The mature sarcocysts depicted in the TEM micrographs in Figs 8 and 9 in that paper have long hair-like protrusions, about 0·3 μm in diameter, whereas the short protrusions of the allegedly immature sarcocysts shown in Figs 6 and 7 could be either early stages of hair-like protrusions or early stages of protrusions with another appearance. Thus, similar short protrusions have been reported from young cysts of S. cruzi of cattle, another species with hair-like protrusions (see Fig. 11 in Mehlhorn et al. Reference Mehlhorn, Heydorn and Gestrich1975b and Fig. 5 in Pacheco, Reference Pacheco, Sheffield and Fayer1978), but also from young cysts of species that develop erect finger-like protrusions e.g., S. tenella of sheep (see Fig. 39c in Dubey et al. Reference Dubey, Speer, Callis and Blixt1982) and S. hominis of cattle (see Fig. 3 in Mehlhorn et al. Reference Mehlhorn, Heydorn and Gestrich1975a). Importantly, throughout the paper by Dubey et al. (Reference Dubey, Jolley and Thorne1983), S. sybillensis is repeatedly described as a species with thick-walled (up to 8 μm) sarcocysts, and the authors point out this feature as a major morphological difference from the thin-walled cysts of S. wapiti, which also occurred in the elk that they examined. Thus, Fig. 1 in the paper by Dubey et al. (Reference Dubey, Jolley and Thorne1983) intends to show the difference between a thick-walled sarcocyst of S. sybillensis and a thin-walled sarcocyst of S. wapiti. Another thick-walled cyst of S. sybillensis shown in Fig. 5 is described as having ‘filamentous’ protrusions. However, in standard histological sections viewed by LM, cysts with hair-like protrusions, like those of S. hjorti, will appear as thin-walled cysts. This has been repeatedly reported in different papers describing both immature and mature cysts of such species e.g., S. cruzi in cattle (Figs 6 and 7 in Mehlhorn et al. Reference Mehlhorn, Heydorn and Gestrich1975b, and Figs 29 and 30 in Dubey et al. Reference Dubey, Speer, Callis and Blixt1982), S. arieticanis in sheep (Fig. 4c–e in Heydorn, Reference Heydorn1985), and S. hircicanis in goats (Fig. 2c, d and f in Heydorn and Unterholzner, Reference Heydorn and Unterholzner1983). Moreover, cysts with hair-like protrusions would be indistinguishable in histological sections from cysts with delicate ribbon-like protrusions like those of S. cervicanis/S. wapiti in red deer and elk, and S. grueneri in reindeer, since all would appear thin-walled. Thus, Gjerde and Bratberg (Reference Gjerde and Bratberg1984) found thin-walled cysts in muscular sections from reindeer (see Fig. 3a in that paper) and subsequent studies by Gjerde using LM of fresh cysts, TEM and SEM, demonstrated that these thin-walled cysts represented 2 species, S. grueneri which had ribbon-like protrusions and S. rangi which had hair-like protrusions (Gjerde, Reference Gjerde1986). Thick-walled sarcocysts seen in histological sections have repeatedly been reported in the literature to represent cysts with fairly upright finger-like protrusions. When these protrusions are fairly thin, they usually appear as filamentous structures in thick sections e.g., protrusions of S. hominis and S. hirsuta of cattle (see Figs 4a–c and 5a, b in Gestrich et al. Reference Gestrich, Heydorn and Baysu1975a, Figs 4 and 5 in Gestrich et al. Reference Gestrich, Mehlhorn and Heydorn1975b, and Fig. 1d–g in Mehlhorn et al. Reference Mehlhorn, Heydorn and Gestrich1975a) and S. tarandi of reindeer (see Fig. 2 in Gjerde and Bratberg, Reference Gjerde and Bratberg1984). Consequently, the thick-walled cyst in Fig. 5 in Dubey et al. (Reference Dubey, Jolley and Thorne1983), is not consistent with a species with hair-like protrusions, but rather with a cyst with finger-like protrusions like those of S. tarandi of reindeer and red deer.
