Introduction
The Atacama Desert in Chile has long been deemed as a Mars-analogue environment because of the long-term hyperaridity, the strong ultraviolet radiation and the paucity of organics (Navarro-Gonzalez et al., Reference Navarro-Gonzalez, Rainey, Molina, Bagaley, Hollen, De La Rosa, Small, Quinn, Grunthaner, Caceres, Gomez-Silva and Mckay2003). The microbial communities in the Atacama Desert thus represent an end-member in their adaptation to extremely dry conditions. In order to study these microbes, we have to investigate their abundance, compositions and environmental adaptations. The membrane of microorganisms is one of the most important constituents of living cells, and phospholipids provide the main components of all viable cell membranes. Thus, membrane phospholipids could be vital biomarkers (Willers et al., Reference Willers, Van Rensburg and Claassens2015).
Phospholipid fatty acid (PLFA) analyses will provide microbiological information such as biomass, biodiversity and functional groups of cell membranes. Normally, PLFAs degrade rapidly after host microorganisms die, so they are hardly able to preserve during the lengthy process of burial in the subsurface (Harvey et al., Reference Harvey, Fallon and Patton1986; Vestal, Reference Vestal1988). Therefore, PLFAs authentically reflect the native microbial organics and viable biomass in subsurface soils of the Atacama Desert (Ewing et al., Reference Ewing, Michalski, Thiemens, Quinn, Macalady, Kohl, Wankel, Kendall, Mckay and Amundson2007; Kuzyakov, Reference Kuzyakov2010; Azua-Bustos et al., Reference Azua-Bustos, Gonzalez-Silva and Corsini2017). Subsurface is also not directly exposed to ultraviolet radiation, which protects organics against irradiative damages (Warren-Rhodes et al., Reference Warren-Rhodes, Lee, Archer, Cabrol, Ng-Boyle, Wettergreen, Zacny, Pointing, Chong, Demargasso, Foil, Tate, Hare, Lacap-Bugler, Moersch, Tanaka, Tebes, Vijayarangan, Wagner, Wang, Wei and Team2019). On the other hand, other organics such as mineral-bound organic carbon can be preserved much longer in soils (Bonani et al., Reference Bonani, Friedmann, Ocampo-Friedmann, Mckay and Woelfli1988; Sun and Friedmann, Reference Sun and Friedmann1999; Wilhelm et al., Reference Wilhelm, Davila, Eigenbrode, Parenteau, Jahnke, Liu, Summons, Wray, Stamos, O'reilly and Williams2017; Brady et al., Reference Brady, Goordial, Sun, Whyte and Slater2018). Therefore, by analysing the relationships between PLFAs and other organics, we can understand the degradation of PLFAs in extremely dry soils. However, it is worthy to note that archaea do not possess PLFAs, but phospholipid ether lipids instead (Quideau et al., Reference Quideau, Mcintosh, Norris, Lloret, Swallow and Hannam2016). The PLFA extraction protocol is not capable of and not developed for hydrolysing archaeal lipids, and hence PLFA analyses cannot be used to unveil archaeal communities.
As for microbial compositions, side chains and biochemical modifications of PLFAs remain distinctive among different taxonomic units. The position and number of unsaturation, hydroxylation and methylation of fatty acids reflect anaerobes, actinobacteria, cyanobacteria, fungi, protozoa, phytoplankton, plants, and sulphate or metal reducers (Vestal and White, Reference Vestal and White1989; Zelles, Reference Zelles1999). For example, normal straight-chain saturated fatty acids indicate all general bacteria; terminally branched fatty acids are indicative of Firmicutes and anaerobic Gram-negative bacteria; monounsaturated fatty acids demonstrate the existence of Proteobacteria (Dong et al., Reference Dong, Zhang, Jiang, Yu, Chapman, Lucas and Fields2006); and polyunsaturated fatty acids are biomarkers of microeukaryotes and fungi (Shi et al., Reference Shi, Becker, Bischoff, Turco and Konopka2002; Jenkins et al., Reference Jenkins, Waite, Blackburn, Husband, Rushton, Manning and O'donnell2009).
