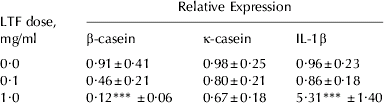Lactoferrin (LTF) is an iron-binding glycoprotein present in milk and exocrine secretions. The concentration of LTF in bovine milk varies throughout lactation. LTF is present at 1–2 mg/ml in mammary secretions during the final prepartum stages, 0·1 mg/ml in milk and rises to 20–50 mg/ml in the mammary gland during involution (Baumrucker & Erondu, Reference Baumrucker and Erondu2000). The iron-binding capacity of LTF imparts antimicrobial properties and it is thought that expression during the lactation cycle confers some protection against infection of the neonate and to the udder during involution. However, the high level of LTF expression during involution may also have other roles as LTF has been reported to act as a transcription factor (Son et al. Reference Son, Park, Chung, Chung, Yu, Lee and Kim2002) and to induce apoptosis in some cell types (Roy et al. Reference Roy, Kuwabara, Hara, Watanabe and Tamai2002; Fujita et al. Reference Fujita, Matsuda, Sekine, Iigo and Tsuda2004; Lin et al. Reference Lin, Chiou, Chen and Kuo2005; Sakai et al. Reference Sakai, Banno, Kato, Nozawa and Kawaguchi2005).
Regulation of bovine LTF in the mammary gland differs from other milk proteins (Schanbacher et al. Reference Schanbacher, Goodman and Talhouk1993). Unlike caseins, LTF secretion in bovine mammary epithelial cells (BMEC) is not dependent on prolactin. Instead, LTF is regulated by cell shape and density (Close et al. Reference Close, Howlett, Roskelley, Desprez, Bailey, Rowning, Teng, Stampfer and Yaswen1997). An inverse relationship in expression patterns for LTF and caseins occurs throughout the lactation cycle; when LTF expression is high, casein expression is low and vice versa (Schanbacher et al. Reference Schanbacher, Goodman and Talhouk1993).
LTF is secreted by epithelial cells in mammary tissue. LTF receptors have been found on mammary epithelial cells (Rochard et al. Reference Rochard, Legrand, Lecocq, Hamelin, Crepin, Montreuil and Spik1992) indicating a role for LTF in autocrine regulation. LTF has a region similar to a nuclear localization signal and binds to DNA (He & Furmanski, Reference He and Furmanski1995) although few target genes have been identified. Human LTF activates transcription of interleukin-1β (IL-1β) in leukaemia cells (Son et al. Reference Son, Park, Chung, Chung, Yu, Lee and Kim2002) and interestingly, IL-1β inhibits casein expression (Baratta et al. Reference Baratta, Motta and Acconero2005). Hence bovine LTF may regulate casein expression in BMEC through IL-1β.
The high level of LTF during involution may have an apoptotic role in BMEC as accumulation of apoptotic factors in milk is a possible contributor to initiation of involution (Wilde et al. Reference Wilde, Knight and Flint1999). LTF or LTF-derived peptides are involved in apoptosis in several cell types (Roy et al. Reference Roy, Kuwabara, Hara, Watanabe and Tamai2002; Fujita et al. Reference Fujita, Matsuda, Sekine, Iigo and Tsuda2004; Lin et al. Reference Lin, Chiou, Chen and Kuo2005; Sakai et al. Reference Sakai, Banno, Kato, Nozawa and Kawaguchi2005). Caspase 3 was induced by bovine LTF in human oral cancer cells (Sakai et al. Reference Sakai, Banno, Kato, Nozawa and Kawaguchi2005) and human leukaemic cells (Roy et al. Reference Roy, Kuwabara, Hara, Watanabe and Tamai2002), and by human LTF in rat neuronal cells (Lin et al. Reference Lin, Chiou, Chen and Kuo2005). Caspase 3 is also induced during bovine mammary gland involution (Motyl et al. Reference Motyl, Gajkowska, Zarzynska, Gajewska and Lamparska-Przybysz2006). Thus the possibility that LTF may be involved in mammary gland involution is worthy of investigation.