Obviously, Saito et al. (Reference Saito, Itagaki, Shibata and Itagaki1995) made a similar interpretation when they stated that the Sarcocystis species they found in sika deer in Japan resembled S. sybillensis of elk, particularly because the cysts from sika deer in histological sections seemed to have a thick wall with hairy protrusions (see Fig. 2 in Saito et al. Reference Saito, Itagaki, Shibata and Itagaki1995). However, the cysts shown in the fresh state in Fig. 1 and the protrusions depicted in the TEM micrograph in Fig. 5 of their paper are consistent with a species that is morphologically indistinguishable from S. tarandi in reindeer and red deer. Based on the above-mentioned facts, we consider S. sybillensis of elk as described by Dubey et al. (Reference Dubey, Jolley and Thorne1983) to represent at least 2 different species, 1 with erect finger-like protrusions and 1 with hair-like protrusions. Since the major feature of S. sybillensis was said to be the thick wall, consistent with a species with finger-like protrusions, as also interpreted by Saito et al. (Reference Saito, Itagaki, Shibata and Itagaki1995), we have chosen a new name, S. hjorti, for the species with hair-like protrusions in red deer (and moose) in Norway, and possibly also elsewhere in the world. Moreover, there are no molecular data available for comparison from either of the 2 species apparently comprised in the taxon S. sybillensis. The name S. sybillensis should probably be considered a nomen dubium until it has been restricted to only 1 species with clearly defined cyst morphology and molecular characteristics.
Compared with Sarcocystis species described from other cervids, S. rangi of reindeer (Gjerde, Reference Gjerde1984c, Reference Gjerde1985a), S. alceslatrans of moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008) and S. capreolicanis of roe deer (Erber et al. Reference Erber, Boch and Barth1978; Kutkienė, Reference Kutkienė2001; Sedlaczek and Wesemeier, Reference Sedlaczek and Wesemeier1995; Spickschen and Pohlmeyer, Reference Spickschen and Pohlmeyer2002) have cysts with similar delicate hair-like protrusions to those of S. hjorti. However, cysts of S. rangi and S. alceslatrans are generally much longer than those of S. hjorti (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008; Gjerde, Reference Gjerde1984a, Reference Gjerde1985a) and molecular data clearly indicate that they are 3 separate species. No molecular data from S. capreolicanis were available in GenBank for a proper comparison with this species, but the cysts of this species seem to be similar in size to those of S. hjorti (Erber et al. Reference Erber, Boch and Barth1978; Sedlaczek and Wesemeier, Reference Sedlaczek and Wesemeier1995).
The ssu rRNA gene sequence of S. hjorti was identical with that of the unnamed Sarcocystis species Type E of moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008). The sequence from moose was obtained from a small cyst isolated from cardiac muscle, for which no morphological data were recorded, except that no protrusions were seen, and the species was therefore not named in that study. However, the identical ssu rRNA gene sequences show that S. hjorti might use both red deer and moose as its intermediate host. Thus, moose may be the intermediate host of 2 species with closely similar cyst morphology i.e., S. hjorti and S. alceslatrans. So far, S. alceslatrans has only been found in Canadian moose, and not in Norwegian moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008).
Phylogenetic analyses based on the ssu rRNA gene placed S. hjorti together with other canine transmitted species, indicating that S. hjorti most likely also uses a canine definitive host. This is also supported by the fact that S. hjorti was a very common species and most likely uses a prevalent definitive host like the red fox.
S. tarandi
The slender and spindle-shaped cysts were morphologically similar to those of S. tarandi of reindeer (Gjerde, Reference Gjerde1984a, Reference Gjerde1985d, Reference Gjerde1986). The general size and shape of the cysts and protrusions were the same in both red deer and reindeer. Previous molecular studies by Dahlgren and Gjerde (Reference Dahlgren and Gjerde2007) indicated a small sequence variation in the ssu rRNA gene within and between isolates of S. tarandi from reindeer, since a few nucleotide positions were difficult to determine due to multiple peaks on DNA sequence chromatograms. By cloning isolates of S. tarandi from both reindeer and red deer we discovered that multiple variants of the ssu rRNA gene of this species existed, which probably caused the multiple peaks on the sequence chromatogram when sequencing was attempted directly after PCR. The variation between the 2 isolates from the same host species (red deer and reindeer) was small and approximately the same as the variation between isolates from different hosts, indicating that all isolates belonged to the same Sarcocystis species. Phylogenetic analyses based on both ssu rRNA gene data and ITS-1 data, seemed to place all clones from both reindeer and red deer at random within the same cluster, providing further evidence that the isolates from the 2 hosts should be considered the same species. In contrast to S. rangiferi, for which several clonal groups were seen, all S. tarandi sequences formed 1 cluster.