Additionally, PLFA molecules change their structures and chemical functional groups to adapt to environmental changes (Quideau et al., Reference Quideau, Mcintosh, Norris, Lloret, Swallow and Hannam2016): cyclopropyl rings (Δ) of fatty acids are usually synthesized by numerous bacteria as a response to oligotrophic conditions (Guckert et al., Reference Guckert, Hood and White1986; Kaur et al., Reference Kaur, Chaudhary, Kaur, Choudhary and Kaushik2005), they can also be indicative of halotolerant species such as Halomonas (Wang et al., Reference Wang, Wu, Ng, Tzeng and Shyu2012); the modification of the hydrophilic hydroxyl group (–OH) on fatty acids indicates these phospholipids are ready to be altered (e.g., hydrolysis to double bonds) by external environments (Wilhelm et al., Reference Wilhelm, Davila, Eigenbrode, Parenteau, Jahnke, Liu, Summons, Wray, Stamos, O'reilly and Williams2017).
Furthermore, results of PLFA analyses have high repeatability. The quantification of biomass and measurement of activity determined by PLFAs is consistent with microbial biomass carbon, substrate-induced respiration and adenosine triphosphate (ATP) approaches (Frostegard et al., Reference Frostegard, Tunlid and Baath1991; Fletcher et al., Reference Fletcher, Conley, Valdivia-Silva, Perez-Montano, Condori-Apaza, Kovacs, Glavin and Mckay2011; White and Ringelberg, Reference White, Ringelberg and Amy2017; Schulze-Makuch et al., Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018). As such, PLFA analyses have previously been successfully applied to quantifying microbial abundance in northern Chilean and southern Peruvian Atacama Desert, and determined the microbial abundance to vary from 105 to 107 cells per gram of soils (Connon et al., Reference Connon, Lester, Shafaat, Obenhuber and Ponce2007; Lester et al., Reference Lester, Satomi and Ponce2007; Shirey, Reference Shirey2013; Valdivia-Silva et al., Reference Valdivia-Silva, Karouia, Navarro-Gonzalez and Mckay2016; Schulze-Makuch et al., Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018). Microbial PLFA data can be further combined with the principal component analysis (PCA), to understand the relationships of microbial communities to environmental variables, e.g., elevation, pH, electrical conductivity, and concentrations of typical salts and organic substances (Kelly et al., Reference Kelly, Haggblom and Tate2003; Michalsen et al., Reference Michalsen, Peacock, Spain, Smithgal, White, Sanchez-Rosario, Krumholz and Istok2007).
Several previous researches studied PLFA profiles in the Atacama Desert. Schulze-Makuch et al. (Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018) found that the abundance and diversity determined by PLFA analyses were anticorrelated with aridity and were highly consistent with soil deoxyribonucleic acid (DNA)-based results (Schulze-Makuch et al., Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018). Knief et al. (Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2020) investigated a longitudinal elevation gradient in the hyperarid region. They found that the PLFA abundance and diversity were positively correlated with elevations, and monosaturated, cyclopropyl and methylated PLFAs were only detected in sites higher than 2000 m elevation (Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019). Shirey (Reference Shirey2013) found that environmental parameters (e.g., pH, soil conductivity, salt concentrations and organic matters) were associated with various classes of PLFAs, without a simple pattern (Shirey, Reference Shirey2013). Ziolkowski et al. (Reference Ziolkowski, Wierzchos, Davila and Slater2013) determined abundant saturated and monounsaturated fatty acids inside of hygroscopic minerals, and less abundant polyunsaturated and branched fatty acids were also detected in some cases (Ziolkowski et al., Reference Ziolkowski, Wierzchos, Davila and Slater2013). However, none of these previous studies have systematically examined the impact of a latitudinal precipitation gradient within the Atacama Desert on microbial community structure after recovery from an unprecedented heavy rainfall.
Based on the points above, PLFA analyses will provide an invaluable tool for understanding the general abundance and diversity of microbial organisms in Mars analogues (e.g., the Atacama Desert), and for comparing environmental parameters with biological diversity. The objective of this study is to (1) combine organic geochemical and statistical techniques to determine the abundance and diversity of microbial membrane phospholipids in Atacama soils across a latitudinal aridity gradient, and to (2) determine the relationships between these lipid biomarkers and environmental variables.
