The present H1N1 swine influenza pandemic and biothreats present public health and global security challenges. The emergence of these novel pathogens highlights the need for rapid diagnostic testing for detection and differentiation of these pathogens and agents. Point-of-care testing (POCT) provides health care professionals with the capability to deliver onsite diagnostic testing wherever medical care is needed. Rapid diagnostic testing would enable health care professionals to take appropriate preventive measures (eg, quarantine, facility closure) and to initiate focused treatment to improve patient outcomes.
Pandemic influenza strains have the potential to increase morbidity and mortality. The 1918 influenza H1N1 subtype resulted in 40 million deaths worldwide, with 675,000 of those deaths occurring in the United States. Reference Kobasa, Takada and Shinya1–Reference Tumpey, Basler and Aguilar3 The 1957 H2N2 subtype caused 70,000 deaths in this country, and the 1968 H3N2 subtype, 34,000 deaths. Reference Monto4–Reference Kawaoka, Krauss and Webster6 With the present 2009 H1N1 subtype, more than 177,457 people worldwide have been infected. The number of deaths was 1462 as of August 6, 2009.7 On June 11, the World Health Organization (WHO) classified H1N1 as a pandemic (phase 6), which is defined as sustained community outbreaks of the disease in 2 or more countries and involving more than 1 WHO region.8Table 1 summarizes these recent disease outbreaks.
TABLE 1 Recent Pandemics and Disease Outbreaks
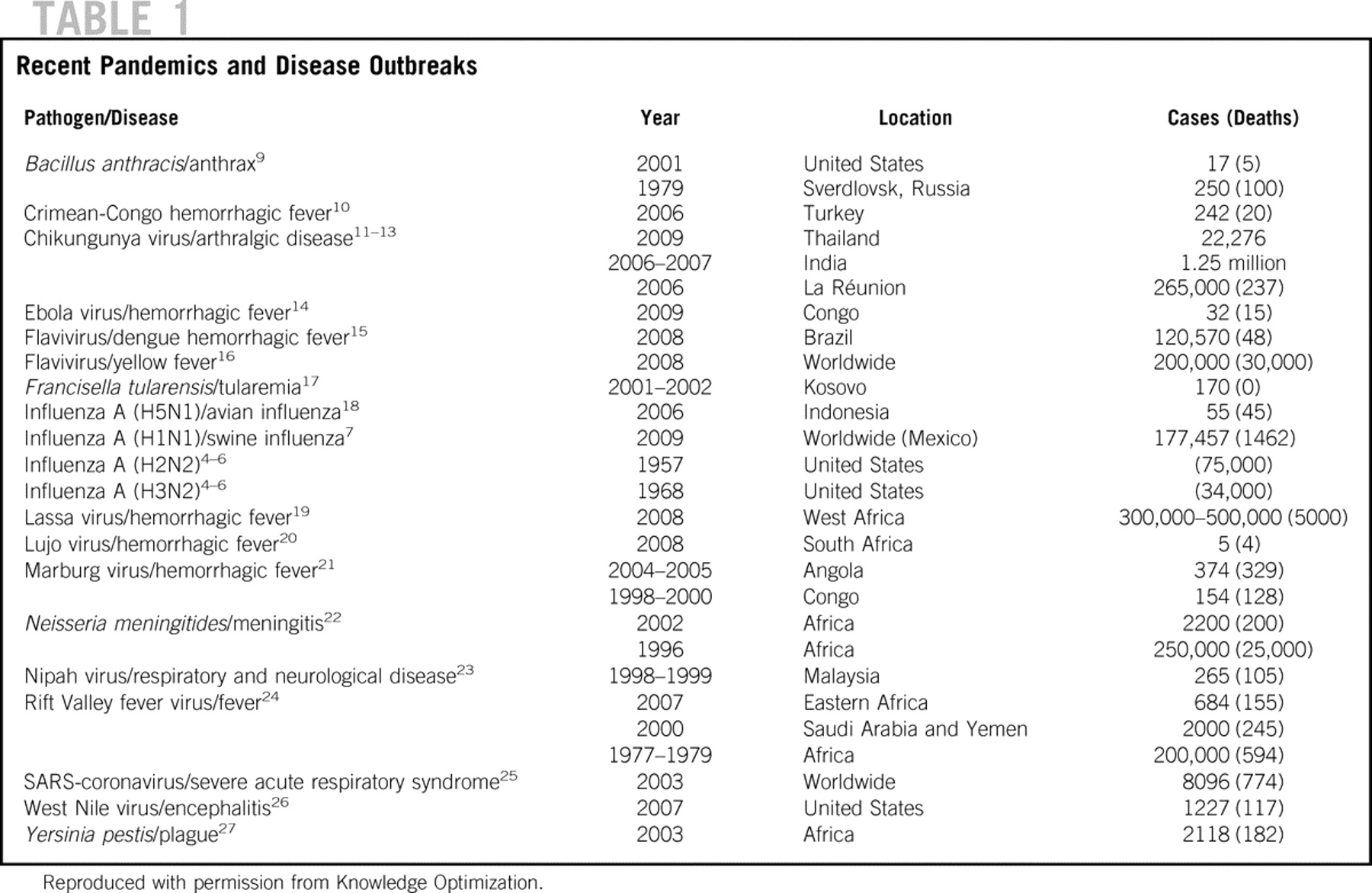
Other recent outbreaks include the reemergence of chikungunya virus in southern Thailand. Although chikungunya generally is not life threatening, the large number of new cases since January 2009 make it an important social issue and generate questions about the reason for the reemergence and virulence of this strain.13 Recently, a new hemorrhagic fever–associated arenavirus, Lujo, was discovered in 5 patients in southern Africa. This aggressive Ebola-like virus has an associated mortality rate of 80%.Reference Briese, Janusz and McMullan20 The emergence of these new viruses highlight the need for flexible and fast diagnostic technologies to prepare for future outbreaks, document sentinel cases, and track local and global dissemination.
DIAGNOSTIC CHALLENGES OF INFLUENZA
More than 200,000 people are hospitalized from flu-related causes in the United States each year, and about 36,000 die from flu-related complications. Reference Thompson, Weintraub and Dhankhar28–30 Early detection and surveillance in the community would enable health care and public health personnel to make evidence-based decisions, such as initiating appropriate antiviral treatment or isolating the patient or victim to prevent spread of the disease, especially if the strain is new and disabling.