Small cysts found in the heart of a few animals were identified as belonging to either S. rangiferi or S. tarandi based on their cyst morphology. However, only by molecular methods could we assign 1 of the cysts to S. tarandi. The other cysts were never molecularly examined and could possibly be of S. rangiferi.
In a previous study of Sarcocystis in moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008), we described the new species Sarcocystis scandinavica, based on molecular and morphological data. The cyst wall of S. scandinavica was similar to that of S. tarandi and S. scandinavica also had several variants of its ssu rRNA gene. However, the molecular data indicated that this was a separate species, and this conclusion was further supported by the results of the phylogenetic analyses in the present study.
S. rangiferi
Ssu rRNA gene sequences of cysts from reindeer and red deer were highly similar, indicating that they were of the same species. As for S. tarandi, the first part of the ssu rRNA gene of S. rangiferi usually resulted in a few multiple peaks on the DNA sequence chromatogram when the gene was sequenced directly after PCR. By cloning S. rangiferi-like isolates obtained from 2 different red deer and 2 isolates of S. rangiferi from different reindeer, we obtained fine single-peak DNA chromatograms with slightly different nucleotide sequences of the different clones. Sequence variation between clones from reindeer isolates was similar to that found between clones from red deer isolates, and sequence variation between clones of isolates from reindeer and clones of isolates from red deer was similar, suggesting that all isolates belonged to the same species. Multiple sequence alignments and phylogenetic analyses based on the ssu rRNA gene indicated that there were 3–4 sequence groups, 1 consisting of all clones from reindeer and 2–3 other sequence types in red deer. Thus, analyses based on ssu rRNA gene sequences could not resolve whether the different clusters of S. rangiferi-type sequences on the phylogenetic tree was due to the occurrence of different paralogous ssu rRNA sequences of S. rangiferi in red deer and reindeer, or if the S. rangiferi-like cysts in red deer should be considered a separate species. Divergent ssu rRNA paralogues have previously been reported from another Sarcocystis species, namely S. singaporensis (Slapeta et al. Reference Slapeta, Kyselová, Richardson, Modrý and Lukeš2002). We could have sequenced more clones of S. rangiferi-like cysts from both red deer and reindeer to see if the ssu rRNA gene sequences from the 2 cervid species would always group separately. However, we choose to further investigate this by examining the more rapidly evolving ITS-1 region to see whether a recent split into 2 species had occurred. Phylogenetic analyses based on ITS-1 sequence data revealed a similar complex pattern in which all but 1 sequence from reindeer formed a cluster, whereas the clones from red deer formed 2 other clusters, of which 1 appeared closely related to the main reindeer cluster. This interwoven pattern of clusters of both red deer and reindeer sequences seems to indicate that they belong to the same species i.e., S. rangiferi. Thus we consider the S. rangiferi-like sequences and cysts in red deer to belong to the previously recognized species S. rangiferi.
A limited examination of the D2-D3 domain of the large subunit (lsu) rRNA gene of the S. ranigferi-like cysts from red deer and reindeer indicated a high degree of variation also at this gene segment (unpublished data). Thus, none of the investigated DNA sequences within the rDNA unit (ssu rRNA, ITS-1, and lsu rRNA) seems to be very suitable for proving unequivocally that S. rangiferi occur in both red deer and reindeer, unless a considerable number of isolates/clones from both hosts are compared. It is probably more convenient to use other genetic markers with less intraspecific variation to resolve this question in future studies, suggestively protein coding genes such as the tubulin genes or the gene for the 70 kDa heat shock protein, could be evaluated.
