Introduction
The whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae), is an important global agricultural pest that is capable of damaging a wide range of hosts, including several greenhouse and field crops, ornamental plants and weeds, worldwide (Muñiz, Reference Muñiz2000; Dinsdale et al., Reference Dinsdale, Cook, Riginos, Buckley and De Barro2010). In addition, whiteflies are also considered to be a super vector, transmitting approximately 300 plant viruses, including viruses of the genera Begomovirus, Carlavirus, Crinivirus, Ipomovirus, and Torradovirus, highlighting the group of Begomovirus, which represents approximately 90% of the viruses transmitted by whitefly (Kanakala and Ghanim, Reference Kanakala and Ghanim2015).
Bemisia tabaci is considered to be a complex of cryptic species, which includes at least 44 identified species (De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011; Kanakala and Ghanim, Reference Kanakala and Ghanim2019). Individual species of this complex are morphologically indistinguishable. However, they differ according to their molecular biology (Boykin and De Barro, Reference Boykin and De Barro2014) and in several biological characteristics, such as their ability to transmit specific begomoviruses (Polston et al., Reference Polston, De Barro and Boykin2014), ability to harbor different sets of bacterial endosymbionts (Czosnek and Ghanim, Reference Czosnek and Ghanim2016), adaptability to different host plants (Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013), insecticide tolerance (Horowitz and Ishaaya, Reference Horowitz and Ishaaya2014) and invasive capability (Liu et al., Reference Liu, De Barro, Xu, Luan, Zang, Ruan and Wan2007).
Among these cryptic species, Middle East-Asia Minor 1 (MEAM1, formerly B biotype) and Mediterranean (MED, formerly Q biotype) are regarded as the most invasive and widespread (Kanakala and Ghanim, Reference Kanakala and Ghanim2015). These cryptic species have attained the global status and are responsible for severe crop damage (Horowitz et al., Reference Horowitz, Denholm, Gorman, Cenis, Kontsedalov and Ishaaya2003; Luo et al., Reference Luo, Jones, Devine, Zhang, Denholm and Gorman2010).
In Brazil, MEAM1 was first reported in the early 1990s (Lourenção and Nagai, Reference Lourenção and Nagai1994), and over the past 20 years, this species has become widespread throughout the country (Moraes et al., Reference Moraes, Muller, Bueno, Santos, Bello, De Marchi, Watanabe, Marubayashi, Santos, Yuki, Takada, de Barros, Neves, da Silva, Gonçalves, Ghanim, Boykin, Pavan and Krause-Sakate2018). With MEAM1 introduction in Brazil, the emergence of begomoviruses also occurred in solanaceous crops (Ribeiro et al., Reference Ribeiro, De Ávila, Bezerra, Fernandes, Faria, Lima, Gilbertson, Maciel-Zambolim and Zerbini1998). More than 20 years later, MED was detected in 2014 in Rio Grande do Sul State (Barbosa et al., Reference Barbosa, Yuki, Marubayashi, De Marchi, Perini, Pavan, Barros, Ghanim, Moriones, Navas-Castillo and Krause-Sakate2015), and a new invasion associated with ornamental plants was reported in São Paulo and Paraná States in 2015 (Moraes et al., Reference Moraes, Marubayashi, Yuki, Ghanim, Bello, De Marchi, Barbosa, Boykin, Krause-Sakate and Agenor Pavan2017). Recently, in 2017, MED was also reported in Santa Catarina and Minas Gerais States in association with ornamental plants (Moraes et al., Reference Moraes, Muller, Bueno, Santos, Bello, De Marchi, Watanabe, Marubayashi, Santos, Yuki, Takada, de Barros, Neves, da Silva, Gonçalves, Ghanim, Boykin, Pavan and Krause-Sakate2018), indicating the importance of ornamental plants for MED invasion in Brazil.
In this study, we report that MED now is also being found on vegetable crops, especially on bell peppers and cucumbers cultivated in greenhouses. However, the insect was also found infesting tomatoes, bell peppers, and eggplants cultivated in open fields. In addition, difficulties have been observed in insect management by spraying insecticides in bell pepper areas, and virus-infected plants were detected in these areas.