Diagnosing influenza A or B infection based on clinical criteria is difficult because viruses associated with upper respiratory tract infection cause overlapping symptoms. Several serious viruses, including adenoviruses, enteroviruses, and paramyxoviruses, may cause influenza-like symptoms initially. The early presentation of mild or moderate cases of flavivirus infections (eg, dengue) may initially mimic influenza. For example, some cases of West Nile fever acquired in New York in 1999 were clinically misdiagnosed as influenza.31
For the present pandemic influenza strain, the Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team32 reported that 94% of patients presented with fever, 92% with cough, and 66% with sore throat. These symptoms are not unlike those of other respiratory viral infections. Rapid triaging and screening of patients for influenza A and B or other communicable diseases (eg, severe acute respiratory syndrome, adenovirus, respiratory syncytial virus, Neisseria meningitides) in the emergency department would be beneficial, because many times patients gather in crowded waiting areas where delays lead to infection of other patients. Table 2 lists POC tests that can be used for POC diagnosis of influenza on site; however, the performance characteristic of these tests for H1N1 virus is not yet known.
TABLE 2 POC Influenza Diagnostic Tests

TABLE 2 POC Influenza Diagnostic Tests (Continued)

INFLUENZA TESTING
Virus Basics
Influenza viruses (A, B, or C) are classified within the family Orthomyxoviridae. These single-stranded RNA viruses are structurally and biologically similar but vary antigenically. The RNA core consists of 8 gene segments. Immunologically, the most significant surface proteins include hemagglutinin (16 subtypes) and neuraminidase (9 subtypes). The viruses are typed based on these proteins.31
Influenza A strains that affect mammals, such as pigs and humans, can undergo genetic reassortment with avian strains. The reassortment can create a chimeric strain that is transmissible to mammals but which may not be recognized by the immune system. The mutation may involve the hemagglutin and neuraminidase proteins.
The present methods for testing influenza include viral culture, nucleic acid testing (polymerase chain reaction), and direct immunofluoresence assays. Specimen types for influenza testing include throat and nasopharyngeal swabs, nasal aspirates, and washes. Aspirates and washes are used less frequently due to high risk for aerosalization.