As regards morphology, the cigar-shaped cysts in red deer were morphologically similar to cysts of S. rangiferi of reindeer (Gjerde, Reference Gjerde1984c; Gjerde, Reference Gjerde1985c; Gjerde, Reference Gjerde1986). The general size and shape of the cysts and protrusions were the same in both red deer and reindeer. However, the cysts of S. rangiferi were generally smaller in red deer than in reindeer, and whereas the host cell of cysts in reindeer were consistently surrounded by fibrous material (Gjerde, Reference Gjerde1984b, Reference Gjerdec), this encapsulation seemed to be absent around cysts in red deer. Further studies of cysts from red deer by TEM will be necessary to confirm or refute this possible difference in host reaction to cysts of S. rangiferi.
S. ovalis and S. hardangeri
This is the first time that Sarcocystis species with such large, ovoid, cysts have been reported from red deer. Cysts found in red deer were morphologically indistinguishable from S. hardangeri of reindeer (Gjerde, Reference Gjerde1984b, Reference Gjerdec; Dahlgren et al. Reference Dahlgren, Gjerde, Skirnisson and Gudmundsdottir2007), S. ovalis of moose (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008), and S. oviformis of roe deer (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2009). In all 4 hosts, cysts of these species seem to induce an encapsulation of their host cells with fibrous material, but this has so far only been clearly demonstrated by TEM for cysts of S. hardangeri in reindeer (Gjerde, Reference Gjerde1985b) and cysts of S. ovalis (‘Type B’ cysts in moose) by Colwell and Mahrt (Reference Colwell and Mahrt1981).
Sequence analyses of the complete ssu rRNA gene from 3 isolates indicated that 2 isolates belonged to S. ovalis and the third isolate to S. hardangeri. Part of the ssu rRNA gene sequence of 10 additional isolates was sequenced to investigate whether a separate species might infect red deer. However, all 10 isolates were of S. ovalis, suggesting that this is the most commonly found species with this cyst type both in red deer and in moose. Sequencing of more cysts will reveal whether there are additional species with this cyst morphology in cervids.
Gjerde (Reference Gjerde1984b) originally detected and described the species S. hardangeri, based on its cyst morphology, from wild reindeer. This reindeer population was located in the high-mountainous area of Hardangervidda in Southern Norway, which lies about 100 kilometres in a straight line, but separated by fjords, from the coastal area from which a single red deer infected with this species originated. Moreover, there are no reindeer or moose in that particular area. In contrast, all red deer infected with S. ovalis originated from a separate costal area which lies about the same distance from Hardangervidda, but with no intervening fjords and with a few reindeer in the vicinity. In addition, a small population of moose has also become established in this region in recent years. Still, the fairly high occurrence of S. ovalis in red deer in this area suggests that this species might easily cycle between red deer via its unknown definitive hosts rather than being occasionally transmitted to red deer from moose.
Comparison of S. rangiferi and S. tarandi in red deer and reindeer, and previous reports of similar species in red deer
S. rangiferi and S. tarandi most likely separated relatively recently, as seen from their phylogenetic positions. Like in reindeer, S. rangiferi and S. tarandi usually occurred concurrently in a given red deer and based on this fact and their phylogenetic position, it is likely that the two species use the same definitive host. In both hosts, S. rangiferi cysts were thicker and usually longer than S. tarandi. S. rangiferi cysts were mostly cigar-shaped to ellipsoidal, whereas S. tarandi cysts were slender and spindle-shaped. Protrusions of S. rangiferi were thicker than the protrusions on S. tarandi cysts, causing a finer striation of the cyst wall in fresh preparations, as seen in the present study, and in histological sections as seen in the study by Gjerde and Bratberg (Reference Gjerde and Bratberg1984) (see Figs 1 and 2 in that paper). Since there are only a few or no reindeer in the area inhabited by the red deer found to be infected with S. rangiferi and S. tarandi in the present study, it is likely that these species cycle within the red deer population via their unknown definitive hosts. Moreover, these species, which are very common in reindeer in Northern Norway, were not found in 7 wild reindeer from the adjacent Hardangervidda area examined by Gjerde in the mid-1980s (Gjerde, Reference Gjerde1984b).
