Published online by Cambridge University Press: 14 September 2004
Major sperm protein (msp) genes were isolated from complementary (cDNA) and genomic DNA libraries prepared from the parasitic nematode, Oesophagostomum dentatum, characterized at the nucleotide and amino acid (aa) levels, and their expression was investigated. Three different msp cDNA and 2 genomic sequences were determinedThe nucleotide sequences reported in this article have been deposited in the EMBL, GenBank and DDJB databases under the Accession numbers AJ627869–AJ627873., each with an open reading frame (ORF) of 381 nucleotides. Nucleotide variation was detected at 30 positions in the ORF among all 5 sequences. Conceptual translation of the full-length msp sequences inferred 4 different MSPs each of 126 aa. These predicted MSPs differed at aa positions 15 (serine<->threonine), 101 (alanine<->glycine), 103 (glutamine<->leucine) and 126 (proline<->leucine). Southern blot analysis of O. dentatum genomic DNA, digested separately with various restriction endonucleases, displayed multiple (n=7–13) bands for each enzyme, providing support for a multigene family. Also, at the genomic level, sequence tracts consistent with a ‘substitute’ TATA box sequence motif were identified within a region (−1 to −123 nt) preceding the 2 msp genes. In contrast to other species of nematode investigated to date, no GATA transcription factor binding motif was detected immediately upstream of the msp coding region. Real-time PCR analysis demonstrated that msp mRNA was expressed exclusively in the males of both fourth-stage larvae (L4s) and adults of O. dentatum (raised in pigs after intragastric inoculation). The magnitude of expression in male O. dentatum raised in pigs in the presence of female worms was the same as in males in the absence of females. Comparative analyses showed aa sequence conservation among MSPs from various nematodes, suggesting similar functional roles for these proteins.
Investigation of the reproductive and developmental processes of socio-economically important parasitic nematodes is of fundamental scientific interest and could, in the longer term, have important implications for developing novel methods for parasite control via the disruption or interruption of such processes (Boag, Newton & Gasser, 2001; Boag et al. 2003a). Central to studying reproductive molecular biology is the identification and characterization of genes with sex-specific expression profiles. Some of the the first sex-specifically expressed proteins of nematodes reported were those of the major sperm protein (MSP) family, originally isolated from the free-living nematode C. elegans and the parasitic nematode Ascaris sp. (see Klass & Hirsh, 1981; Ward & Klass, 1982; Bennett & Ward, 1986). The MSPs described thus far are small (~15 kDa), nematode-specific, cytoskeletal proteins which account for ~10–15% of the total cellular protein in spermatozoa (e.g., Klass & Hirsch, 1981; Nelson & Ward, 1981). MSPs have been shown to be involved in sperm motility (e.g., King et al. 1994a,b; Roberts & Stewart, 2000; Italiano et al. 2001; Buttery et al. 2003) and in oocyte maturation and sheath cell contraction (Miller et al. 2001, 2003; Kuwabara, 2003). The msp genes characterized for animal-parasitic nematodes include those of Ascaris sp., the bovine lungworm Dictyocaulus viviparus and the filarioids Mansonella ozzardi and Onchocerca volvulus (see Bennett & Ward, 1986; Scott et al. 1989b; Schnieder, 1993; Hojas & Post, 2000). For instance, 2 genes (Ovgs-1 and Ovgs-2) have been isolated from O. volvulus, and both have ~80% identity to Ascaris msp cDNA and ~79% to the msp-3 cDNA sequence of C. elegans. However, there is only limited DNA sequence similarity between the putative promoters of the O. volvulus msp genes and those of C. elegans, although 2 GATA transcription factor binding motifs have been identified for O. volvulus, suggesting that they are involved in regulating msp gene expression in this nematode. Surprisingly, O. volvulus and Globodera rostochiensis are currently the only parasitic nematodes for which the genomic organization of msp has been established (Scott et al. 1989b; Novitski et al. 1993). Also, in spite of the functional significance of MSPs (see Theriot, 1996; Miller et al. 2001, 2003; Bottino et al. 2002; Buttery et al. 2003) and the availability in public gene databases of expressed sequence tags (ESTs) representing msp genes, there is a paucity of information on their expression and genomic organization for the majority of parasitic nematodes.