Materials and methods
Sampling of whiteflies
Bemisia tabaci samples were collected from several regions of São Paulo and Paraná States in 2017, 2018, and 2019. The samplings were performed in commercial greenhouses with Capsicum annuum Linnaeus (Solanaceae), Solanum lycopersicum Linnaeus (Solanaceae), Cucumis sativus (Cucurbitaceae), Solanum melongena Linnaeus (Solanaceae) and weeds; and in open field containing tomatoes, bell peppers, and eggplants. The insects (adults) were collected with a hand-held aspirator, and the specimens were immediately transferred to a tube containing 100% ethanol and stored at −4°C until further analyses. For each population, analyses of ten adults were carried out for microsatellite and mitochondrial cytochrome oxidase I (mtCOI) regions. Table 1 summarizes the populations collected and analyzed in this study. Additionally, one B. tabaci sample was collected from ornamental plants in Formosa/Goiás.
Table 1. Information of whitefly Bemisia tabaci Middle East-Asia Minor 1 (MEAM1) and Mediterranean (MED) species collected in São Paulo and Paraná State, Brazil, between 2017 and 2019, and viruses identification

GH, greenhouse; OF, open field; nt, not tested; ToSRV, Tomato severe rugose virus; ToCV, Tomato chlorosis virus.
In bold samples collected in the field.
a Samples highlighted in bold belong to OF samples for a better visualization.
MED and MEAM1 species identification
The molecular analyses were carried out following the total DNA extraction from individual specimens following a modified Chelex method. Whitefly adults were macerated and homogenized in 50 μl of Chelex 5% solution in a 2.0 ml Eppendorf® tube. The tube was agitated for a few seconds and then incubated at 56°C for 15 min and at 99°C for 3 min. After centrifugation at 14,000 rpm for 5 min, the supernatant was then collected and used as a template for PCR amplification.
All individual DNA samples per population were first analyzed for an initial PCR using the BEM23 primer pair: Bem23F (5′-CGGAGCTTGCGCCTTAGTC-3′) and Bem23R (5′-CGGCTTTATCATAGCTCTCGT-3′) (De Barro et al., Reference De Barro, Scott, Graham, Lange and Schutze2002). This primer pair differentiates the MED and MEAM1 based on the microsatellite locus of approximately 200 and 400 bp for each species, respectively (Kontsedalov et al., Reference Kontsedalov, Abu-Moch, Lebedev, Czosnek, Horowitz and Ghanim2012; Škaljac et al., Reference Škaljac, Anić, Hrnčić, Radonjić, Perović and Ghanim2013). Furthermore, the whitefly species samples were confirmed by PCR and sequencing using the primer pair 2195Bt (5′-TGRTTTTTTGGTCATCCRGAAGT-3′) and C012/Bt-sh2 (5′-TTTACTGCACTTTCTGCC-3′), which amplifies a fragment from the mtCOI gene (Mugerwa et al., Reference Mugerwa, Seal, Wang, Patel, Kabaalu, Omongo, Alicai, Tairo, Ndunguru, Sseruwagi and Colvin2018). Subsequently, the mtCOI amplicons were submitted to a restriction fragment length polymorphism analysis (Bosco et al., Reference Bosco, Loria, Sartor and Cenis2006), and 6.5 μl of each PCR (800 bp) was digested with one unit of TaqI at 65°C for 2 h in a final volume of 16.5 μl. The restricted DNA was observed through electrophoresis in a 1.8% agarose gel stained with ethidium bromide.
The mtCOI amplicons of B. tabaci were purified (QIAquick Gel Extraction Kit, Qiagen, Hilden, Germany) and sequenced in both directions using the primers 2195Bt and C012/Bt-sh2. The curated dataset of mtCOI (Boykin and De Barro, Reference Boykin and De Barro2014) was used to confirm the species. Nucleotide sequences of mtCOI were deposited in GenBank, and accession numbers are available in table 1.
Endosymbionts detection
The same DNA extracted from the individual MED whitefly specimens for identification was used for the screening of the six secondary endosymbionts Hamiltonella, Rickettsia, Wolbachia, Arsenophonus, Cardinium, and Fristchea using genus-specific primers targeting the 16S and 23S rDNA genes (Marubayashi et al., Reference Marubayashi, Kliot, Yuki, Rezende, Krause-Sakate, Pavan and Ghanim2014). Endosymbiont presence was confirmed by sequencing representative individuals. The nucleotide sequences were deposited in GenBank, and accession numbers are available in Supplementary table S1.