Materials and methods
Sampling approach
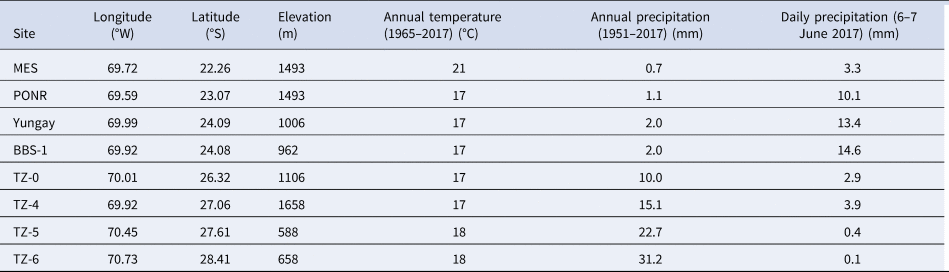
Details of sampling sites in the Atacama Desert, Chile, were itemized in Shen et al. (Reference Shen, Zerkle, Stueken and Claire2019). Briefly, samples were obtained in sterilized foils using aseptic techniques along a latitudinal transect from the north hyperarid sites María Elena South (MES), Point of No Return (PONR), Yungay, Badwater Basin South (BBS-1) to the south transition zone (TZ) sites TZ-0, TZ-4, TZ-5 and TZ-6 (Fig. 1). The mean annual precipitation rises from 0.7 to 30 mm towards the south (Table 1). Sites Yungay and BBS-1 were from the same location (Fig. 2), but the difference was that BBS-1 received more precipitation than Yungay during the unprecedented June 2017 rain event (Azua-Bustos et al., Reference Azua-Bustos, Fairen, Gonzalez-Silva, Ascaso, Carrizo, Fernandez-Martinez, Fernandez-Sampedro, Garcia-Descalzo, Garcia-Villadangos, Martin-Redondo, Sanchez-Garcia, Wierzchos and Parro2018) (6 months before sampling). Except for the BBS-1 moist soil sample collected beneath a 4.5 cm thick layer of evaporites, all other seven samples were dry sandy samples (~300 g) collected in the subsurface, 10–20 cm of depth, after removing the top rapidly transportable soils.

Fig. 1. Sampling sites across a latitudinal precipitation gradient in the Atacama Desert, Chile.

Fig. 2. Landscapes of (a) Yungay site and (b) BBS-1 site, only 8 km apart.
Table 1. Site information: latitude (determined by GPS), annual temperature, annual precipitation (data from the Center of Climate and Resilience Sciences, http://explorador.cr2.cl/) and the daily precipitation during the unprecedented heavy rainfall in June 2017 (data from Ventusky, https://www.ventusky.com/)
Phospholipid extraction, analysis and nomenclature
Solvents used in PLFA analyses were all high-performance liquid chromatography grade. All samples for PLFA analysis were freeze-dried within 2 weeks after collection. PLFAs were extracted from ~2 g aliquots of each freeze-dried sample (Turpin-Jelfs et al., Reference Turpin-Jelfs, Michaelides, Biederman and Anesio2019) together with a blank using a modified Bligh and Dyer method (1959) as described by Frostegard et al. (Reference Frostegard, Tunlid and Baath1991) by mixing with phosphate-buffered water:chloroform:methanol (v:v:v 4:5:10) (Bligh and Dyer, Reference Bligh and Dyer1959; Frostegard et al., Reference Frostegard, Tunlid and Baath1991). Total lipid extracts (TLEs) were fractionated and derivatized according to methods by Dickson et al. (Reference Dickson, Bull, Gates and Evershed2009) and Christie (Reference Christie1993), respectively. In short, TLEs in the organic phase were separated on a silicic gel column into simple lipid (SL), glycolipid (GL) and phospholipid (PL) fractions by eluting with chloroform, acetone and methanol, respectively (Dickson et al., Reference Dickson, Bull, Gates and Evershed2009). After lipid fractionation, 0.1 mg ml−1 n-nonadecane (C19H40) was added into the PL fraction as an internal standard to quantify the fatty acid methyl esters (FAMEs). The PL fraction was then further methylated with methanolic HCl (Christie, Reference Christie1993).
Individual phospholipids were analysed in duplicate on a Thermo Scientific TRACE 1300 gas chromatograph (GC) equipped with an Agilent VF-23 ms column (60 m × 0.32 mm × 0.15 μm; helium carrier gas at constant 2 ml min−1 flow rate) using the temperature program: 1 min at 50°C, followed by a 10°C min−1 ramp to 100°C, and a final heating rate of 4°C min−1 to 250°C that was maintained for 15 min. The mass spectra of individual phospholipids were obtained using a Thermo Scientific ISQ LT single quadrupole mass spectrometer in the scan range of 50–650 m z−1. The current was maintained at 50 μA with an electron impact voltage of 70 eV and an ion source temperature of 240°C.