Culture Method
Viral culture involves the propagation of the viruses using several cell lines or by “shell vial” technique.Reference Kowalski, Karenchak and Romanowski39 Propagated viruses are detected by direct fluorescence antibodies, which target the matrix protein or nucleoprotein. Viral culture is labor intensive, time consuming, and may take several days to provide a definitive result. At our institution, shell vial culture coupled with direct fluorescence antibody testing (SimulFluor Flu A/FluB, Temecula, CA) provides results in 24 hours, and cultures are retested in 48 hours if initially negative. Furthermore, subtyping may not be available at local hospital laboratories, which is the case at our institution. Specimens often are sent to the county or state laboratory for characterization of H1, H3, and H5 subtypes.
POCT
POCT offers simple and rapid alternatives to the laboratories for diagnosis of influenza A and B. WHO recommends the use of rapid testing (POCT) for influenza diagnosis.40 The Centers for Disease Control and Prevention (CDC) recommends that testing be prioritized for those with severe respiratory illness and those with highest risk of complications from influenza. Test sensitivity may vary depending on when in the course of illness the specimen is collected. False-negative test results are more likely during peak activity when the prevalence of disease is high. False-positive test results are more likely during periods of low influenza activity when prevalence is moderate to low, such as at the beginning or end of an outbreak. The CDC recommends that the decision to test patients with rapid influenza diagnostic tests should be based upon the patient’s presenting symptoms, whether cases of novel H1N1 have been confirmed in the area, and the patient’s risk for severe disease or other complications.
Table 2 lists commercially available POC tests for influenza A and B. The list was compiled based on Internet searches, using search terms such as “rapid influenza tests” and “POCT influenza.” We also referred to WHO and CDC lists for rapid influenza tests. These tests typically are immunoassays that target the nucleoprotein or matrix protein. Many of these tests do not perform subtyping. However, Hx Diagnostics (Emeryville, CA) is developing a POC test, the fluID Rapid Influenza Test, that subtypes A/H1 and A/H3. The analysis time for POC influenza tests is about 10 to 15 minutes, much faster than culture. For the most part, rapid tests are qualitative and limited to reporting whether the test sample is positive for either influenza A or B, and in some cases no differentiation between A or B.
POC tests for influenza A and B have relatively high specificity 90% to 100%, but sensitivity as low as 10% (Table 2). Low sensitivity makes it difficult for clinicians to rule out influenza in a patient who has typical symptoms. Therefore, rapid (<1 hour) yet more sensitive tests, such as nucleic acid testing, are needed.
Nucleic Acid Testing
Table 2 lists assays for nucleic acid recognition of influenza viruses. Compared with immunoassays, nucleic acid testing offers a more sensitive method for detection of influenza A and B and can provide subtyping data. The Luminex xTag respiratory viral panel (Austin, TX) multiplex polymerase chain reaction (PCR) assay demonstrated a clinical sensitivity of 98.0% and specificity of 99.8% for influenza A and a clinical sensitivity of 94.4% and specificity of 100% for influenza B.Reference Pabbaraju, Tokaryk and Wong38 Other technologies including ArrayTube (Clondiag, Jena, Germany),Reference Gall, Hoffmann and Harder41 FluChip-55,Reference Townsend, Dawson and Mehlmann42 and electronic microarrayReference Huang, Tang and Duffy43 are being developed to enable simultaneous typing and subtyping of the influenza virus.
Subtyping allows health care personnel and public health workers to identify the viral strain circulating among the population and to start surveillance of new strains in the community. Rapid POC subtyping of influenza virus has the potential to guide antiviral treatment. For example, in fall 2008, most strains of H1N1 influenza were resistant to oseltamivir.44 However, the spring 2009 H1N1 subtype (swine flu) was sensitive to oseltamivir.Reference Gubareva, Okomo-Adhiambo and Deyde45
Food and Drug Administration (FDA) Approval of H1N1 (Swine Flu) Tests
The FDA gave approval on April 27, 2009 to public health and other qualified laboratories to make emergency use of a real-time reverse transcriptase (RT)-PCR swine flu diagnostic panel test for presumptive diagnosis of the new H1N1 strain.46 The panel is authorized for use in individuals who have been diagnosed as having influenza A caused by a virus not subtypable by current FDA-cleared devices. Primer Design (Southampton, England) has produced a commercial RT-PCR test kit for the new H1N1 strain based on genetic sequence data for the virus published by the CDC.47 This assay is suited for the laboratory setting because it requires a trained laboratorian to perform the test. The test is performed on extracted nucleic acid from a respiratory sample and results are obtained in ≈2.5 hours. The test kits (for 150 reactions) cost between £295 and £395 ($485–$649US).