The potential of LTF to play a role in involution led us to investigate whether LTF could influence casein expression and apoptosis of BMEC. Factors that influence milk composition and lactational persistence are of economic importance to the dairy industry.
Materials and Methods
All reagents and oligonucleotides were from Sigma-Aldrich, Castle Hill, NSW 2154, Australia, unless stated otherwise.
Primary bovine mammary epithelial cell isolation
Mammary biopsies were taken from multiparous Holstein-Friesian cows as previously described (Sheehy et al. Reference Sheehy, Della-Vedova, Nicholas and Wynn2004). Mammary tissue was washed in phosphate-buffered saline (pH 7·4), extraneous tissue excised and mammary tissue placed into 1× Hanks Balanced Salt Solution (HBSS) containing 4·2 mm-NaHCO3, 50 U/ml penicillin/50 μg/ml streptomycin (Invitrogen, Mount Waverley, VIC 3149, Australia), 50 μg/ml kanamycin (Invitrogen) and 625 ng/ml Fungizone (Invitrogen), pH 7·4. Typically, 8–10 g was collected.
Tissue was weighed, minced with scissors and incubated in 10 ml/g predigest medium [1×HBSS, 1×minimal essential medium (MEM), 5·5 mm-glucose, 4% bovine serum albumin, 20 μm-CaCl2, 20 μm-MgSO4, 5 mm-4-(2-hydroxyethyl)piperazine-1-ethanesulphonic acid (HEPES), 200 μm-glutamine, 10 U/ml penicillin/10 μg/ml streptomycin, 10 μg/ml kanamycin, 250 ng/ml Fungizone, 1 μg/ml insulin, 1 μg/ml cortisol, 50 U/ml DNase I (Roche Molecular Biochemicals, Castle Hill, NSW 2154, Australia) pH 7·4] at 37°C for 10 min at 200 rpm. Media was aspirated and replaced by digest media (predigest media+200 U/ml collagenase Type II, 250 U/ml hyaluronidase) and incubated at 37°C, at 225 rpm for 2 h. Digested tissue was filtered through a 150-μm nylon mesh. Retained tissue was further incubated in fresh digest media. The filtrate was centrifuged at 200 g for 5 min and the cell pellet was washed three times in 20 ml wash media (1×HBSS, 1×MEM, 5·5 mm-glucose, 20 μm-CaCl2, 20 μm-MgSO4, 5 mm-HEPES, 2 mm-glutamine, 100 U/ml penicillin/100 μg/ml streptomycin, 100 μg/ml kanamycin, 2·5 μg/ml Fungizone, 100 U/ml DNase I, 100 μg/ml trypsin inhibitor, 5 μg/ml insulin, 1 μg/ml cortisol, pH 7·4). The final wash was filtered through a 50-μm nylon mesh and the filtrate centrifuged at 200 g for 5 min. The cell pellet was resuspended in 2·5 ml 1×HBSS and overlayed onto a 17·5 ml 1·01–1·07 g/ml Percol-Ficoll step gradient. The gradient was centrifuged at 800 g for 20 min. BMEC were subsequently collected from the 1·03/1·04 density layers and washed in 20 ml 1×HBSS. Cells were then cryopreserved.