Insignificant peaks of the chromatogram were filtered out based on the manual noise region using Thermo Xcalibur 3.0. FAME structures of the remaining valid peaks were identified by comparing their relative retention times and mass spectra with authentic laboratory reference standards, a mixture of 26 bacterial acid methyl esters (BAMEs) (Fig. S1 and Table S1). FAMEs were designated in terms of the total numbers of carbon atoms:numbers of double bonds. The ω indicates the double bonds on the nth carbon from the CH3- end. Prefixes i- and a- refer to iso- and anteiso-branched fatty acids, respectively. ‘cy’ and n-OH refer to cyclopropyl and hydroxy fatty acids, respectively. ‘cis’ and ‘trans’ differentiate the isomeric configurations of the carbon chain at double bond (Fan et al., Reference Fan, Zhang and Morrill2017).
Environmental variables and principal component analysis
Environmental variables such as elevation, pH, electrical conductivity, salt concentrations, total organic carbon (TOC) and total organic nitrogen (TON) were reported in Shen et al. (Reference Shen, Zerkle, Stueken and Claire2019). δ 13C of TOC and δ 15N of TON were determined together with TOC and TON by Elemental Analyzer-Isotope Ratio Mass Spectrometry (EA-IRMS) (Shen et al., Reference Shen, Zerkle, Stueken and Claire2019). Concentrations of microbial P extracted using the sequential soil phosphate extraction method (Tamburini et al., Reference Tamburini, Pistocchi, Helfenstein and Frossard2018) were reported in Shen et al. (Reference Shen, Smith, Claire and Zerkle2020b). Viable cells were determined using trypan blue assay, and the viability hereafter is defined as the percentage of viable cells. Trypan blue assay was performed within one month of sample collection: in duplicate, soils were suspended in 10× serially diluted two to four times to clear out any sand particles; 0.4% trypan blue stain was added to 9× of the dilute solution; viable (unstained) and non-viable (blue) microbial cells were mounted on a Hirschmann Instruments™ Counting Chamber and counted using oil immersion light microscopy (AmScope).
The proportions of individual phospholipids within each dry soil sample were analysed together with 11 environmental variables – elevation, pH, conductivity, TOC, TON, chloride (Cl−), nitrate (NO3−), sulphate (SO42−), microbial viability (Table S2), δ 13C-TOC and δ 15N-TON (Shen et al., Reference Shen, Zerkle and Clairein review) – by PCA using OriginPro 2019 (OriginLab Corp., 2019).
Results
Microbial biomass estimation and lipid compositions
Common classifications of phospholipids, e.g., normal straight-chain saturated fatty acids (nsat), hydroxy fatty acids, terminally branched saturated fatty acids (terbrsat), cyclopropyl fatty acids, monounsaturated fatty acids (monounsat) and polyunsaturated fatty acids (polyunsat), were determined within sampling sites in the Atacama Desert. Identified normal saturated fatty acids included C14:0, C15:0, C16:0, C17:0, C18:0, C19:0 and C20:0. Identified hydroxy fatty acids were 2-OH C12:0 and 3-OH C14:0. Identified terminally branched fatty acids were a-C15:0 and i-C16:0. Identified monounsaturated fatty acids were C16:1ω9 (cis), C18:1ω9 (cis) and C18:1ω9 (trans). The only identified cyclopropyl fatty acid was cyC19:0, and the only identified polyunsaturated fatty acid was C18:2ω9,12 (cis) (Table 2 and Fig. S2).
Table 2. Concentrations (means ± standard errors) of identified phospholipid fatty acids (PLFAs) in Atacama soils at 10–20 cm depth

n.d.: not detected or below limit of detection; nsat (saturated n-fatty acid): C14:0, C15:0, C16:0, C17:0, C18:0, C19:0, C20:0. hydroxy (hydroxy fatty acid): 2-OH C12:0, 3-OH C14:0; terbrsat (terminally branched saturated fatty acid): a-C15:0, i-C16:0. cyclopropyl (cyclopropane fatty acid): cyC19:0; monounsat (monounsaturated fatty acid): C16:1ω9 (cis), C18:1ω9 (cis), C18:1ω9 (trans); polyunsat (polyunsaturated fatty acid): C18:2ω9,12 (cis). Cell conversion: 2 × 104 cells pmol−1 FAME.
