Advanced age and multiple comorbidities are risk factors for severe outcomes from coronavirus disease 2019 (COVID-19) Reference Panagiotou, Kosar and White1–Reference Wang, Zuo and Liu3 ; hence, outbreaks in geriatric facilities could be devastating. With multiple outbreak reports from healthcare settings Reference Lucey, Macori and Mullane4–Reference Chan, Jones and Redmond10 and healthcare-associated COVID-19 infections (HCAI) documented in 5%–59% of hospitalized patients, Reference Carter, Collins and Barlow-Pay2,Reference Wang, Zuo and Liu3,Reference Hall, Clement and Farrow11–Reference Jewkes, Zhang and Nicholl13 the need for effective infection prevention and control (IPC) is evident. Long-lasting, close interactions with infected individuals Reference Lee, Wada, Grabowski, Gurley and Lessler14–Reference Meyerowitz, Richterman, Gandhi and Sax16 and cluster infections are important drivers of the pandemic. Reference Sun, Wang and Gao15,Reference Liu, Gong and Xiao17 Although social and physical distancing are not practicable within hospitals, measures to prevent and contain cluster infection may be of particular importance. Early case detection is key for outbreak prevention, and presymptomatic or asymptomatic transmission must be considered. Reference Sun, Wang and Gao15,Reference He, Lau and Wu18 Healthcare workers (HCWs) are also important IPC targets because they represent the interface between the healthcare environment and the community and may act as reservoirs, vectors, or victims of transmission. Reference Arons, Hatfield and Reddy5,Reference Asad, Johnston and Blyth7,Reference Sikkema, Pas and Nieuwenhuijse19–Reference Ran, Chen, Wang, Wu, Zhang and Tan21
Whole-genome sequencing (WGS) has been used in epidemiological investigations of nosocomial transmission of influenza Reference Sansone, Andersson, Gustavsson, Andersson, Norden and Westin22 and, recently, for severe acute respiratory coronavirus virus 2 (SARS-CoV-2). Reference Lucey, Macori and Mullane4,Reference Klompas, Baker and Rhee9,Reference Chan, Jones and Redmond10,Reference Meredith, Hamilton and Warne12,Reference Paltansing, Sikkema, de Man, Koopmans, Oude Munnink and de Man20,Reference Seemann, Lane and Sherry23 This approach may be challenging for SARS-CoV-2 due to a lower mutation rate than other RNA viruses, Reference Abdelrahman, Li and Wang24 especially if conducted during widespread community transmission. Here, we investigated the usefulness of including WGS and phylogeny for a detailed outbreak analysis of COVID-19 in a geriatric hospital setting.
Methods
Setting
The outbreak occurred during the first wave of the pandemic in a 2,000-bed tertiary-care hospital in western Sweden that serve a population of ˜700,000. The affected 30-bed unit was assigned for orthogeriatric patients without COVID-19 infection and comprised 6 single-bed rooms, 6 two-bed rooms, and 3 four-bed rooms. During the outbreak period, 166 patients received inpatient care at the ward.
Definitions and data collection
The outbreak period was set from the sampling day of the index case until 14 days had passed without any newly discovered cases, considering an incubation period of 2–14 days. A patient or HCW from the ward with a nasopharyngeal sample (NPS) positive for SARS-CoV-2 RNA was considered an outbreak case. Patient data were retrospectively collected from medical records, and information regarding contact tracing and ward occupancy from the IPC team and the hospital administrative unit. No individual data were available for HCWs.
HCAIs were classified according to Meredith et al Reference Meredith, Hamilton and Warne12 as true when confirmed >14 days after admission, days 7–14 after admission (suspected), days 3–6 after admission (indeterminate), and ≤2 days after admission (community associated).
Infection prevention and control measures
In accordance with the Swedish National Health Authority, personal protective equipment (PPE) was recommended only when within 1 m from a suspected or confirmed case of COVID-19 and included a plastic apron and a full-face visor (stretching below the chin) or a surgical mask (IIR) and face shield or googles. A respirator (FFP2-3) was added if aerosol-generating procedures were performed in the room. Gloves and a long-sleeve apron were recommended for those at risk of contact with bodily fluids.