Cysts with similar finger-like protrusions as in S. rangiferi and S. tarandi have been reported previously from red deer and elk. In Germany, Wesemeier and Sedlaczek (Reference Wesemeier and Sedlaczek1995a) found such sarcocysts in both a free-ranging red deer and a captive red deer and an elk in a zoo. They referred to these cysts as S. cf. hofmanni, due to their resemblance with sarcocysts in the badger (Odening et al. Reference Odening, Stolte, Walter and Bockhardt1994). Some of the LM- and TEM pictures in their publication (Figs 13–16) depict sarcocysts with thick protrusions similar to those of S. rangiferi, whereas others (Figs 11, 17 and 18) show cysts with more slender protrusions similar to those of S. tarandi. The protrusions were reported to be 6·7–8·3 μm long and 1·7–3·4 μm wide, which also suggests a mixture of 2 species similar to S. tarandi and S. rangiferi. The cysts from the captive wapiti were also depicted in Plate IV of a paper by Stolte et al. (Reference Stolte, Bockhardt and Odening1996) and referred to as cysts of an unnamed Sarcocystis sp. In 1997, Stolte et al. described typical encapsulated S. rangiferi cysts from a reindeer that had lived its entire life in a zoo in Germany. Free-living reindeer do not occur in Germany and the finding by Stolte et al. (Reference Stolte, Bockhardt and Odening1997) therefore suggests that other cervids, like red deer, harboured this species and spread the infection, via a definitive host, to the captive reindeer. In Lithuania, Kutkienė (Reference Kutkienė2003) also found cysts with villar protrusions, consistent with those of S. rangiferi. No information regarding cyst size or shape is given in her publication; however, she also referred to the species as S. cf. hofmanni.
It thus seems that S. rangiferi- and S. tarandi-like species also are present in red deer in other European countries, but it remains to be determined whether they are genetically identical to the species found in red deer and reindeer in Norway.
Comparison of Sarcocystis species described in this study with Sarcocystis species from sika deer in Japan
Sarcocystis species with similar cyst morphology to 3 of the species we found in red deer have also been reported from sika deer, another host species of the genus Cervus.
Saito et al. (Reference Saito, Itagaki, Shibata and Itagaki1995) examined sarcocysts from a farmed sika deer (Cervus nippon centralis) in Japan by LM and TEM. The size (510×57 μm) and shape (elongated, slender, with pointed tips) of these cysts and the appearance of their protrusions when viewed in the fresh state (Fig. 1 in their paper) make these cysts indistinguishable from those of S. tarandi of red deer and reindeer. The TEM micrograph (Fig. 5) is of poor quality, and mostly shows obliquely sectioned protrusions, but they appear to be finger-like. As already mentioned, the authors thought these cysts closely resembled those of S. sybillensis described by Dubey et al. (Reference Dubey, Jolley and Thorne1983), probably because S. sybillensis was reported to have thick-walled cysts.
Saito et al. (Reference Saito, Itagaki, Shibata and Itagaki1995) also fed Sarcocystis infected muscle tissue from the same sika deer to 2 raccoon dogs, 2 dogs and 2 cats. The raccoon dogs and the dogs shed sporocysts, whereas the 2 cats did not shed any sporocysts or oocysts. They therefore concluded that raccoon dogs and dogs were definitive hosts for this Sarcocystis species with S. tarandi-like cysts, a conclusion that was repeated in a subsequent paper which compares the ultrastructure of cysts of canine transmitted Sarcocystis species (Saito et al. Reference Saito, Shibata, Kobayashi, Kobayashi, Kubo and Itagaki1996). The conclusion is understandable since they believed they had found S. sybillensis, which had been reported by Dubey et al. (Reference Dubey, Jolley and Thorne1983) to be transmitted by dogs. No molecular data were obtained from the sporocysts shed by the canines, and therefore the species identity of those cannot be determined. If the S. tarandi-like species in sika deer was genetically similar to S. tarandi, it was probably not transmitted by canines, as suggested from the phylogenetic position of S. tarandi in our study. We therefore believe that the sika deer muscle tissue fed to the canines in the study by Saito (Reference Saito, Itagaki, Shibata and Itagaki1995) also was infected with microscopic sarcocysts of a truly canine transmitted species.