Effect of exogenous lactoferrin and lactoferrin short interfering RNA on casein gene expression
Mammospheres capable of milk protein expression, were formed by culturing BMEC on an extracellular matrix (Matrigel; BD Biosciences, North Ryde, NSW 2113, Australia) in the presence of lactogenic hormones (German & Barash, Reference German and Barash2002; Rose et al. Reference Rose, Aso, Yonekura, Komatsu, Hagino, Ozutsumi and Obara2002). BMEC were plated onto 6-well plates that had been coated with Matrigel (25 μl/cm2) according to the manufacturer's instructions, at 2×106 cells/well in attachment media [4·75 g/l M199, 5·32 g/l Hams F12, 2·38 g/l HEPES, 1·875 g/l NaHCO3, 0·164 g/l sodium acetate, 20% horse serum (Invitrogen), 5% fetal calf serum (FCS; Invitrogen), 100 U/ml penicillin/100 μg/ml streptomycin, 100 μg/ml kanamycin, 1·25 μg/ml Fungizone, 5 μg/ml insulin, 1 μg/ml cortisol, 3 μg/ml prolactin (NHPP, Torrance CA 90509, USA) 10 μg/ml transferrin] containing 0, 0·1 and 1 mg/ml bovine LTF. BMEC from the same batch were used to plate an entire experiment and treatments were performed in triplicate to minimize any well to well variation. One day (24 h) after plating, media was changed to differentiation media (4·75 g/l M199, 5·32 g/l Hams F12, 2·38 g/l HEPES, 1·875 g/l NaHCO3, 0·164 g/l sodium acetate, 100 U/ml penicillin/100 μg/ml streptomycin, 100 μg/ml kanamycin, 1·25 μg/ml Fungizone, 5 μg/ml insulin, 1 μg/ml cortisol, 3 μg/ml prolactin, 10 μg/ml transferrin) containing 0, 0·1 and 1 mg/ml bovine LTF. Media was changed every 2 d. After 4 d in differentiation media cells were harvested into RNA extraction lysis buffer (QIAGEN, Doncaster, VIC 3018, Australia).
Short interfering RNA (siRNA) studies were performed to investigate the effect of LTF knockdown on casein mRNA expression. BMEC were plated onto Matrigel-coated 6-well plates at 2×106 cells/well in attachment media. After 24 h media was replaced with differentiation media. After a further 48 h media was replaced with differentiation media lacking antibiotics or Fungizone and mammospheres were transfected with 250 pmol LTF siRNA duplex (5′-GCCUUUGCCUUGGAAUGUATT-3′; BLOCK-iT RNAi designer, Invitrogen) per well, or an off target control siRNA duplex (5′-[Fluo]GAAGCUGACCCUGAAGUUCTT-3′) using 5 μl lipofectamine RNAiMAX (Invitrogen) per well. Cells were harvested into RNA extraction lysis buffer (QIAGEN) 48 h after transfection.
RNA was extracted using an RNeasy Mini kit (QIAGEN) with some modifications. Following homogenization, samples were made up to 800 μl in RNase-free water and incubated with 0·2 mg Proteinase K (Roche Molecular Biochemicals) at 55°C for 10 min. Then 0·5 vol of 100% ethanol was added prior to addition to the column. An on-column DNaseI treatment was included. RNA yield and purity was determined by spectrophotometry and 1·2% agarose/formaldehyde gel electrophoresis.
cDNA was generated from 1 μg of purified and Dnase-treated RNA using SuperScript III MMLV RT (Invitrogen) according to the manufacturer's instructions. Expression levels of β-casein, κ-casein, LTF, IL1β and a housekeeping gene, ribosomal protein, large P0 (RPLP0) were determined by real-time polymerase chain reaction (PCR) analysis using gene specific primer sets (Table 1). Each gene was amplified in a separate reaction and each reaction performed in quadruplicate. Each reaction contained 20 ng cDNA for the casein genes, 50 ng for IL1β or 10 ng cDNA for LTF and RPLP0, 0·3 μm each primer, 0·2 mm-dNTPs (Invitrogen), 0·05 U Platinum Taq polymerase (Invitrogen), 1×Platinum Taq buffer, 3 mm-MgCl2 and 0·4X SYBR green. Amounts ranging from 0·5 fg to 50 pg of purified β-casein, κ-casein, LTF and RPLP0 PCR product were also amplified under the same conditions to generate standard curves from which the copy number of each gene was determined. Reactions were incubated at 95°C for 2 min then 35 cycles were performed as follows: 95°C 20 s, 60°C 20 s, 72°C 20 s (Rotor-Gene 3000; Corbett Life Science, Mortlake, NSW 2137, Australia). Finally, a 72–99°C gradient at 5 s per degree was performed to test product specificity by melt curve analysis. Expression levels of genes of interest were normalized to RPLP0 expression levels. Data from multiple experiments was then expressed as relative normalized expression by setting the control in each experiment to 1-fold expression.