Recent studies have emphasized that Oesophagostomum dentatum (Strongylida: subfamily Oesophagostominae), a nodule worm of the large intestine of pigs, provides a unique model system for investigating reproductive processes in parasitic nematodes (see Boag et al. 2003b). The parasite has a direct life-cycle (with a pre-patent period of ~18–24 days; Talvik et al. 1997), produces large numbers of progeny and can be maintained readily as a laboratory line. Also, ‘uni-sex’ or ‘mixed-sex’ infection of the parasite can be established readily by rectal transplantation to naïve pigs, thus allowing studies of sexual maturation and mating behaviour in vivo as well as molecular investigations of the parasite produced under well-controlled experimental conditions. As part of a study investigating reproductive processes, Boag et al. (2000) isolated 10 male-specific and 2 female-specific ESTs from adult O. dentatum by differential display analysis, of which 2 molecules (inferred to represent a trypsin inhibitor-like serine protease inhibitor and a serine/threonine phosphatase) have been characterized in detail (Boag et al. 2002, 2003b). The present study extends this work to elucidate aspects of the molecular biology of MSPs of O. dentatum. The specific aims were to characterize msp gene(s) from this nematode, to determine whether msp is a single gene or whether it belongs to a gene family, to study the genomic structure and organization of the msp gene(s), to characterize msp gene expression in adults of O. dentatum raised in pigs under different experimental conditions and to discuss the findings in relation to information available for other nematodes.
Infective 3rd-stage larvae (L3) of O. dentatum were produced by incubating faeces from monospecifically infected pigs for 14 days at 20–22 °C (cf. Talvik et al. 1997), harvested using the Baermann funnel technique (Ash & Orihel, 1987) and then stored at 10 °C for a minumum of 21 days. Helminth-free pigs (Danish Landrace×Yorkshire×Duroc; both sexes; 20 kg live weight) were inoculated via intragastric intubation with 6×103 L3s. Stages of O. dentatum were collected from the large intestinal content at day 15 (L4s), 22, 51 or 60 (adults) after inoculation, harvested using an agar gel method (Slotved et al. 1996), the sexes separated using a dissection microscope and then snap-frozen in liquid nitrogen. Also, L4s of O. dentatum were collected in the same manner from the large intestinal content at day 15 after inoculation, and the sexes separated. Male or female L4s were immediately transplanted rectally to helminth-free recipient pigs as described previously (Christensen et al. 1996a,b). Each uni-sex O. dentatum infection established in this way was maintained for 45 days prior to the euthanasia of the pigs, and the collection and freezing of adult nematodes in liquid nitrogen. The specific identity of adult nematodes was verified according to the descriptions by Haupt (1966).
Total RNA was extracted from ‘pooled’ worms representing different stages and sexes using Tripure™ (Roche Molecular Biochemicals), and mRNA was purified using Poly(A)Pure (Ambion). Genomic DNA from O. dentatum or pig musculature (host control) was extracted using a sodium dodecyl-sulphate (SDS)-proteinase K method (Gasser et al. 1993) and then column-purified (Wizard® DNA Clean-Up, Promega).
A PCR product (~200 bp) representing msp was amplified from an O. dentatum genomic λFIX® II library (cf.Boag et al. 2003b) using degenerate oligonucleotide primers MSPF1 (5′-ATYGGWTGGGCVATHAAAAC-3′) and MSPR1 (5′-CGGAAYTGYTTGGCMGCACC-3′) under the following cycling conditions: 1 cycle of 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 sec, 50 °C for 30 sec, 72 °C for 30 sec. The amplicon was then column-purified (Wizard® PCR Prep, Promega) and labelled by random priming (Prime-a-gene, Promega) with α32P dCTP (GeneWorks). This probe was used to screen a λZapII cDNA-library (containing ~5×105 independent clones; constructed from mRNA from 22 day-old male O. dentatum; see Boag et al. 2002) and a genomic DNA library (Boag et al. 2003b). Plaque lifts on to positively-charged nylon membranes (Roche Molecular Biochemicals) were performed in duplicate and the membranes were pre-hybridized at 60 °C for 1 h in 0·25 M NaHPO4, pH 7·5, 1 mM EDTA, 7% (w/v) sodium dodecyl-sulphate (SDS) with 100 μg/ml sonicated herring sperm DNA (Roche Molecular Biochemicals). The denatured probe was added to the pre-hybridization solution and incubated overnight at 60 °C. The filters were washed 3 times in 2×SSC (0·3 M NaCl, 30 mM Na citrate, pH 7·0), 0·1% SDS at 60 °C for 20 min and then subjected to autoradiography for 12 h. Positively hybridizing plaques were re-screened to ensure that they were clonal, plaques were picked and phage was eluted into 500 μl of SM buffer (Sambrook, Fritsch & Maniatis, 1989).