Patients were triaged for COVID-19–associated symptoms upon arrival at the emergency department and were considered suspected cases if they presented at least 2 of the following symptoms: cough, sore throat, fever, and shortness of breath (or upon judgment by the treating physician). Suspected cases were isolated at a quarantine ward until the diagnosis was confirmed or averted. Confirmed cases were transferred to assigned COVID-19 wards. Visitor restrictions were enforced throughout the hospital and admitted patients were restricted to their rooms.
Contact tracing was performed around all confirmed cases. Patients sharing a room with a case during their infectious phase were considered close contacts and were isolated and monitored for symptoms for 14 days. HCWs were considered close contacts when exposed to an infectious case without PPE and continued to work if asymptomatic during the incubation period. HCWs with symptoms of COVID-19 self-quarantined at home for at least 7 days unless they tested negative. Testing resources were limited and prioritized for suspected patient cases requiring in-hospital care.
Laboratory methods and details on bioinformatics and phylogenetic analysis are provided in the Supplementary Material (online).
Ethical statement
Approval for this study was granted by the Swedish Ethical Review Authority (protocol no. 2020-03276).
Results
Outbreak description
In total, 32 patients and 15 HCWs were included in this study (Fig. 1). The index case (patient 1) developed COVID-19 symptoms and was sampled 8 days after admission (outbreak day 0). The first secondary case (patient 2) tested positive on outbreak day 5, 11 days after admission. Close contact between them was excluded. Several staff members reported illness during this period, and HCW–patient transmission was suspected. An outbreak investigation initiated by staff management and the IPC team identified several possible factors contributing to transmission: difficulties in symptom interpretation, crowding in workspaces, PPE shortage and insufficient IPC training. Testing of HCWs was available from outbreak day 14. IPC training and PPE resource allocation was initiated in week 3. By week 5, HCWs used full-face visors in all patient care activities and social gatherings were limited during breaks. The ward closed for new admissions on day 30, and screening of the remaining patients (n = 17) identified 8 cases, of whom 5 were asymptomatic. Repeated screening (n = 9) on day 32 identified 1 additional case.
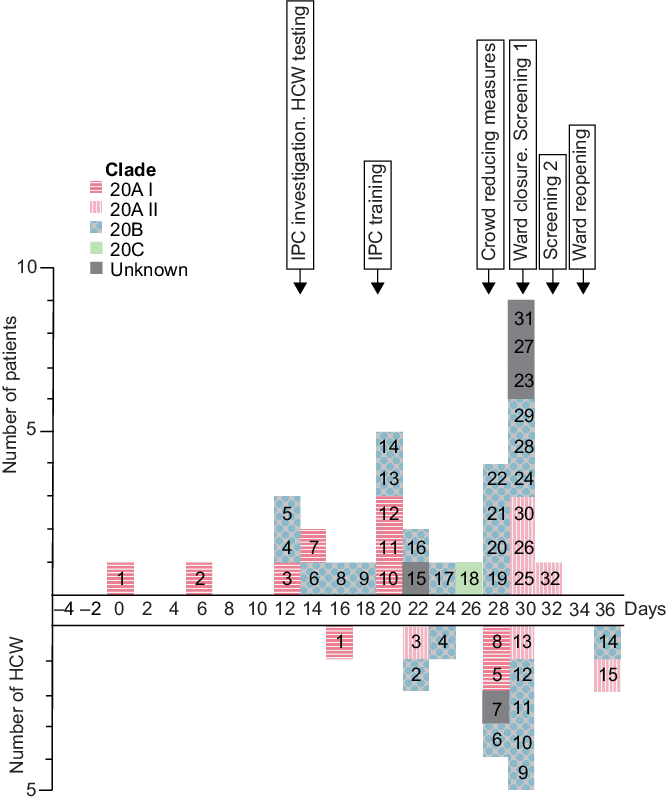
Fig. 1. Epidemic curve of COVID-19 cases in a hospital ward outbreak. Day of positive SARS-CoV-2 nasopharyngeal samples from 32 patients (above x-axis) and 15 healthcare workers (HCW; below x-axis) are displayed according to timeline throughout the outbreak period. Individual case numbers are shown for patients and HCWs separately. Colors indicate viral clades and arrows the implementation of outbreak control measures.