Table 1. Primers used for real-time PCR analyses

Cell viability assays
BMEC were plated in 96-well plates at 1×104 cells/well in proliferation media (4·75 g/l M199, 5·32 g/l Hams F12, 2·38 g/l HEPES, 1·875 g/l NaHCO3, 0·164 g/l sodium acetate, 20% horse serum, 5% FCS, 100 U/ml penicillin/100 μg/ml streptomycin, 100 μg/ml kanamycin, 1·25 μg/ml Fungizone, 5 μg/ml insulin, 1 μg/ml cortisol, 10 ng/ml epidermal growth factor) or on Matrigel-coated 96-well plates at 1×105 cells/well in attachment media and incubated at 37°C in 5% CO2. Bovine lactoferrin was added to cell monolayers at 0–1 mg/ml in proliferation media 24 h after plating. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay (Promega Corporation, Annandale, 2038 NSW, Australia) was performed 2 d later. For cells on Matrigel, 0, 0·1 or 1 mg/ml bovine LTF in differentiation media was added 24 h after plating and a MTT cell proliferation assay performed 4 d later. Absorbances were read at 560 nm and 690 nm in a plate reader (POLARstar OPTIMA, BMG Labtech; Mount Eliza, VIC 3930, Australia). Data from multiple experiments were normalized by setting the control to 100% cell viability.
Caspase assay
BMEC were cultured on Matrigel in 96-well white-walled plates at 1×105 cells/well. On day 4 of culture LTF was added to 1 mg/ml. Etoposide (5 μm) was used as a positive control. Mammospheres were incubated with LTF in differentiation media for 6 h or overnight and then assayed for caspase activity using the Caspase Glo 3/7 Assay (Promega). Luminescence was measured after 30 min (POLARstar OPTIMA, BMG Labtech).
Statistical analyses
All data were analysed using restricted maximum likelihood (REML) procedures in GenStat (Release 10, VSN Intl., Hemel Hempstead, UK) using the following model:
where Yij is the measure being modelled (either % cell viability or relative normalized gene expression). Treatment (either LTF dose or siRNA used) was designated as a fixed effect while Experiment was a random effect, with εij being the residual random error. A natural logarithm was applied to the gene expression data to normalize the variance, while cell viability data did not require transformation. Model-based means were calculated and presented in the graphs and in instances where the data were log-transformed, were back-transformed (exponentiated) to facilitate interpretation of the results. Standard errors shown in tables and figures are in the form mean±se, using model-based se, with approximate se calculated for the back-transformed means.
Results
To investigate the potential regulatory role of LTF in milk protein yield, the effect of adding LTF to bovine mammosphere cultures on casein mRNA expression was measured. Exogenously added LTF decreased casein mRNA expression in mammospheres. β-Casein mRNA expression was reduced by 80% in the presence of 1 mg/ml LTF (P<0·001; Table 2) and by 46% in the presence of 0·1 mg/ml LTF compared with untreated mammospheres. κ-Casein mRNA expression was not significantly altered although a downward trend was observed (Table 2). We hypothesized that IL-1β was involved in the reduction of casein mRNA expression so the gene expression level of IL-1β was also measured. IL-1β gene expression in mammospheres was not altered by the presence of 0·1 mg/ml LTF but was increased 6-fold by treatment with 1 mg/ml LTF (P<0·001; Table 2).
Table 2. Effect of LTF on β-casein, κ-casein and IL-1β expression (n⩾6 for each treatment). Values are predicted mean±se
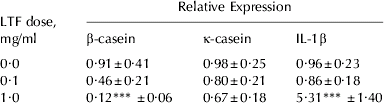
*** Values significantly different from the control (0 mg/ml) dose; P<0·001
To exclude the possibility that the effect of the exogenously added LTF was due to a contaminant in the commercial preparation, or to a non-specific effect, an siRNA targeting the LTF gene was transfected into mammospheres to knockdown LTF gene expression. As addition of LTF decreased casein mRNA expression, we expected that a decrease in LTF expression could lead to an increase in casein mRNA expression. The siRNA reduced LTF gene expression by 11% (P<0·05; Fig. 1). This had the expected effect of increasing casein mRNA expression, with β-casein expression increased 2·6-fold (P<0·01; Fig. 1) and κ-casein expression increased 1·6-fold (P<0·01, Fig. 1).