Virus identification
At least one plant sample for each locality was analyzed to confirm begomovirus and crinivirus presence. Begomoviruses were detected by DNA extraction (Dellaporta et al., Reference Dellaporta, Wood and Hicks1983) and PCR using the degenerate primer pair PAL1v1978/PAR1c496 (Rojas et al., Reference Rojas, Gilbertson, Russell and Maxwell1993). For crinivirus Tomato chlorosis virus (ToCV) detection, total RNA was extracted with a Total RNA Purification Kit® (Norgen, Canada), followed by RT-PCR with the primers HS-11/HS-12 and nested PCR specific for ToCV using the primer pair ToCV-5/ToCV-6 (Dovas et al., Reference Dovas, Katis and Avgelis2002). Representative PCR products amplified from samples were purified (QIAquick Gel Extraction Kit, Qiagen) and sequenced using the degenerate primers PAL1v1978/PAR1c496 for begomoviruses or the specific primers ToCV-5/ToCV-6 for ToCV. Respective sequences were analyzed and compared with other sequences in GenBank using BLAST tools (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Additionally, the same DNA extracted from individual whiteflies for identification was used to identify begomoviruses, and these identifications were confirmed by sequencing. All nucleotide sequences were deposited in GenBank, and accession numbers are available in table 1.
Results
Samples collected under greenhouse conditions
A total of 360 individual whiteflies collected from bell peppers (C. annuum) were analyzed at 14 localities, and 100% were identified as MED. From cucumber crops (C. sativus), 110 individual whiteflies were tested, and only MED was detected. Among the tomato crops (S. lycopersicum), 110 individual whiteflies were analyzed, 107 were identified as MED, and three were identified as MEAM1.
MED was also detected infesting weeds associated with bell peppers in greenhouses (table 1, fig. 1) and was identified in Goiás state for the first time in association with ornamental plants collected from a flower shop.
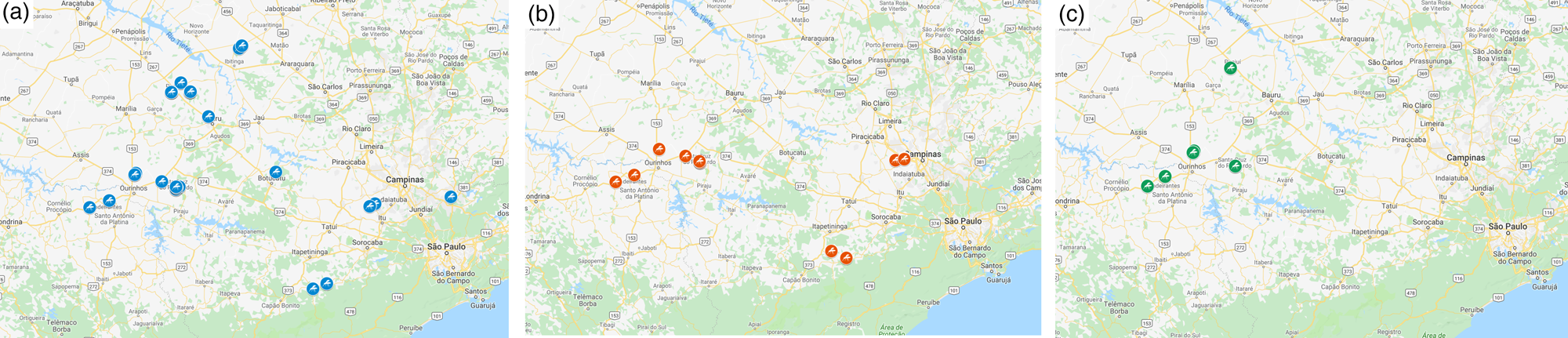
Figure 1. Sampling locations for Bemisia tabaci Mediterranean (MED) species in São Paulo and Paraná States, Brazil. These points represent places where MED was found in (a) bell pepper, (b) tomato, and (c) cucumber crops.
Samples collected under open field conditions
From 40 individuals collected on tomatoes, 16 were identified as MED, and 24 as MEAM1. On eggplants (S. melongena), 40 specimens were analyzed and 31 were identified as MED and nine as MEAM1. On bell peppers, ten specimens were analyzed and 100% were identified as MED.