Takano (Reference Takano, Hamada, Ogiwara and Yagi2006) examined another subspecies of sika deer (Cervus nippon (j)ezoensis) from another area of Japan (Hokkaido). They isolated cysts from 1 animal and described 1 cyst by its morphology as being spindle-shaped, 3×1 mm long, and with a 5 μm thick, ‘hairy’ wall. This description is consistent with the morphology of S. rangiferi cysts. However, Figs 2 and 3 in their paper depict macroscopically visible oval cysts, which could be of either a S. hardangeri/S. ovalis-like species or of a S. rangiferi-like species. They sequenced the complete ssu rRNA gene, but unfortunately the sequence was not deposited in GenBank and therefore it could not be compared with our sequences from red deer. From the placement of this species in their phylogenetic tree (Fig. 5) it is difficult to determine whether this species represents a S. rangiferi-like or a S. hardangeri/S. ovalis-like species. However, the authors compared their sequence(s) with other Sarcocystis species, and based on the given identity values, the Sarcocystis species from sika deer differed by approximately the same percentage from Sarcocystis fusiformis and S. hominis as our isolates of S. rangiferi differed from those two species. It is therefore likely that the species described by Takano et al. (Reference Takano, Hamada, Ogiwara and Yagi2006) is closely related to S. rangiferi.
In 2008, Narisawa et al. described 3 different species from the same subspecies of sika deer (Cervus nippon yesoensis) in Hokkaido, Japan. Light microscopy pictures and TEM micrographs in their publication illustrate cysts which are similar to those of S. rangiferi (Type 1), S. tarandi (Type 2), and S. hjorti (Type 3). Thus, Fig. 1 shows a large, cigar-shaped cyst with broad protrusions, Fig. 2 shows a thick-walled cyst (in histological section) with thin finger-like protrusions, whereas Fig. 3 shows a thin-walled cyst with hair-like protrusions. Figs 2 and 3 demonstrate, as already discussed, that cysts with hair-like, in contrast to those with finger-like protrusions, have a thin wall in histological sections. The species with hair-like protrusions (Type 3) is probably transmitted by canines, since this seems to be a common feature of species with this cyst type, and it is therefore likely that the muscle tissue used by Saito et al. (Reference Saito, Itagaki, Shibata and Itagaki1995) to infect dogs and raccoon dogs had an undetected infection with such a species.
Molecular data of Sarcocystis isolates from sika deer (Cervus nippon yesoensis), are available in GenBank, but no morphological data are linked to these sequences (Accession nos. AB251926, AB257085-86, AB257155-62). According to GenBank data 2–3 isolates (sarcocysts) seem to have been cloned and sequenced at the ssu rRNA gene. The slightly different gene sequences are highly similar to ssu rRNA gene sequences of S. rangiferi, and phylogenetic analyses suggest that the Sarcocystis sp. of sika deer, our S. rangiferi-like species from red deer, and S. rangiferi of reindeer belong to the same species. Presumably these sarcocysts from sika deer had a similar morphology to our S. rangiferi cysts and probably to those described as Type 1 cysts by Narisawa et al. (Reference Narisawa, Yokoi, Kawai, Sakui and Sugawara2008).
Comparison of S. rangiferi and S. tarandi with Sarcocystis species described from other Cervidae
Because S. rangiferi and S. tarandi were seen to occur in both red deer and reindeer, and because S. rangiferi, or a closely related species, also occur in sika deer, we have reviewed reports of S. rangiferi-like and S. tarandi-like cysts in other cervid hosts as well. Molecular data are lacking for all these species, so as yet it can not be conclusively determined whether they are identical to S. rangiferi and S. tarandi.
Fallow deer (Cervus dama/Dama dama)
Hernandez-Rodriguez et al. (Reference Hernandez-Rodriguez, Acosta and Navarrete1992) found large, spindle-shaped sarcocysts in a fallow deer in Spain, which they described as the new species Sarcocystis jorrini. By TEM, the cysts of S. jorrini were morphologically indistinguishable from cysts S. rangiferi in reindeer, and the cysts of this species were also encapsulated by a thick fibrous layer, identical to the capsule seen around S. rangiferi cysts in reindeer, but not around cysts in red deer.