Fig. 1 Effect of LTF siRNA on gene expression in mammospheres. White bars=control siRNA; black bars=LTF siRNA. Values are predicted means±se.
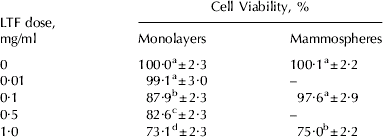
As there are reports of LTF inducing death of several cell types (Roy et al. Reference Roy, Kuwabara, Hara, Watanabe and Tamai2002; Fujita et al. Reference Fujita, Matsuda, Sekine, Iigo and Tsuda2004; Lin et al. Reference Lin, Chiou, Chen and Kuo2005; Sakai et al. Reference Sakai, Banno, Kato, Nozawa and Kawaguchi2005), and MEC death is known to occur during involution when LTF concentrations rise, we investigated the effect of LTF on BMEC viability using an MTT proliferation assay. Viability of BMEC was reduced in a dose-dependent manner by LTF (P<0·001). Growth of cells was reduced by 12% in the presence of 0·1 mg/ml LTF, by 17% in the presence of 0·5 mg/ml LTF and by 26% in the presence of 1 mg/ml LTF (Table 3). The effects of all doses were significantly different from each other, except for the control and 0·01 mg/ml treatment which showed there was no reduction in growth rate in the presence of 0·01 mg/ml LTF.
Table 3. Effect of LTF on BMEC viability (n⩾7 for each treatment). Values are predicted mean±se
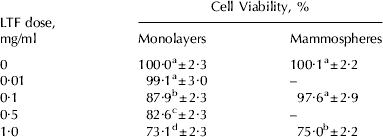
a Different letter superscripts in the same column denote significant differences between doses
Since differentiated BMEC are a better model for the lactating mammary gland, the effect of LTF on mammosphere viability was also measured. A reduction in viability was also observed in mammospheres (Table 3; P<0·001). The viability of mammospheres cultured in the presence of 0·1 mg/ml LTF was not significantly different from the control. However, in the presence of 1 mg/ml LTF mammosphere viability was reduced by 25%.
A caspase assay was employed to determine whether apoptosis was responsible for the decreased cell viability observed in the mammosphere assays. No Caspase 3/7 activity was detected after exposure of mammospheres to LTF for 6 h or 24 h, whereas etoposide, which was used as a positive control, induced caspase activity in both time frames (data not shown).
Discussion
During lactation the concentration of LTF in milk is ~0·1 mg/ml; however, at involution the concentration rises rapidly. At day 2–4 of bovine mammary gland involution the LTF concentration is ~1·6 mg/ml and at 21–28 d the concentration is ~20 mg/ml (Welty et al. Reference Welty, Smith and Schanbacher1975). Exogenous LTF had no significant effect on casein mRNA expression in secretory BMEC at concentrations found in milk during lactation; however, at concentrations equivalent to those found during early involution, β-casein mRNA expression was reduced by 80% and κ-casein mRNA expression was also reduced. β-Casein mRNA abundance is reduced by ~85% 1 week after cessation of milking in dairy cattle (Wilde et al. Reference Wilde, Addey, Li and Fernig1997; Baumrucker & Erondu, Reference Baumrucker and Erondu2000). Our results are consistent with in-vivo observations during involution, where LTF expression increases and casein expression decreases (Wilde et al. Reference Wilde, Addey, Li and Fernig1997; Baumrucker & Erondu, Reference Baumrucker and Erondu2000) but no direct link between the two has previously been reported.
The suppression of endogenous LTF expression by a targeted siRNA had the opposite effect to that of adding exogenous LTF to mammospheres, supporting the hypothesis that LTF can regulate casein expression. It is generally thought that high levels of gene knockdown are required to detect a physiological effect, but a modest reduction in LTF gene expression resulted in increased casein mRNA expression. LTF is highly expressed in mammospheres (unpublished microarray studies) but there is no evidence that highly expressed genes are more difficult to silence (Krueger et al. Reference Krueger, Bergauer, Kaufmann, Wolter, Pilk, Heider-Fabian, Kirch, Artz-Oppitz, Isselhorst and Konrad2007). From these results we suggest that casein mRNA expression may be sensitive to small changes in LTF expression levels.