Complementary DNA was amplified by PCR from individual clones (5 μl of a 1[ratio ]1000 dilution of phage suspension) using the vector-specific primers T7 (5′-TAATACGACTCACTATAGGG-3′) and T3 (5′-AATTAACCCTCACTAAAGGG-3′), employing the following cycling conditions: 1 cycle of 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 sec, 60 °C for 30 sec, 72 °C for 1 min, followed by a final cycle of 72 °C for 5 min. Following amplification, PCR primers and dNTPs were removed using shrimp alkaline phosphatase and exonuclease I, and the purified amplicon was subjected to sequencing, employing BigDye terminator II chemistry (PE Applied Biosystems) and an automated sequencer (ABI 377; via the Griffith University DNA Sequencing Facility, Queensland, Australia).
For each genomic clone isolated, 15 μl of phage suspension were added to 500 μl of Escherichia coli XL1-blue culture and then to 100 ml of Luria Bertani (LB) medium and incubated at 37 °C (shaking at 200 rpm) until cell lysis (~6 h). Lambda DNA was purified using the Wizard™ λPrep DNA purification system (Promega). Regions of the genomic clones were amplified by PCR using the Expand 20kbPLUS System (Roche Molecular Biochemicals), employing the vector-specific primer LPCRT7 (5′ GCTCTAATACGACTCACTATAGGGCGTC 3′) and an msp-specific primer LPMSP2/R (5′-TCTGCGGACCATACCGTCACCTTGGAAC-3′), or rtMSP1/F (5′-AAGATCGTCTTCAACGCTCC-3′) and LPMSP3/R (5′-TCAAGGATTGTATTCGATGGG-3′). Cycling conditions for the PCR were 1 cycle at 92 °C for 2 min, followed by 10 cycles at 92 °C for 10 sec, 60 °C for 30 sec, 68 °C for 10 min, then 20 cycles at 92 °C for 10 sec, 60 °C for 30 sec, 68 °C for 10 min (plus an additional 10 sec extension for each cycle), and 1 cycle of 68 °C for 7 min. Amplicons were column-purified (Wizard® PCR Preps, Promega), ligated into the pGEM®T-Easy vector (Promega) and then sequenced (both strands). Also, purified λDNA was digested with EcoRI and XbaI (Promega), excised from agarose gels (following Southern blot analysis), column-purified (Qiagen), subcloned, and then sequenced.
Nucleic acid and protein non-redundant databases were interrogated using the Basic Local Alignment Search Tool (BLAST), available at the National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/blast/) or Parasite Genome database from the European Bioinformatics Institute (EMBL). Conceptual translations (in 6 different reading frames) of msp cDNA were conducted at the BCM search launcher (Human Genome Centre, Baylor College of Medicine) (http://searchlauncher.bcm.tmc.edu/seq-util/Options/sixframe.html). Nucleic acid and protein sequences were aligned using the program Clustal W (Thompson, Higgins & Gibson, 1994), available at the Network Protein Sequence Analysis server (http://npsapbil.ibcp.fr/cgibin/npsa_automat.pl?page=npsa_server.html), and alignments corrected manually.
Genomic DNA (~20 μg) isolated from O. dentatum adults was digested to completion with endonuclease ClaI, EcoRI, HindIII or PstI (Promega). Digested DNA samples, and undigested O. dentatum and porcine genomic DNA (~20 μg each) were resolved in a 0·8% (w/v) agarose gel using 40 mM Tris-Acetate, 2 mM Na2EDTA, pH 8·5 (TAE) buffer and then transferred overnight to a positively-charged nylon membrane (Roche Molecular Biochemicals) under alkaline conditions (Sambrook et al. 1989). The membrane was pre-hybridized at 60 °C for 1 h, prior to hybridization at 65 °C for 12 h with an α-32P-labelled cDNA (from clone OdmspC06 containing a full-length msp gene). The membrane was then washed 3 times with 0.1×SSC, 0.1% SDS (w/v) at 60 °C for 20 min and subjected to autoradiography for 1–5 days at −70 °C.
The MSP amino acid (aa) sequences inferred for O. dentatum were compared with those published for A. suum (see Accession number P27439; Bennett & Ward, 1986; Smith & Ward, 1998), C. elegans (msp-56) (Accession number NM_069361; Klass, Ammons & Ward, 1988), Onchocerca volvulus (Accession number P13262; Scott et al. 1989b), Dictylocaulus viviparus (Accession number S64873; Schnieder, 1993). The latter sequence was amended at nucleotide position 204 to correct the ORF (see Setterquist & Fox, 1995). Also, selected MSP sequences from other species of nematodes were obtained from the parasite genome database (http://www.ebi.ac.uk/blast2/parasites.html) by interrogating with the sequence of cDNA OdmspC06 (see Results section).