The microbial lipid analyses revealed the presence of even-numbered saturated, hydroxy, cyclopropyl and monounsaturated phospholipids in all sites. The absolute concentrations and numbers of all fatty acids generally increased from the hyperarid core toward the southern transition zone. The fatty acids were present in the range from C12 to C20, while the straight-chain even-numbered phospholipids were the predominant compounds. Several fatty acids existed only at the wettest sites, such as the terminally branched a-C15:0 and i-C16:0 in TZ-5, and the hydroxy 3-OH C14:0 in TZ-6.
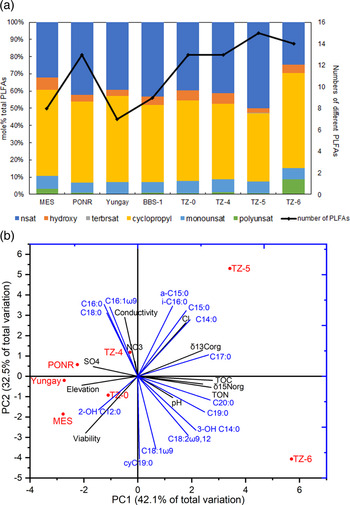
Using the conversion coefficient 2 × 104 cells pmol−1 FAME (Balkwill et al., Reference Balkwill, Leach, Wilson, Mcnabb and White1988; Connon et al., Reference Connon, Lester, Shafaat, Obenhuber and Ponce2007; Willers et al., Reference Willers, Van Rensburg and Claassens2015; Schulze-Makuch et al., Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018; Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019), viable cell numbers ranged from 3 × 106 cells g−1 in the hyperarid core to 5 × 107 cells g−1 in the transition zone (Table 2). The number of different individual PLFAs varied between 7 and 15 (Fig. 3a).

Fig. 3. (a) Mole per cent of PLFAs in Atacama soils. (b) Principal component analysis (PCA) plot of PLFA proportion profiles. Red dots represent sampling sites in the Atacama Desert, blue lines represent the PLFA species, and black lines represent measured environmental parameters. TOC, total organic carbon; TON, total organic nitrogen; δ 13Corg, δ 13C of TOC; δ 15Norg, δ 15N of TON.
The relative proportions of normal saturated phospholipids (25–50%) and the cyclopropyl cyC19:0 (40–55%) were the most abundant in all sites. Monounsaturated phospholipids C16:1 and C18:1 remained stable at around 6.8 ± 0.7%. The percentage of hydroxy phospholipids varied between 3 and 7%. The polyunsaturated C18:2ω9,12 was more dominant in TZ-6 than the other sites (Fig. 3a). Terminally branched saturated phospholipids were only detected in TZ-5 (Table 2).
PLFAs and environmental variables
PCA generated two primary principal components that took over 42.1 and 32.5% of total variation for combining and reducing the dimension of multiple variables to allow the plotting of these variables all in one graph. Hydroxy fatty acid 2-OH C12:0 was associated with elevation and viability. C16:0, C18:0 and C16:1ω9 were associated with conductivity, nitrate and sulphate concentrations. All the remaining PLFAs were associated with higher organic C and organic N (as total organic content), and increasing δ 13C-TOC and δ 15N-TON (as indicative of biological metabolisms (Ewing et al., Reference Ewing, Michalski, Thiemens, Quinn, Macalady, Kohl, Wankel, Kendall, Mckay and Amundson2007, Reference Ewing, Macalady, Warren-Rhodes, Mckay and Amundson2008; Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019)), which characterized TZ-5 and TZ-6 sites of the highest annual precipitations. Moreover, terminally branched saturated PLFAs were also associated with chloride concentration; and 3-OH C14:0, C18:2ω9,12, C18:1ω9 and cyC19:0 were also associated with alkalinity (Fig. 3b).
Discussion
Since the composition of Atacama soils are highly heterogeneous due to the absence of homogenization such as dissolution, results and their interpretations reported in this article only suggest the findings from these eight sampled single points in this study but not conclusive universally. Across the latitudinal precipitation gradient of the Mars-like Atacama soils, the diversity of PLFAs increased toward the wetter sites in the transition zone, although no universal trend was observed in the compositional structure of different classes of phospholipids across the precipitation gradient (Fig. 3a). Different individual PLFAs demonstrated their adaptations to the extreme environment in the Atacama Desert:
(1) Even-numbered saturated fatty acids generally dominated the phospholipid pool in all Atacama Desert sites (Fig. 3a) as in other Earth ecosystems (Moss et al., Reference Moss, Smith, Tavernier, Burrows and Stohner1997).