LTF expression is regulated by different mechanisms from other milk proteins. Casein expression is induced by lactogenic hormones, prolactin, cortisol and insulin via the JAK-STAT pathway whereas lactoferrin expression is independent of lactogenic hormones and has been shown to be induced by changes in cell shape and density (Schanbacher et al. Reference Schanbacher, Goodman and Talhouk1993; Close et al. Reference Close, Howlett, Roskelley, Desprez, Bailey, Rowning, Teng, Stampfer and Yaswen1997). These independent mechanisms of regulation may allow LTF to regulate casein expression. Although the casein genes are found at a common locus, there are differences in their promoter regions (Alexander et al. Reference Alexander, Stewart, Mackinlay, Kapelinkskaya, Tkach and Gorodetsky1988; Rijnkels et al. Reference Rijnkels, Kooiman, Krimpenfort, de Boer and Pieper1995; Rijnkels et al. Reference Rijnkels, Kooiman, de Boer and Pieper1997). In the bovine mammosphere model studied here, κ-casein mRNA was more highly expressed than β-casein, but less susceptible to changes in LTF expression levels whereas β-casein and κ-casein mRNA levels occur in equal proportions in bovine mammary tissue (Bevilacqua et al. Reference Bevilacqua, Helbling, Miranda and Martin2006). In a similar manner to our results, expression from the equine κ-casein gene promoter is higher than from the β-casein promoter (Lenasi et al. Reference Lenasi, Kokalj-Vokac, Narat, Baldi and Dovc2005). Unlike the other three bovine caseins, κ-casein is not calcium dependent but is glycosylated and has a unique function in stabilizing casein micelles (Dagleish, Reference Dagleish, Welch, Burns, Davis, Popay and Prosser1997). The level of κ-casein in milk has major consequences for processing efficiency into dairy products (Dagleish, Reference Dagleish, Welch, Burns, Davis, Popay and Prosser1997). Hence a greater understanding of itsexpression is of commercial importance and warrants further investigation.
LTF may act at the transcriptional level to indirectly regulate casein expression through IL-1β. LTF is a highly basic protein and the N-terminus binds to DNA (He & Furmanski, Reference He and Furmanski1995). Human LTF has been reported to act synergistically with a phorbol ester to stimulate transcription of IL-1β in human leukaemia cells (Son et al. Reference Son, Park, Chung, Chung, Yu, Lee and Kim2002). We found that addition of 1 mg/ml LTF to mammosphere cultures led to a 6-fold increase in IL-1β mRNA expression whereas at lower concentration again there was no effect, suggesting that IL-1β would only be activated during involution. IL-1β treatment has been reported to inhibit expression of a reporter gene from a β-casein promoter in mouse mammary epithelial cells (Baratta et al. Reference Baratta, Motta and Acconero2005). IL-1β activates nuclear factor kappa beta (NF-κB), a transcription factor that is a negative regulator of the JAK-STAT pathway and hence lowers casein expression (Bonizzi et al. Reference Bonizzi, Piette, Merville and Bours2000; Beaton et al. Reference Beaton, Broadhurst, Wilkins and Wheeler2003). Here we report that bovine LTF suppressed casein mRNA expression in secretory BMEC and we propose that this also occurs by a mechanism involving IL-1β. Interestingly, intramammary infusion of IL-1β into cows after the last milking increases LTF levels in the gland and accelerates involution (Wedlock et al. Reference Wedlock, McCarthy, Doolin, Lacy-Hulbert, Woolford and Buddle2004). There have been other reports of IL-1β both inducing, and being induced by, NF-κB (Wu & Kral, Reference Wu and Kral2005). Taken together, these results suggest that IL-1β and LTF may regulate each other.