Double-stranded cDNA was synthesized from total RNA from each stage and sex of O. dentatum using Superscript II reverse transcriptase (Invitrogen). Briefly, 5 μg of total RNA were added to 14 μl of H2O and 1 μl of oligo d(T)n=12–18 primer (0.5 μg/μl), heated to 70 °C for 10 min and chilled on ice. First and second strand cDNAs were synthesized via the addition of 4 μl of first strand cDNA buffer (250 mM Tris–HCl, pH 8·3, 375 mM KCl and 15 mM MgCl2), 2 μl of 0·1 M dithiothreitol, and 1 μl of 10 mM of each dNTP, followed by an incubation at 25 °C for 10 min, 42 °C for 50 min and 70 °C for 15 min. Double-stranded cDNA was purified using a QIAquick gel extraction column (Qiagen) prior to analysis. Primers rtMSP1/F and rtMSP2/R (5′-ACCATACCGTCACCTTGGAA-3′, spanning nucleotide positions 46 to 344) were employed to PCR-amplify a 298 bp region of the O. dentatum msp cDNA (Fig. 1). A portion (187 bp) of the large subunit (28S) of ribosomal RNA was used as a control for the different developmental stages and genders, employing the primers rt28S1/F (5′-GCATAAGCTCTCGCGTTACC-3′) and rt28S2/R (5′-GAGAGGGACAGCAGGTTCAC-3′) in the PCR. The PCR analyses were conducted using a LightCycler™ (Roche Molecular Biochemicals). Each reaction contained 0·5 μM of each primer, 2 μl of LightCycler™FastStart DNA Master SYBR Green I, 5 mM (for msp) or 3 mM (for ribosomal RNA) of MgCl2, 2 μl (~150 ng/μl) of O. dentatum cDNA, and H2O to a final volume of 20 μl. Complementary DNA from male O. dentatum from a uni-sex infection was used as the ‘calibrator’ for quantification. Negative (no-DNA) and positive (plasmid containing the DNA target) controls were also included. Cycling parameters were: 1 cycle of 95 °C for 10 min, 40 cycles of 95 °C for 1 sec, 60 °C for 5 sec, 72 °C for 13 sec, followed by 1 cycle of each 95 °C for 1 sec, 65 °C for 15 sec, increasing by increments of 0·1 °C per sec until reaching 95 °C, followed by a cooling cycle of 40 °C for 30 sec. Numerical values of msp gene expression were calculated using RelQuant software (Roche, version 1.01). All analyses were performed in triplicate, and relative expression levels were determined statistically by ANOVA.

Fig. 1. Alignment of msp nucleotide sequences isolated from complementary (designated with C) and genomic (designated with G) DNA libraries. A stop codon is denoted by a large dot. Respective amino acid sequences are aligned below the nucleotide sequences. The alignment positions are above each respective block of sequences.

Fourteen positively hybridizing clones were isolated from the male-specific O. dentatum cDNA library. Amplicons produced from these clones contained inserts of ~650 bp. Sequence analysis showed that 7 of the clones contained putative full-length msp sequences, and conceptual translation of all sequences revealed an ORF of 381 nucleotides. Of the 14 ESTs, 12 had identical sequences (OdmspC06 was a full-length representative of these sequences), whereas clones OdmspC01 and OdmspC11 varied in nucleotide sequence from all other clones (see Fig. 1). Nucleotide variation was detected at 22 positions among the 3 representative Odmsp cDNA sequences (OdmspC01, OdmspC06 and OdmspC11), and these sequences had a similarity of >95%. The sequences of variants OdmspC01, OdmspC06 and OdmspC11 were aligned with msp sequences of 2 clones isolated from the genomic DNA library (see Fig. 1). Of the 30 nucleotide alterations detected in the ORF among all 5 sequences, most were concordant (i.e., at least 1 base was common) between cDNA and genomic DNA, indicating that they did not represent artefacts generated by the reverse transcriptase used during cDNA library construction. This conclusion was also supported by a comparison of other msp ESTs (sequences available from primary author) with the 2 genomic sequences.
The nucleotide sequence of each cDNA OdmspC01, OdmspC06 and OdmspC11 and of each genomic clone OdmspG23 and OdmspG33 was conceptionally translated into a polypeptide of 126 aa. Alignment of the 5 predicted peptides revealed that the variation for all 5 msp nucleotide sequences resulted in 4 aa residue changes at positions 15, 101, 103 and 126 (Fig. 1). For example, the change from a T to an A at position 43 in the nucleotide sequence of OdmspC01 resulted in a change from a serine to a threonine at aa position 15. Similarly, the change from a C to a T at position 377 resulted in the change from a proline to a leucine at aa residue 126. Nucleotide changes other than those at positions 43, 302, 308 and 377 did not result in any alterations in aa sequence (Fig. 1).