Case characteristics
Patient characteristics are shown in Supplementary Table 1 (online). The median age was 84 years and 17 (53%) of 32 patients were male. The overall 30-day mortality was 44% (death occurring in median 8 days after sampling). No additional mortality was observed within 90 days, and no cases were lost to follow-up. Viral load was significantly higher among the deceased. Also, 5 asymptomatic patients were identified, of whom 4 developed symptoms within the following 4 days (Supplementary Fig. 1 online). One patient remained asymptomatic within 5 days of follow-up and had a possibly false-positive test due to a very low viral load (Ct value, 39).
Outbreak analysis
WGS was successful in 28 (88%) of 32 patient samples and 14 (93%) of 15 HCW samples. Patient 31 was excluded from phylogenetic analysis due to low viral load. Patients 15 and 23 and HCW 7 were excluded due to lack of material and patient 27 was excluded due to low genomic coverage. Viral strains from 3 genetically distinct clades (20A–C) were found among both outbreak and community sequences, although outbreak sequences showed greater internal genetic similarity (Supplementary Fig. 2a online). Two outliers among patient sequences were found in clade 20B and clade 20C. Based on the phylogenetic clustering, clade 20A was separated into subclades I and II, which also appeared during different phases of the outbreak (Fig. 1). Clade 20AI-II and 20B were dispersed among both patient and HCW sequences (Supplemental Fig. 2b online). These phylogenetic analyses suggest 4 separate introductions, of which 3 (20AI-II and 20B) resulted in secondary transmission.
Contact tracing revealed an epidemiological link (close contact) between 22 of 32 patient cases (Fig. 2). The phylogenetic analysis did not support transmission in 5 of these cases due to clade differences (patients 4, 18, and 21) or sequence differences (patients 9 and 13) (Supplementary Fig. 2b online and Fig. 2). In contrast, for 6 of the 17 remaining cases (patients 11, 17, 20, 25, 27, and 28), a patient–patient transmission event between close contacts was supported by a positive nasopharyngeal sample (NPS) or symptom onset occurring within 2–14 days of each other (Fig. 2).

Fig. 2. Patient–patient transmission of SARS-CoV-2 in a hospital ward outbreak. The panel display 22 patient cases defined as close contacts due to sharing a room with another case. Individual case numbers and letters indicating shared room are seen on the y-axis, and timeline of the outbreak period (days) seen on the x-axis. Bars show day of admission until discharge from the affected ward. Colors represent duration of shared room with another case and viral clade. Dots indicate time point for sampling and stars represent symptom onset. A patient–patient transmission event was probable if: sequence differences did not exclude a genetic relationship, and day of sampling or symptom onset for 2 close contacts occurred within 2–14 days of each other.
Based on the case definitions, Reference Meredith, Hamilton and Warne12 4 of 32 patients had true HCAIs. The phylogenetic analysis revealed a close relationship to other outbreak sequences for 27 of 28 patients (Supplemental Fig. 2a online). The single finding of clade 20C (patient 18, NPS 4 days after admission) supported community transmission. Patient 13 (NPS 6 days after admission, day 20) and HCW 9 (day 30) were genetically similar although more closely related to community sequences than other outbreak sequences. However, the direction of transmission between them is unclear due to lack of clinical data for HCWs. Subclade 20AII was first identified in HCW 3, and patient 1 may have introduced 20AI (Fig. 1). In contrast, clade 20B was unlikely introduced by either patient 4 or 5 because they had no close contact and were sampled the same day (Supplementary Fig. 1 online). Altogether, strong support of true HCAI was found for 25 (78%) of 32 patients.