Virus identification
The crinivirus ToCV was detected on tomatoes (sampling site 12), and the begomovirus Tomato severe rugose virus (ToSRV) was found infecting bell pepper (sampling sites 41, 47, and 49) (table 1). In addition, ToSRV was also detected by PCR in MED specimens. Both ToCV and ToSRV were found in areas where only MED was detected.
Endosymbionts detection
The endosymbiont analysis revealed that MED harbors Hamiltonella, Rickettsia, Wolbachia, and Arsenophonus, with a predominance of Hamiltonella in most of the individuals sampled (Supplementary table S1; fig. 2).

Figure 2. Individual infections by secondary endosymbionts found harbored Bemisia tabaci Mediterranean (MED) populations from São Paulo and Paraná States, Brazil. Numbers inside the columns represent the individual positive for the respective endosymbiont by total individuals analyzed. H: Hamiltonella, R: Rickettsia, W: Wolbachia, and A: Arsenophonus.
Discussion
Our results showed that outbreaks of whiteflies in vegetable crops in two eastern states in Brazil (São Paulo and Paraná) associated with the MED species. High populations of MED infesting bell peppers, cucumbers, and eggplants not only under greenhouses, but also under open field conditions lead to the appearance of sooty mold growth (fig. 3). Difficult to manage the insects was also a common complaint on the visited areas. Moreover, ToCV-infected tomatoes and ToSRV-infected bell pepper plants were detected in greenhouses, where only MED was identified, indicating that MED may be contributing to virus transmission on these vegetables.

Figure 3. Outbreaks of Bemisia tabaci Mediterranean (MED) populations on bell pepper and cucumber crops. (a) Adults and nymphs of MED infesting bell pepper plants and causing the sooty mold (b). Adults of MED on cucumber leaves (c).
Bemisia tabaci MED was first reported in Brazil in Rio Grande do Sul State (Barbosa et al., Reference Barbosa, Yuki, Marubayashi, De Marchi, Perini, Pavan, Barros, Ghanim, Moriones, Navas-Castillo and Krause-Sakate2015). After 1 year, a new MED introduction was reported in São Paulo and Paraná States (Moraes et al., Reference Moraes, Marubayashi, Yuki, Ghanim, Bello, De Marchi, Barbosa, Boykin, Krause-Sakate and Agenor Pavan2017) and more recently in Minas Gerais and Santa Catarina States associated to ornamental plants (Moraes et al., Reference Moraes, Muller, Bueno, Santos, Bello, De Marchi, Watanabe, Marubayashi, Santos, Yuki, Takada, de Barros, Neves, da Silva, Gonçalves, Ghanim, Boykin, Pavan and Krause-Sakate2018). In this study, we also detected MED on ornamental plants collected in the Midwest region (Goiás state) of Brazil, where the extensive agriculture is practiced, including vegetable and field crops.
The MED species is often found infesting greenhouses in two eastern states in Brazil. Studies have shown that MED is more adapted to greenhouse conditions than MEAM1. Kontsedalov et al. (Reference Kontsedalov, Abu-Moch, Lebedev, Czosnek, Horowitz and Ghanim2012) reported that MED predominated over MEAM1 under nethouse and greenhouse conditions, not only when insecticides spraying were used but also in the absence of insecticides application. It likely occurs because MED is more resistant to high temperatures than MEAM1. A study from Israel has shown that the mortality of MEAM1 is much higher at 37 and 40°C than that of MED (Mahadav et al., Reference Mahadav, Kontsedalov, Czonesk and Ghanim2009). Another study from China also showed that MEAM1 has a higher mortality than MED under high-temperature conditions (Xiao et al., Reference Xiao, Pan, Zhang, Sham and Liu2016). In general, the temperature between greenhouses and open field production is different, being higher and lower, respectively. Thus, as MED is more tolerant to high temperature, greenhouses can be a great environment to increase the population of this species. However, in this study, we have also found MED under open field conditions in mix infestation with MEAM1 on tomatoes and eggplants.