A Southern blot of O. dentatum genomic DNA digested with restriction endonuclease ClaI, EcoRI, HindIII or Pst I was probed with radio-isotope labelled cDNA OdmspC06. The resultant autoradiograph displayed multiple bands for each of the digests (~7 bands for ClaI, 13 for EcoRI, 8 for HindIII, and 10 for PstI; Fig. 2), providing evidence for a multigene family. There was no hybridization of the probe to porcine genomic DNA.

Fig. 2. Southern blot analysis of Oesophagostomum dentatum DNA digested with restriction endonuclease ClaI, EcoRI, HindIII or PstI. Undigested DNA samples from O. dentatum (U) and pig musculature (Upm) were included as controls.
In order to characterize any putative promoter elements, msp genes OdmspG23 and OdmspG33, isolated from the O. dentatum genomic library, were sequenced (Fig. 1). To do this, a 1 kb region from clone OdmspG23 was amplified by PCR using the primer set LPCRT7-LPMSP2/R or rtMSP1/F-LPMSP3/R.
Using the same primers, a PCR product was amplified from OdmspG33 λDNA (previously subcloned). Sequence analysis revealed no nucleotide variation between clones OdmspG23 and OdmspG33 in the 123 bp region preceding the initiation codon. The sequence tracts CTAATCT and TCAGACC (commencing at positions −122 and −104, respectively, from the putative initiation codon; Fig. 1), were consistent with the ‘substitute’ TATA box sequence PyPyANT/APyPy, in which Py (pyrimidine) is either a C or a T (see Smale, 1997). Neither of the 2 genomic sequences contained a GATA transcription factor binding motif characteristic for the msp genes of other species of nematodes, including O. volvulus (see Scott et al. 1989b) and C. elegans (see Klass et al. 1988), and other nematode genes which are gender- and spatially-regulated (see MacMorris et al. 1992, 1994; Kuwabara, 1996; Boag et al. 2003b).
Transcripts of msp were detected exclusively in males of the late L4 and adult stages of O. dentatum (Fig. 3), suggesting that msp is not expressed in male zygotes or eggs or in sperm in the uterus of the female nematode. Real-time PCR analyses were conducted to establish whether gene expression in O. dentatum was altered when either male or female L4s were transplanted to pigs and maintained as ‘single-sex’ infections for 45 days. No statistical difference in msp expression was detectable between the male nematodes (collected 45 days after transplantation) and males from a normal, ‘mixed-sex’ infection (collected at day 51 after intragastric inoculation) (Fig. 3). Northern blot analysis of O. dentatum total RNA using radio-isotope labelled OdmspC06 as a probe demonstrated that msp was expressed as a single, male specific transcript of ~600 bp (not shown).

Fig. 3. Expression of msp mRNA at different stages of development of Oesophagostomum dentatum measured by real-time PCR. Expression was detectable exclusively in males of the late L4 (LL4) and adult stages (A) of the parasite, but not in the ensheathed L3 (EnL3), artificially exsheathed L3 (ExL3) or early L4 stage (ExL4). In adult nematodes, there was no statistical difference in expression values between males (Am+) raised together with females (Af+) for 51 days (after intragastric inoculation of pigs with L3s) and males (Am−) maintained without females for 45 days (following transplantation of male-only L4s to a recipient pig). No expression was detected in females (Af−) maintained without males for 45 days (following transplantation of female-only L4s to a recipient pig). The calibrator represents a reference control.
Database comparisons revealed that the peptide predicted from cDNA OdmspC06 had significant sequence identity with other nematode MSPs. Greatest identity (89% identity and 92% similarity over 127 aa) was recorded to the protein sequences MSP1 (P=4×10−61, Accession number P13263) and MSP2 (P=1×10−60, Accession number A45528) of the filarioid nematode O. volvulus. Comparison with MSP of A. suum (P=5×10−60, Accession number P27440; Smith & Ward, 1998) revealed an 88% identity and 93% similarity over 127 alignment positions, and similar levels of identity/similarity were determined against the sequence of MSP-113 from C. elegans (Accession number NP_500773.1). An alignment of selected MSP sequences representing C. elegans and a number of parasitic nematodes of human and/or veterinary importance (Fig. 4) revealed that they were either 126 or 127 aa in length. Interestingly, the MSPs of parasites from clade V (cf. Blaxter et al. 1998) were all 126 aa in length, whilst the MSPs of nematodes from other clades were 127 residues in length. Comparison of the N-terminal domain (aa positions 1–106) (Miller et al. 2001) among all 13 nematode MSPs revealed ~67% identity. In the C-terminal domain (aa positions 106–126), there was ~95% identity.