The current study found that LTF only affected mammosphere viability at concentrations present during early involution. LTF may assist in the removal of some of the epithelial cell population that occurs during involution (Wilde et al. Reference Wilde, Addey, Li and Fernig1997). The viability of BMEC monolayers was also reduced by LTF, with cells in this conformation being more susceptible than in mammospheres. BMEC undergo extensive proliferation during late pregnancy (Cowie et al. Reference Cowie, Forsyth and Hart1980). LTF concentrations rise to 1–2 mg/ml in the final prepartum secretions and may have some effect on reducing proliferation rate prior to initiation of lactation. While LTF, or LTF-derived peptides, have been reported to induce cell death in a number of cell lines, there have not been any reports of LTF affecting growth of primary BMEC. LTF has been reported to reduce proliferation of MAC-T cells, an immortalized BMEC line (Rejman et al. Reference Rejman, Oliver, Muenchen and Turner1992). IL-1β also reduced proliferation of MAC-T cells (Rejman et al. Reference Rejman, Turner and Oliver1993), suggesting that IL-1β may mediate both the reduced casein expression and cell viability observed in BMEC in the presence of 1 mg/ml LTF. NF-κB, a target of IL-1β, is activated during involution and has been associated with survival of specific epithelial subpopulations of the mammary gland (Green & Streuli, Reference Green and Streuli2004); however, in some studies NF-κB is associated with cell death (Baxter et al. Reference Baxter, Neoh and Tevendale2007). It is possible that other IL-1β targets are involved.
Loss of mammary epithelial cells during involution is primarily due to apoptosis (Green & Streuli, Reference Green and Streuli2004). Regulation of apoptosis during involution occurs via several pathways, many converging on activation of Caspase 3, which is considered to be a marker of involution (Motyl et al. Reference Motyl, Gajkowska, Zarzynska, Gajewska and Lamparska-Przybysz2006; Sutherland et al. Reference Sutherland, Lindeman and Visvader2007). LTF or LTF-derived peptides induce cell death by various mechanisms, including apoptosis by induction of caspase 8 and/or 3 in neuronal cells (Lin et al. Reference Lin, Chiou, Chen and Kuo2005), colon mucosa (Fujita et al. Reference Fujita, Matsuda, Sekine, Iigo and Tsuda2004) and oral cancer cells (Sakai et al. Reference Sakai, Banno, Kato, Nozawa and Kawaguchi2005) and by generation of reactive oxygen species in human leukaemic cells (Yoo et al. Reference Yoo, Watanabe, Koike, Mitobe, Shimazaki, Watanabe and Azuma1997; Roy et al. Reference Roy, Kuwabara, Hara, Watanabe and Tamai2002). Caspase 3 was activated by an LTF peptide after 3-h exposure and ceased at 24 h in an oral squamous cell carcinoma cell line (Sakai et al. Reference Sakai, Banno, Kato, Nozawa and Kawaguchi2005). In the present study, there was no evidence that LTF activated caspases 3 or 7 after 6-h or 20-h exposure. It may be that caspase 3 was active at a different time point or that LTF reduced cell viability by a caspase independent mechanism such as autophagy, which also occurs during involution (Motyl et al. Reference Motyl, Gajkowska, Zarzynska, Gajewska and Lamparska-Przybysz2006).
The antimicrobial properties of LTF make it a protein of interest to the biopharmaceutical industry. Human LTF has been expressed in transgenic dairy cows (van Berkel et al. Reference van Berkel, Welling, Geerts, van Veen, Ravensbergen, Salaheddine, Pauwels, Pieper, Nuijens and Nibbering2002; Hyvonen et al. Reference Hyvonen, Suojala, Haaranen, von Wright and Pyorala2006) and goats (Han et al. Reference Han, Li, Zhang, Xiao, Gao, Wu, Li, Zhao, Jiang and Hu2007) without any reported adverse effects on casein levels or milk yield. However, the concentration of human LTF produced was ~2-fold the concentration found during human lactation and hence may not have been deleterious at those levels (Lonnerdal & Atkinson, Reference Lonnerdal, Atkinson and Jensen1995). Our results suggest that attempts to overexpress bovine LTF in cows may have adverse side effects; however, there may be other factors that modulate LTF activity in vivo.
In addition to the known antimicrobial properties of LTF, believed to confer a protective effect on the udder during involution, it appears that the high concentrations of LTF during involution may assist the involution process by reducing BMEC viability and inhibiting synthesis of caseins by a mechanism involving IL-1β.
We thank Peter Thomson for advice on statistical analyses.