In 1 of 3 fallow deer examined by Wesemeier and Sedlaczek (Reference Wesemeier and Sedlaczek1995b) in Germany, they found sarcocysts that by TEM are consistent with the morphology of S. tarandi cysts (see Figs 5–7 in their paper). The authors believed this species was identical to S. cf. hofmanni of roe deer and red deer, which again they believed were identical to S. hofmanni of the badger. But, since no molecular data were recorded, it cannot be determined which species the cysts in fallow deer truly belonged to.
Thus it seems that the fallow deer might also harbour 2 species that are morphologically similar to and probably closely related to S. rangiferi and S. tarandi of reindeer and red deer.
Roe deer (Capreolus capreolus)
Sarcocysts consistent with those of S. tarandi and S. rangiferi have been reported from roe deer in several studies from Europe. Bergmann and Kinder (Reference Bergmann and Kinder1976) examined tissues from roe deer by LM and TEM and found thick-walled cysts with finger-like protrusions, which they referred to as Typ Rh 1. These cysts are similar to those of S. tarandi.
Blažek et al. (Reference Blažek, Schramlova, Ippen and Kotrly1978) examined muscle tissue samples using histological sections and found thick-walled cysts with the same morphology as S. rangiferi-cysts.
Erber et al. (Reference Erber, Boch and Barth1978) examined fresh sarcocysts from roe deer after tryptic digestion of muscle tissues and found 3 cyst types, 2 of which were given the names S. gracilis and S. capreolicanis. The third species was never named, however, those cysts (Type 3) are similar in size and shape to those of S. tarandi and they also have similar finger-like protrusions.
Schramlová and Blažek (Reference Schramlová and Blažek1978) examined tissues by TEM which initially had been fixed for histology. Although the TEM micrographs are of poor quality, the thick walled-cysts in Fig. 2a–d appear to be similar to those of S. tarandi.
Entzeroth (Reference Entzeroth1982) found 6 types of cyst walls by TEM examination of sarcocysts from roe deer. The protrusions of Type 1, 2 and 3 cysts differed slightly in size and shape, but were similar to those of S. tarandi. The author believed that Type 1, 2 and 3 cysts represented cysts of the same species at different stages of development. Entzeroth (Reference Entzeroth1985) probably described 2 species in this publication, although he believed that all cysts belonged to the same species i.e., S. gracilis. Thus, the LM- and TEM photos (Figs 2 and 3) show cysts with finger-like protrusions like those of S. tarandi.
Sedlaczek and Wesemeier (Reference Sedlaczek and Wesemeier1995) found 3 types of cysts by TEM in tissue samples from roe deer. Thick-walled cysts with finger-like protrusions were seen in all types of muscle tissue examined, including the heart. They referred to all cysts with this morphology as cysts of S. cf. hofmanni. However, the given size descriptions of the cysts indicate a mixture of thin, slender cysts and fairly thick cysts, suggesting a mixed infection with S. rangiferi-like and S. tarandi-like cysts. Stolte et al. (Reference Stolte, Bockhardt and Odening1996) compared the sarcocyst morphology of 11 species as seen in fresh preparations by LM and SEM. Plate V shows S. tarandi-like cysts from roe deer, which were classified as S. hofmanni.
Kutkienė (Reference Kutkienė2001) examined tissue samples from roe deer by a glass compressor and isolated some cysts for examination of the cyst wall in wet mounts. She identified 4 cyst types, 1 of which was similar to those of S. tarandi. She referred to the S. tarandi-like cyst as S. cf. hofmanni. Spickschen and Pohlmeyer (Reference Spickschen and Pohlmeyer2002) examined muscle tissues from roe deer and found 3 cyst types. The Sarcocystis sp. depicted in Fig. 3a–b is similar to S. tarandi.
Thus, roe deer in several European countries seem to be commonly infected with sarcocysts similar to those of S. tarandi, and possibly also S. rangiferi, in red deer and reindeer. However, no such cysts were detected in a recent study of sarcocysts in Norwegian roe deer (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2009).