Discussion
We present an integrated epidemiological and genetic analysis of a COVID-19 hospital outbreak resulting in the discovery of not 1 but multiple separate viral clusters. We also found that instead of 4 patients, 25 patients likely had true HCAI, highlighting both the uncertainties of nosocomial COVID-19 case definitions and the power of WGS.
The mortality in our study was in line with the national mortality rate for COVID-19 in the group aged 80–89 years during this period, 25 although it was higher than previous reports of ˜30% among elderly hospitalized COVID-19 patients. Reference Carter, Collins and Barlow-Pay2,Reference Lucey, Macori and Mullane4,Reference Hagg, Jylhava and Wang26,Reference Rickman, Rampling and Shaw27 COVID-19 has been reported as a significant risk factor of death within 30 days for patients with hip fractures, Reference Hall, Clement and Farrow11,Reference Hall, Clement and Farrow28,Reference Egol, Konda and Bird29 which may have influenced our results. The short median survival time (8 days) corresponds with previous findings, Reference Carter, Collins and Barlow-Pay2,Reference Rickman, Rampling and Shaw27 supporting the finding that death was caused by acute infection. Significantly higher viral load was seen among the deceased patients, previously reported in geriatric patients. Reference De Smet, Mellaerts and Vandewinckele30 The severe outcome stresses the importance to protect this patient group from COVID-19.
Asymptomatic transmission has been suggested a key factor in hospital outbreaks. Reference Asad, Johnston and Blyth7,Reference Lesho, Walsh and Gutowski8 Interpreting symptoms in elderly patients with COVID-19 may be difficult Reference Roxby, Greninger and Hatfield6,Reference Graham, Junghans and Downes31 and asymptomatic cases are more common among patients aged >80 years. Reference Singanayagam, Patel and Charlett32 The asymptomatic cases identified in our study support the insufficiency of a strictly symptom-based testing strategy. We recommend screening combined with serial testing, especially during significant community transmission or when hospital outbreaks are suspected. Reference Klompas, Baker and Rhee9,Reference Taylor, Carter and Lehnertz33
We identified only 6 events of probable patient–patient transmission. This finding suggests that transmission from HCWs might have occurred, which has been reported previously. Reference Lucey, Macori and Mullane4,Reference Asad, Johnston and Blyth7,Reference Klompas, Baker and Rhee9,Reference Meredith, Hamilton and Warne12,Reference Paltansing, Sikkema, de Man, Koopmans, Oude Munnink and de Man20,Reference Taylor, Carter and Lehnertz33 However, the direction is often unclear, and exposure from HCWs seldom appears to result in infection. Reference Baker, Fiumara and Rhee34 The close genetic relationship between sequences from HCWs and patients support the hypothesis that transmission occurred between them; hence, breeches in IPC measures were identified. The symptom-based recommendations overlooked silent transmission from pre- or asymptomatic individuals, and PPE recommendations may have been insufficient. Adherence to IPC measures was unknown. Therefore, the cause of infection could have been inadequate IPC or PPE recommendations, insufficient adherence, or all of these.
The main limitation of this study was the restricted testing policy, which resulted in unrecognized cases among patients and HCWs that might have influenced the course of the outbreak. Establishing the direction of transmission was complicated due to lack of clinical information for HCWs. Patients and HCWs had multiple contacts in other units and transmission outside of the ward may have been overlooked.
The details provided by WGS and phylogeny emphasize the limitations of classic outbreak investigations and the potential of molecular characterization. We recommend increasing the use of WGS for outbreak investigations to clarify transmission links, to identify nosocomial infections, and to evaluate IPC measures.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2021.374
Acknowledgments
We thank the staff at the Department of Clinical Microbiology and Geriatric Medicine, Sahlgrenska University Hospital for their aid in data collection and preparation and all laboratories that originated and submitted sequences to the GISAID database.
Financial support
This work was supported by the Region Vastra Gotaland research fund (grant nos. ALFGBG-67131114 and -719911) and AFA Forsakring (grant no. 200245).
Conflicts of interest
All authors report no conflicts of interest relevant to this article.