(2) The two terminally branched saturated fatty acids (a-C15:0 and i-C16:0) were only detectable in TZ-5, indicating more Firmicutes there (Dong et al., Reference Dong, Zhang, Jiang, Yu, Chapman, Lucas and Fields2006), which was consistent with DNA taxonomic analysis, 34.9% of Firmicutes in TZ-5 compared to 0.0–5.7% in other sites (Ben-David et al., Reference Ben-David, Zaady, Sher and Nejidat2011; Shen et al., Reference Shen, Shirey, Wyness, Claire and Zerkle2020a).
(3) Similarly, in consistence with taxonomic analysis, the proportion of Proteobacteria did not change much between sites: 11–13% Proteobacteria as observed in identified taxa (Shen et al., Reference Shen, Shirey, Wyness, Claire and Zerkle2020a) and 5.8–7.7% Proteobacteria-derived monounsaturated fatty acids as found in this study.
(4) 3-OH C14:0 was only detectable in TZ-6, indicating a higher diversity but not necessarily higher abundance in hydroxy fatty acid (Table 2 and Fig. 3a).
(5) The abundant cyclopropyl cyC19:0, occasionally even more than normal saturated PLFAs, in all sites reflected that Atacama microbiomes endeavoured to resist the infertile and hypersaline environments (Guckert et al., Reference Guckert, Hood and White1986; Kaur et al., Reference Kaur, Chaudhary, Kaur, Choudhary and Kaushik2005).
(6) The high proportion of the eukaryotic and fungal biomarker C18:2ω9,12 in site TZ-6 implied a larger proportion of eukaryotes (e.g., higher plants, protozoa and fungi) (Shi et al., Reference Shi, Becker, Bischoff, Turco and Konopka2002; Jenkins et al., Reference Jenkins, Waite, Blackburn, Husband, Rushton, Manning and O'donnell2009) but a smaller proportion of bacteria within this wettest sampling site (Fig. 3a).
At about 20 cm depth, Schulze-Makuch et al. (Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018) determined similar compositions of PLFAs but with more monounsaturated (Proteobacteria) and terminally branched (Firmicutes and anaerobic Gram-negative bacteria) fatty acids in the hyperarid core (Schulze-Makuch et al., Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018). The microbial abundance in Schulze-Makuch et al. (Reference Schulze-Makuch, Wagner, Kounaves, Mangelsdorf, Devine, De Vera, Schmitt-Kopplin, Grossart, Parro, Kaupenjohann, Galy, Schneider, Airo, Frosler, Davila, Arens, Caceres, Cornejo, Carrizo, Dartnell, Diruggiero, Flury, Ganzert, Gessner, Grathwohl, Guan, Heinz, Hess, Keppler, Maus, Mckay, Meckenstock, Montgomery, Oberlin, Probst, Saenz, Sattler, Schirmack, Sephton, Schloter, Uhl, Valenzuela, Vestergaard, Wormer and Zamorano2018) was between 5 × 104 and 5 × 106 cells per gram of soils, generally lower than this study. The lower biomass and different PLFA compositions might be a result of the timing of sampling: they sampled 1 month after an even more massive rainfall (33.3 mm) in 2015 that could damage most microbial communities in a short period, with Proteobacteria and Firmicutes being the dominant survivors (Azua-Bustos et al., Reference Azua-Bustos, Fairen, Gonzalez-Silva, Ascaso, Carrizo, Fernandez-Martinez, Fernandez-Sampedro, Garcia-Descalzo, Garcia-Villadangos, Martin-Redondo, Sanchez-Garcia, Wierzchos and Parro2018; Fernandez-Martinez et al., Reference Fernandez-Martinez, Severino, Moreno-Paz, Gallardo-Carreno, Blanco, Warren-Rhodes, Garcia-Villadangos, Ruiz-Bermejo, Barberan, Wettergreen, Cabrol and Parro2019). Sampling of this study was performed 6 months after the heavy rainfall in 2017, and microbial communities could partly recover (Orlando et al., Reference Orlando, Alfaro, Bravo, Guevara and Caru2010; Armstrong et al., Reference Armstrong, Valverde, Ramond, Makhalanyane, Jansson, Hopkins, Aspray, Seely, Trindade and Cowan2016; Uritskiy et al., Reference Uritskiy, Getsin, Munn, Gomez-Silva, Davila, Glass, Taylor and Diruggiero2019) to pre-disturbance abundance (Connon et al., Reference Connon, Lester, Shafaat, Obenhuber and Ponce2007; Lester et al., Reference Lester, Satomi and Ponce2007; Shirey, Reference Shirey2013; Valdivia-Silva et al., Reference Valdivia-Silva, Karouia, Navarro-Gonzalez and Mckay2016; Finstad et al., Reference Finstad, Probst, Thomas, Andersen, Demergasso, Echeverria, Amundson and Banfield2017; Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019).