Mule deer (Odocoileus hemionus)
Dubey and Speer (Reference Dubey and Speer1985) examined samples from mule deer by LM and TEM and found 3 types of sarcocysts. Type 2 cysts were very similar to those of S. tarandi (Figs 8–11 in their paper), and Type 3 cysts resembled those of S. rangiferi (Fig. 12 in their paper). In a subsequent paper, Dubey and Speer (Reference Dubey and Speer1986), named the species with Type 2 cysts, Sarcocystis hemioni and the species with Type 3 cysts, Sarcocystis youngi. They also named a third species with hair-like villar protrusions, Sarcocystis americana. However, these cysts were only observed in histological sections, and their appearance is consistent with cysts of S. tarandi in such sections. Thus, it is likely that the authors misinterpreted their sections and that Fig. 1 A–C in their paper, reportedly showing Sarcocystis hemionilatrantis, S. hemioni, and S. youngi probably represents only 1 species, the S. rangiferi-like species S. youngi, whereas Fig. 4, allegedly showing S. americana, represents the S. tarandi-like species S. hemioni.
White tailed deer (Odocoileus virgnianus)
Dubey and Lozier (Reference Dubey and Lozier1983) examined muscle tissues from white tailed deer by LM of histological sections and by TEM and found 3 types of cysts. They named the species with Type 2 cysts, Sarcocystis odoi. Type 3 cysts were never assigned a species name. The sarcocyst morphology, including the outline of the surface protrusions, of S. odoi cysts, is consistent with the cyst morphology of S. tarandi. However, the fairly large and thick cysts they described for S. odoi, probably represent cysts of both S. odoi and the unnamed species with Type 3 cysts. The cyst morphology of the unnamed species is consistent with that of S. rangiferi. Interestingly, a cat fed tissues from a white-tailed deer got infected and shed sporocysts.
Preparation of sarcocysts for SEM
Air-drying the cysts from HMDS at room temperature (all cysts depicted in Figs 2, 4 and 6) gave at least as good results by SEM as critical-point drying from CO2 (cysts shown in Fig. 8), as also reported by Stolte et al. (Reference Stolte, Bockhardt and Odening1996). Moreover, using HMDS allowed us to fixate, dehydrate and dry the tiny cysts while they were kept within the same Eppendorf tube, which prevented mechanical damage to, or loss of cysts during processing. In addition, drying from HMDS requires no expensive equipment and is easy to do in an ordinary lab.
Conclusions
This study has revealed that red deer (in Norway) share 5 Sarcocystis species with reindeer and moose, and that consequently these particular species are not intermediate host specific. A sixth cyst type or species of red deer and elk, variously referred to as S. cervicanis, S. wapiti and S. cf. grueneri, was not found in the present study, even though the morphologically indistinguishable species S. grueneri is very common in reindeer in Norway (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2007). Although S. grueneri-like cysts have been reported from moose and roe deer elsewhere in Europe, neither were S. grueneri-like cysts nor DNA sequences of a similar species detected in moose and roe deer in Norway (Dahlgren and Gjerde, Reference Dahlgren and Gjerde2008, Reference Dahlgren and Gjerde2009). This might indicate that S. grueneri of reindeer does not infect other cervids, or that the small cysts of this species have been overlooked. The present study clearly demonstrates the importance of using molecular methods to identify cysts and to combine molecular and morphological data when describing new species. Today, any name given to a Sarcocystis species that is not accompanied by relevant molecular data is more or less a nomen dubium, since it is almost impossible to determine whether a sarcocyst found in one host species is identical to morphologically similar cysts found in another related intermediate host species. Only after a comprehensive molecular study of numerous sarcocysts isolated from different intermediate hosts will it be possible to determine whether a given morphological cyst type represents 1 or more species using 1 or more intermediate hosts. Alternatively, muscle tissues can be fed to definitive hosts and their feces/intestines screened for sporocysts of different species by molecular methods.
ACKNOWLEDGEMENTS
We would like to thank all hunters who provided material from red deer for this investigation, and particularly Arne Gravelsæter at ‘Fjordvilt’, Etne, who collected, stored and forwarded the majority of the samples, originating from deer carcasses delivered by hunters for processing at his abattoir. We also thank Steinar Stølen at the University of Oslo, Department for Oral Biology, Faculty of Dentistry, for excellent technical assistance with the SEM study.