DETECTION OF BIOTHREAT AGENTS
Intentional release of biological agents may present diagnostic dilemmas for clinicians and hospital laboratorians. Detecting the first case (“index case”) from a biological attack represents a pivotal strategy. Table 3 summarizes high-priority biothreat agents.Reference Khan and Sage48 The CDC defines 3 categories (A, B, and C) of biological agents based on ease of dissemination or transmission, major health impact (eg, high mortality), public panic and social disruption, and requirements for public health preparedness.Reference Khan and Sage48
TABLE 3 Priorities of Biothreat Agents

The challenge of identifying an index case and then taking action is whether the positive result represents a biological attack or a sporadic infection resulting from natural presence of the organism in the environment. For example, Bacillus anthracis, Francisella tularensis, and Yersinia pestis may cause sporadic infections. Bacillus anthracis normally is found in soil. False alarms may generate more harm than good, such as public panic, unnecessary use of antivirals or antimicrobials, and adverse drug reactions.
Highly sensitive and specific biothreat detectors, such as the Autonomous Pathogen Detection System developed by the Lawrence Livermore National Laboratory, collect air samples and perform nucleic acid and protein testing for select biothreat agents. Reference McBride, Masquelier and Hindson49–Reference Regan, Makarewicz and Hindson51 Biothreat detectors have the potential to detect pathogens that occur naturally in the environment. Clinicians and public health personnel will need to decide after considering the frequency and pattern of data how best to respond to a biothreat warning. Recent testimony of public health officials focuses on the index case.
Table 4 summarizes commercial biothreat detectors. The instruments were identified by Internet searches using search terms such as “biothreat detectors,” “biothreat detection,” and “biothreat assays,” and referrals from colleagues in the field of biothreat assay development. Bacillus anthracis, Francisella tularensis, and Yersinia pestis appear in the test clusters of several of the biothreat detection devices listed. These devices are designed for detection of biothreat agents in environmental samples that are not approved for clinical diagnostic testing. These devices are suited for mobile laboratory or field setting for environmental surveillance. There is a lack of POC devices available for testing clinical samples. Portable and handheld devices enable emergency and disaster responders to triage victims at the point of care (eg, mobile hospital, ambulance, field) and to deliver appropriate treatment early.
TABLE 4 Biothreat Instrument Systems

TABLE 4 Biothreat Instrument Systems (Continued)

The Bacillus anthracis (anthrax) release by domestic terrorists in 2001 demonstrated how unprepared our nation was for biothreats. Jernigan et alReference Jernigan, Raghunathan and Bell52 reported that about 32,000 individuals initiated prophylaxis antimicrobial therapy in response to the anthrax events. Ciprofloxacin was initially used and then switched to doxycycline once antimicrobial sensitivity data were available. Adverse events from prophylaxis therapy were reported in 57% of cases.Reference Baillie53, Reference Shepard, Soriano-Gabarro and Zell54 Rapid diagnostic testing, especially if it identifies multidrug-resistant strains of a pathogen, could optimize treatment care paths.
GAP ANALYSIS
Pandemics and emerging biothreats highlight gaps in POCT technology. Pandemics place all nations, rich and poor, at risk and can interrupt sustainable development. Medical innovations and health initiatives, and scientific knowledge bases for research and development represent high-priority avenues to reduce global disease burden.Reference Kost55, Reference Jamison, Breman and Measham56
Technological deficiencies (“gaps”) in workflow prevent POCT from being a fully effective tool for disaster care. Technological gaps include the complexity of performing the test, multiplex test cluster capability, flexibility of the test panel, size of the instrument, susceptibility to environmental stresses, and performance tradeoffs such as sensitivity and specificity versus cost. The complexity of nucleic acid testing tends to confine it to laboratory settings where tests are performed by highly trained personnel in controlled environments. Preanalytical processing steps, such as DNA extraction, need to be automated or, if possible, removed entirely. A recent study showed that POC glucose and blood gas measurements were compromised when reagent test strips or test cartridges were thermally stressed.Reference Louie, Sumner and Belcher57 Hence, once miniaturized and automated, highly portable POCT devices and reagents must be environmentally robust to withstand thermal (eg, high and low temperature, humidity) and physical (eg, pressure, shock, vibration) stresses in disaster scenarios.