MED is also known to develop resistance much faster than other whitefly species, such as pyriproxyfen (Horowitz et al., Reference Horowitz, Denholm, Gorman, Cenis, Kontsedalov and Ishaaya2003, Reference Horowitz, Kontsedalov, Khasdan and Ishaaya2005), acetamiprid (Horowitz and Kontsedalov, Reference Horowitz and Kontsedalov2004; Horowitz and Ishaaya, Reference Horowitz and Ishaaya2014), imidacloprid (Karunker et al., Reference Karunker, Benting, Lueke, Ponge, Nauen, Roditakis, Vontas, Gorman, Denholm and Morin2008), thiamethoxam (Horowitz and Kontsedalov, Reference Horowitz and Kontsedalov2004), and cyantraniliprole (Yao et al., Reference Yao, Zheng, Huang, Ding, Zhao, Desneux, He and Weng2017). In Israel, the dynamics of the whitefly populations on cotton were studied for several years in terms of management with insecticide and the predominant species. It was observed that in areas where MEAM1 was the predominant species, MED displaced MEAM1 with high insecticide use. However, after spraying cessation, MEAM1 returned as the predominant species, indicating that the insecticide directly influenced the prevalence of the species (Horowitz and Ishaaya, Reference Horowitz and Ishaaya2014). Similarly, it was verified in China that on eggplant and tomatoes on which MEAM1 was the predominant species, MED became the main species with high insecticide use (Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013). This lower susceptibility to several insecticides is often regarded as a main cause of the faster establishment and dominance of MED in several regions worldwide (Yao et al., Reference Yao, Zheng, Huang, Ding, Zhao, Desneux, He and Weng2017).
The introduction of an exotic pest into a country is always of great concern for its agriculture, mainly if it is a vector of viruses. The great concern started in the 1990s, when the exotic MEAM1 was first detected in Brazil. Ever since, several losses caused by this pest have been reported in practically all regions of Brazil (Chintkuntla, 2015), and outbreaks of begomoviruses infecting Solanaceae have occurred (Faria et al., Reference Faria, Bezerra, Zerbini, Ribeiro and Lima2000). Some years later, the crinivirus ToCV was first reported in Brazil associated with MEAM1 (Barbosa et al., Reference Barbosa, Teixeira, Moreira, Camargo, Bergamin Filho, Kitajima and Rezende2008) and nowadays the begomovirus ToSRV and the crinivirus ToCV are the most important viruses transmitted by B. tabaci cryptic species infecting tomatoes in Brazil (Inoue-Nagata et al., Reference Inoue-Nagata, Lima and Gilbertison2016).
We also verified in this work that ToCV-infected tomato plants and ToSRV-infected bell pepper plants were associated with the presence of MED specimens. The interactions between ToCV and MED in tomato plants are well known because MED fecundity and developmental time are increased by the presence of this virus, indicating that the virus is beneficial to MED (Orfanidou et al., Reference Orfanidou, Pappi, Efthimiou, Katis and Maliogka2016; Shi et al., Reference Shi, Tang, Zhang, Zhang, Li, Yan, Zhang, Zhou and Liu2018). The ability of B. tabaci MED to efficiently transmit ToSRV, a native Brazilian begomovirus and the main species infecting tomato in southwest Brazil, was recently demonstrated (Bello et al., Reference Bello, Watanabe, Santos, Marubayashi, Yuki, De Marchi, Pavan and Krause-sakate2019a). It is expected that changes in the epidemiology of whitefly-transmitted viruses may occur and this has been recently observed for cucumber plants found naturally infected with ToCV in association with MED specimens (Bello et al., Reference Bello, Gorayeb, Watanabe, De Marchi, Ribeiro-Junior, Vicentin, Barreto and Krause-sakate2019b). Cucumber plants have never been reported before as the host of ToCV, but what we could observe is that MED specimens are well adapted to cucumbers cultivated under greenhouse conditions in Brazil and ToCV in this crop can become an important problem to this host.