Fig. 4. Alignment of the MSP amino acid sequences of Oesophagostomum dentatum (Accession numbers AJ627869–AJ627873) with selected sequences for Caenorhabditis elegans (NM_069361), Ascaris sp. (P27439), Onchocerca volvulus (P13262), Dictyocaulus viviparus (S64873), Ancylostoma ceylanicum (CB176395), Haemonchus contortus (CB191457), Teladorsagia circumcincta (CB037657) and Nippostrongylus brasiliensis (BQ529557). Amino acid identity among all species of nematode is indicated by an asterisk.
The cDNA sequences of the O. dentatum msp had a high degree (80–87%) of identity to those of C. elegans, A. suum, D. viviparus, M. ozzardi and O. volvulus (see Bennett & Ward, 1986; Klass et al. 1988; Scott et al. 1989b; Schnieder, 1993; Hojas & Post, 2000). Intraspecific sequence comparisons revealed that the majority of nucleotide variability detectable was at the third codon position (i.e. was silent), not usually associated with aa alterations. At the genomic level, no introns were detected in the msp sequences (clones OdmspG23 and OdmspG33) of O. dentatum. This finding was in accordance with genomic msp sequences from C. elegans (see Klass et al. 1988) but in contrast to the Ovgs-1 and Ovgs-2 sequences from O. volvulus which both have an intron of 153 bp in length (Scott et al. 1989b). Conceptual translation of msp cDNA sequences obtained from O. dentatum and comparison among the predicted aa sequences revealed 4 distinct ‘isoforms’ with multiple aa differences. The significance of the different MSP isoforms is presently unclear, but, based on evidence for A. suum (see King et al. 1992; Bullock et al. 1996), it is possible that each has a distinct structure and/or biological role. It may be possible to dissect their functional roles using the cell-free in vitro systems developed recently (Buttery et al. 2003; Miao et al. 2003).
The different isoforms of MSP and the Southern blot results for O. dentatum provided evidence that msp constitutes a multigene family, which is consistent with findings for most nematodes studied to date (Scott et al. 1989a). For example, similar hybridization patterns for msp have been displayed on Southern blots for other bursate nematodes, Nippostrongylus brasiliensis and Ancylostoma caninum, the insect parasite, Steinernema carpocapsae, and the plant root-knot parasite, Meloidogyne incognita (see Scott et al. 1989a). However, in free-living nematodes, the number of different msp variants seems to be markedly greater. For example, in C. elegans a major proportion of the large family of msp genes located to chromosomes II and IV appears to be transcribed (Ward et al. 1988). This contrasts the situation for Ascaris in which only a single gene is transcribed (Bennett & Ward, 1986; Scott et al. 1989a). Most parasitic nematodes studied to date have 5–13 msp genes, while free-living nematodes usually have a greater number, ranging from 15–50 (Scott et al. 1989a). It is suggested that the high concentration of MSP in C. elegans sperm is due to simultaneous expression of the numerous msp genes, resulting in the availability of a large pool of msp mRNA for translation. In this nematode, the accumulation of sperm-associated proteins occurs within a period of ~90 min during spermatogenesis, as the endoplasmic reticulum, Golgi complex and ribosomes are abandoned when the spermatocyte separates from the residual body (see L'Hernault, 1997; Muhlrad & Ward, 2002). Hence, a gene family may be required for the rapid synthesis and accumulation of MSP, since spermiogenesis (pseudopod extension and initiation of crawling in utero) has been shown to commence without the synthesis of new mRNA or protein (cf.Muhlrad & Ward, 2002). It has been proposed that MSP production is a rate-limiting step in sperm production in C. elegans and, thus, a relatively high number of msp genes is maintained (Scott et al. 1989a). It is possible that parasitic nematodes have a smaller rate of sperm production compared with C. elegans due to longer life-cycles, and that the required msp mRNA concentration can be produced from fewer msp genes. Alternatively, msp genes of parasitic nematodes may be transcribed under the control of a more efficient promoter, or the transcript may have increased stability.