POC devices need to have flexible test panels and modularity for given clinical scenarios such as critical care, pandemics, and biothreats. These modular POC tests could serve as sentinels for emerging diseases and outbreaks. For pandemics (eg, swine flu) inexpensive veterinary POC devices are needed to detect sentinel cases in animals (eg, pigs). For example, Smiths Detection (Edgewood, MD) has developed a POC veterinary diagnostic device, Bio-Seeq Vet, that detects the foot-and-mouth disease virus. This device could be adapted to test for swine flu. In addition, quantitative testing is lacking on the immunoassays listed in Table 2. Determining viral or pathogen load would allow clinicians to monitor the status of the disease, provide insight on pathogenesis, and help guide treatment. Multiplex testing (eg, multiple pathogens, multiple markers) would enhance diagnostic resolution, such as providing speciation (subtyping) and characterizing the drug-resistant profile of the pathogen. Bioinformatics will be integral in identifying unique genetic markers for development of these diagnostic technologies. The CDC weekly influenza surveillance report (FluView) ending on June 6, 2009 stated that 89% of all influenza strains reported to CDC were the novel influenza A (H1N1).58 This highlights the need for POC tests that are able to subtype the novel H1N1 strain.
Optimal technology would entail simple operation, short analysis time, and accuracy. Technology developers strive to produce a test that has both high clinical sensitivity and specificity, but must address tradeoffs, such as cost, portability, and complexity. Clinicians need to describe and prioritize acceptable compromises in performance and utility tradeoffs for future diagnostic tests.
CONCLUSIONS AND POLICY RECOMMENDATIONS
Medical innovation and health care initiatives help prepare the nation for pandemics and biothreats. New POC technologies must have high impact and address unmet clinical needs to deliver value cost effectively. They provide valuable qualitative and quantitative data that help guide clinicians in making appropriate evidence-based treatment decisions.
New diagnostic tests should strive to provide a level of diagnostic detail necessary for clinicians to make an impact on treatment. For example, multiplex testing with a portable or handheld POC device will allow emergency responders not only to type viruses but also to obtain subspeciation (subtyping) data (eg, influenza A or H1N1 subtype). This information will aid in antiviral selection and provide valuable community surveillance including sentinel cases in both humans and animals.
Technology developers should explore innovative approaches (eg, nucleic acid testing, surface acoustic wave technologies,Reference Bisoffi, Hjelle and Brown59 DNA/protein microarraysReference Zhu, Stybayeva and Macal60) to engineer portable, rapid (<1 hour), highly sensitive, and specific tests for nonpandemic, pandemic, and biothreat agents. Funding agencies (eg, National Institutes of Health) are investing more in POCTReference Kost, Korte and Beyette61 and should continue to do so in view of several strategic gaps identified in diagnostic–therapeutic cycles.
Specifically, funding must encourage POC technology development for nonculture-based drug susceptibility results. Multidrug-resistant pathogens (eg, Staphylococcus aureus, Mycobacterium tuberculosis, and Bacillus anthracis) are becoming more prevalent in association with increased use of antimicrobials and antivirals.
Integration and placement of POCT technologies at strategic levels of patient care, such as emergency departments, urgent care settings, clinics in rural settings, and alternative care facilities,Reference Lam, Waldhorn and Toner62 will provide appropriate accessibility to diagnostic testing and national preparedness.
Authors' Disclosures
The authors report no conflicts of interest.
Acknowledgments
The work was supported by award number U54EB007959 from the National Institute of Biomedical Imaging and Bioengineering.
The authors thank Ms Kristin Hale for critical review and input on the manuscript, and Dr John Dzenitis at Lawrence Livermore National Laboratory for contributing information on biothreat detection.