Bemisia tabaci MED has also been shown to be an excellent vector of the most important begomovirus in Europe and Asia: Tomato yellow leaf curl virus (TYLCV) (Ning et al., Reference Ning, Shi, Liu, Pan, Wei, Zeng, Sun, Xie, Wang, Wu, Cheng, Peng and Zhang2015; Czosnek et al., Reference Czosnek, Hariton-Shalev, Sobol, Gorovits and Ghanim2017), which has not yet been reported in Brazil but was detected in the neighboring country of Venezuela (Zambrano et al., Reference Zambrano, Carballo, Geraud, Chirinos, Fernández and Marys2007). In China, TYLCV became an emergent virus after MED introduction (Pan et al., Reference Pan, Chu, Yan, Su, Liu, Wang and Wu2012; Ning et al., Reference Ning, Shi, Liu, Pan, Wei, Zeng, Sun, Xie, Wang, Wu, Cheng, Peng and Zhang2015). The transmission of TYLCV by MED is more efficient than that by MEAM1 (Ning et al., Reference Ning, Shi, Liu, Pan, Wei, Zeng, Sun, Xie, Wang, Wu, Cheng, Peng and Zhang2015), and the fecundity of MED increases in the presence of this virus, while the developmental time decreases (Chen et al., Reference Chen, Pan, Xie, Wang, Wu, Fang, Shi and Zhang2013; Fang et al., Reference Fang, Jiao, Xie, Wang, Wu, Shi, Chen, Su, Yang, Pan and Zhang2013). In this way, all results together point to the need for TYLCV monitoring in Brazil.
In Brazil, the variability in the secondary endosymbionts found in MED is quite large and may be explained by the recent and different introductions of this species into the country (Moraes et al., Reference Moraes, Muller, Bueno, Santos, Bello, De Marchi, Watanabe, Marubayashi, Santos, Yuki, Takada, de Barros, Neves, da Silva, Gonçalves, Ghanim, Boykin, Pavan and Krause-Sakate2018). MED populations typically harbor Hamiltonella, Rickettsia, Wolbachia, Cardinium, and Arsenophonus (Gueguen et al., Reference Gueguen, Vavre, Gnankine, Peterschmitt, Charif, Chiel, Gottlieb, Ghanim, Zchori-Fein and Fleury2010; Czosnek and Ghanim, Reference Czosnek and Ghanim2016). Our work has also shown that MED collected in this study harbors Hamiltonella, Rickettsia, Wolbachia, and Arsenophonus (fig. 2) and that Hamiltonella is the predominant secondary endosymbiont. The influence of endosymbionts on virus transmission is well described for B. tabaci MED, in which the absence or low frequency of Hamiltonella implies low transmission efficiency of TYLCV (Gottlieb et al., Reference Gottlieb, Zchori-Fein, Mozes-Daube, Kontsedalov, Skaljac, Brumin, Sobol, Czosnek, Vavre, Fleury and Ghanim2010). Additionally, individuals that harbor this bacterium show high benefits in reproduction factors that increase the populations of B. tabaci MEAM1 species (Kliot et al., Reference Kliot, Kontsealadov, Lebedev, Czonesk and Ghanim2019). In addition, populations of MED harboring high frequency of Hamiltonella proved to be better vectors of ToCV and ToSRV (Bello et al., Reference Bello, Watanabe, Santos, Marubayashi, Yuki, De Marchi, Pavan and Krause-sakate2019a).
In conclusion, our results suggest that in São Paulo and Paraná States, Brazil, MED is now found on vegetable crops both in greenhouses and under open field conditions, and ToCV-infected and ToSRV-infected plants were associated to this species. These results reinforce that MED is a real concern for Brazilian agriculture, because it has a great performance and can displace MEAM1 in some important crops (Watanabe et al., Reference Watanabe, Bello, De Marchi, Silva, Machado, Sartori, Pavan and Krause-Sakate2019), is a good vector of important viruses found in Brazil, and is less susceptible to insecticides than MEAM1 (Horowitz and Kontsedalov, Reference Horowitz and Kontsedalov2004; Kontsedalov et al., Reference Kontsedalov, Abu-Moch, Lebedev, Czosnek, Horowitz and Ghanim2012; Yao et al., Reference Yao, Zheng, Huang, Ding, Zhao, Desneux, He and Weng2017). Then, the management of B. tabaci should be performed according to which whitefly species is present in the area, in order to ensure a better control as well as to limit the spreading of MED species. Thus, the monitoring and identification of the B. tabaci cryptic species are essential for helping to choose the greatest management strategy, including not only the chemical control, but also other integrated pests, as biological control.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485319000841.
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001; FAPESP (processes number 2017/21588-7, 2017/50222, and 2018/18274-3) and CNPq (405684/2018-5 and 303411/2018-0). RKS, RCOFB, and MAP received CNPq fellowships.