In O. dentatum, the msp gene family was expressed as a male-specific transcript of ~600 bp (not shown), similar to those identified for C. elegans and A. suum (see Burke & Ward, 1983; Bennett & Ward, 1986). Also, real-time PCR analysis demonstrated that O. dentatum msp was expressed exclusively in the male of the late L4 and the adult. This is consistent with Northern blot analyses of C. elegans males (Burke & Ward, 1983), and corresponds to the developmental stage (L4) in which sperm production is initiated (L' Hernault, 1997). Interestingly, analysis of male O. dentatum raised in vivo (from L4) in the absence of females did not reveal a significant reduction of the level of msp expression (over a 45-day period). Previous information has indicated that males of O. dentatum are required for the ‘normal’ development of the female parasite through to sexual maturity. Preliminary observation suggests that female worms raised in the absence of males are not reproductively mature or fecund (cf.Christensen, 1997). There is clear evidence for the dioecious trematode, Schistosoma mansoni, that female worms regress to a ‘sexually immature state’ when raised separately from males, whereas the reproductive tissues of the males raised separately is not affected (Erasmus, 1973; Popiel, Cioli & Erasmus, 1984; Kunz, 2001). If a similar process operates in O. dentatum, the absence of ‘morphological regression’ in the male (raised in separation from the female) would explain the lack of a difference in msp expression between ‘mated males’ and ‘unmated males’ of O. dentatum. Further work is required to investigate the proposal put forward by Christensen (1997) that females of O. dentatum ‘stunt’ in the absence of males. If this proposal is supported, expression profiling of ESTs isolated from suppressive subtractive hybridization gene libraries constructed from O. dentatum males or females raised (over a long period; e.g., 60–100 days) in the presence or absence of their sexual partner would provide insights into the effect(s) of mating or pairing on gene expression linked specifically to the sexual maturation of the parasite.
Like the msp genes of O. dentatum, those of A. suum and C. elegans are expressed in a gender-specific and developmentally-regulated manner (e.g., Burke & Ward, 1983; Bennett & Ward, 1986). Studies have demonstrated regulatory elements upstream of the msp genes of both O. volvulus and C. elegans (see Klass et al. 1988; Scott et al. 1989b). Specifically, 2 GATA transcriptional factor binding motifs have been recorded in the first 100 nucleotides of the 5′ region prior to the initiation codon (representing methionine) in the latter two species (Klass et al. 1988; Scott et al. 1989b). Other genes known to be regulated in a sex- and tissue-specific manner, such as those of the serine/threonine phosphatase of O. dentatum (see Boag et al. 2003b), vitellogenins (MacMorris et al. 1992; Boag et al. 2001) and tra-2 of C. elegans (see Kuwabara, 1996), also contain 2 GATA transcription factor binding elements within this upstream region. Surprisingly, such elements were not identified in the −1 to −123 bp region of the O. dentatum msp genes. The reason for this is presently unclear, but it is possible that the msp genes of O. dentatum are arranged in tandem arrays and are controlled via a promoter which is located further upstream. This proposal warrants investigation. Current genome sequence and gene expression data for C. elegans (The C. elegans Sequencing Consortium, 1998; Kim et al. 2001) indicate that 15–25% of genes are arranged in bacterial-like operons of 2–8 genes (Blumenthal, 1998; Lercher, Blumenthal & Hurst, 2003), which also provides some support for such a proposal. Such an organization of regulatory elements would allow gene products to be expressed in a temporal and spatial manner, and different isoforms could be expressed from a single operon via alternative splicing of a polycistronic sequence (see Blumental, 1998).
A number of MSP sequences were compared in a pairwise manner. The sequence identity among them ranged from 82 to 98%. The level of identity between O. dentatum and C. elegans (82–83%) was essentially the same as between A. suum and C. elegans (83%). The MSP sequences of all other nematodes (including Ancylostoma ceylanicum, Haemonchus contortus, Teladorsagia circumcincta, D. viviparus and N. brasiliensis, of the order Strongylida) had a higher degree of identity (84–87%) to C. elegans, suggesting that they all share functional roles. This statement is supported by the finding that A. suum MSP functions biologically in C. elegans (see Miller et al. 2003).
Given that there is evidence that Ascaris MSP functions effectively in C. elegans (see Miller et al. 2003), it is likely that the function(s) of O. dentatum MSPs could also be investigated in this free-living nematode. In addition to their role in maintaining sperm mobility, MSPs of C. elegans have been shown to act as signalling molecules to stimulate the production and maturation of oocytes in the uterus of hermaphrodites. A recent, detailed study of sperm-less C. elegans hermaphrodites (Miller et al. 2001) demonstrated that MSP signalling was retained after C. elegans MSP had been injected into the uterus. The role of MSPs in the production and maturation of oocytes in C. elegans has been linked to their N- and C-termini. It was shown that injection of deletion mutants of the N-terminal domains of MSP-38 and MSP-77 (aa positions 1–106) into the uterus of hermaphrodites of the fog-2 strain of C. elegans promoted the ‘re-entry’ of oocytes (from G2/M arrest) into meiosis (Miller et al. 2001). Conversely, injection of deletion mutants of the C-terminal domains of these 2 MSPs (aa positions 106–126) into the same organ was demonstrated to stimulate the contraction of sheath cells, culminating in the release of oocytes for subsequent fertilization (Miller et al. 2001). Given the aa sequence conservation (87–95% identity) in both the N-terminal and the C-terminal domains (for MSP-38 and MSP-77) between O. dentatum and C. elegans, it is possible that O. dentatum MSPs have a similar signalling function. Clearly, this proposal warrants testing.
Using well-defined criteria, Miller et al. (2003) identified 6 potential receptors for MSP from 258 oocyte-enriched genes of C. elegans (see Reinke et al. 2000). The gene vab-1 (variable abnormal morphology) has been shown to encode an ephrin receptor, expressed on the surface of C. elegans oocytes and sheath cells. When sperm is present, MSP binds to the ephrin receptor, stimulating the mitogen-activated protein kinase (MAPK) pathway leading to oocyte production (Miller et al. 2003). In the absence of sperm, the gene product VAB-1 represses MAPK, and oocytes remain developmentally arrested. In spite of the numerous EST sequencing projects currently under way for parasitic nematodes, interestingly, vab-1 homologues have not yet been reported for parasites. However, the relative conservation of MSPs at the aa level for a wide range of nematode species, coupled with the ability of A. suum MSP to activate oocyte production in C. elegans (see Miller et al. 2003), would suggest that the control of this process is mediated via homologous receptors in parasitic nematodes. This hypothesis should be tested.
In conclusion, the present study has provided some insights into msp genes and their expression in O. dentatum. Comparison of MSPs from O. dentatum with those from various other nematodes showed aa sequence conservation, suggesting similar functional roles for the proteins. Further study focussed specifically on the interaction of MSPs with receptors (such as VAB-1) in female nematodes should improve our understanding of the role(s) of this interesting family of proteins and may reveal a target for chemotherapeutic intervention.
The authors sincerely thank Niels-Peter Hansen of the Danish Centre of Experimental Parasitology for producing O. dentatum and for consistent support. Thanks also to Xingquan Zhu for undertaking preliminary work, and to Youssef Abs EL-Osta and Min Hu for their discussions and help. Assistance from Alison Bant and Debbie Donald of the Molecular Vaccines group (Victorian Institute of Animal Science, Attwood, Victoria, Australia) with expression analysis is gratefully acknowledged. This work was supported by the Australian Research Council (Linkage Project ID LP0346983), the Australian Academy of Science, Meat and Livestock Australia, GTG and The University of Melbourne.

Fig. 1. Alignment of msp nucleotide sequences isolated from complementary (designated with C) and genomic (designated with G) DNA libraries. A stop codon is denoted by a large dot. Respective amino acid sequences are aligned below the nucleotide sequences. The alignment positions are above each respective block of sequences.

Table 1. Pairwise comparison of amino acid sequence differences (%) among the representative MSPs of Oesophagostomum dentatum (designated OdMSPG23, OdMSPG33, OdMSPC01 and OdMSPC06; see accession numbers AJ627869–AJ627873), Caenorhabditis elegans (NM_069361), Ascarissp. (P27439), Onchocerca volvulus (P13262), Dictyocaulus viviparus (S64873), Ancylostoma ceylanicum (CB176395), Haemonchus contortus (CB191457), Teladorsagia circumcincta (CB037657) and Nippostrongylus brasiliensis (BQ529557). The amino acid sequence of OdMSPC06 was the same as OdMSPC11 (cf. Fig. 1)

Fig. 2. Southern blot analysis of Oesophagostomum dentatum DNA digested with restriction endonuclease ClaI, EcoRI, HindIII or PstI. Undigested DNA samples from O. dentatum (U) and pig musculature (Upm) were included as controls.
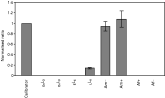
Fig. 3. Expression of msp mRNA at different stages of development of Oesophagostomum dentatum measured by real-time PCR. Expression was detectable exclusively in males of the late L4 (LL4) and adult stages (A) of the parasite, but not in the ensheathed L3 (EnL3), artificially exsheathed L3 (ExL3) or early L4 stage (ExL4). In adult nematodes, there was no statistical difference in expression values between males (Am+) raised together with females (Af+) for 51 days (after intragastric inoculation of pigs with L3s) and males (Am−) maintained without females for 45 days (following transplantation of male-only L4s to a recipient pig). No expression was detected in females (Af−) maintained without males for 45 days (following transplantation of female-only L4s to a recipient pig). The calibrator represents a reference control.

Fig. 4. Alignment of the MSP amino acid sequences of Oesophagostomum dentatum (Accession numbers AJ627869–AJ627873) with selected sequences for Caenorhabditis elegans (NM_069361), Ascaris sp. (P27439), Onchocerca volvulus (P13262), Dictyocaulus viviparus (S64873), Ancylostoma ceylanicum (CB176395), Haemonchus contortus (CB191457), Teladorsagia circumcincta (CB037657) and Nippostrongylus brasiliensis (BQ529557). Amino acid identity among all species of nematode is indicated by an asterisk.