PLFA studies in Negev Desert, Israel, supported that cyclopropyl fatty acids were more abundant in more arid locations by comparing intershrub arid and semi-arid regions (Ben-David et al., Reference Ben-David, Zaady, Sher and Nejidat2011). Knief et al. (Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019) have also detected abundant but less dominant cyclopropyl fatty acids in a more northern region of the Atacama Desert in 2014 before massive rainfall input, and they found much more cyC17:0 than cyC19:0 (Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019). The fatty acid cyC19:0 was detected more in Proteobacteria (Wang et al., Reference Wang, Wu, Ng, Tzeng and Shyu2012), indicating a more proportion of Proteobacteria in soil samples of this study as the microbial post-rainfall recovery was still in progress.
The increase of C18:2ω9,12 from dry soils of Yungay (0.0%) to PONR (1.1%) and to MES (3.3%) (Table 2) implicated that an appropriate rather than a massive amount of daily rainfall (Table 1) was more beneficial to eukaryotic and fungal growth. Similarly, Ziolkowski et al. (Reference Ziolkowski, Wierzchos, Davila and Slater2013) did not find C18:2ω9,12 in the Yungay region, but they detected C18:2ω9,12 and much more monounsaturated fatty acids from the interiors of hygroscopic minerals in a salt flat and meteorite impact crater regions of Atacama (Ziolkowski et al., Reference Ziolkowski, Wierzchos, Davila and Slater2013). Compared to Yungay and MES, PONR had more cell counts and numbers of different PLFAs; MES had the lowest biomass, while Yungay had the least numbers of different PLFAs in the hyperarid core (Table 2 and Fig. 3a). In summary, different precipitations of desert rare heavy rainfall (Table 1) made different impacts on bacteria, eukaryotes, biomass and PLFA diversity. Especially, the relatively low daily precipitation at hyperarid MES stimulated eukaryotic growth under more alkaline conditions that were less electrically conductive and saline (Fig. 3); the PONR sample was characterized by high sulphate repository (Fig. 3b) that could have stored moisture in the form of hydration water (Palacio et al., Reference Palacio, Azorin, Montserrat-Marti and Ferrio2014) after rainfall events, which might result in its high biomass that was even equivalent to the southern transition sites (Table 2).
Although phospholipid studies in Gurbantunggut Desert, a temperate desert in northwestern China, suggests that rainwater addition does not affect local microbial composition and the bacteria to fungi ratios (Huang et al., Reference Huang, Li and Su2015a, Reference Huang, Li and Su2015b), results of this study do not wholly agree with this finding. Comparing the dry sandy soils in Yungay and wet soils in BBS-1 (Fig. 2) after the rare rainfall event 6 months ago, we can determine how the conformation of membrane fatty acids have adapted to sudden changes in the Mars-analogous Atacama settings, from a long-lasting extremely dry rocky environment to an oxidative hypersaline brine induced by a flash flood. The viable cell numbers and the number of individual PLFAs were not significantly different between Yungay and BBS-1 (Table 2 and Fig. 3a), since heavy rainfall not only brought moisture but also caused osmotic damages to microbial communities (Armstrong et al., Reference Armstrong, Valverde, Ramond, Makhalanyane, Jansson, Hopkins, Aspray, Seely, Trindade and Cowan2016; Azua-Bustos et al., Reference Azua-Bustos, Fairen, Gonzalez-Silva, Ascaso, Carrizo, Fernandez-Martinez, Fernandez-Sampedro, Garcia-Descalzo, Garcia-Villadangos, Martin-Redondo, Sanchez-Garcia, Wierzchos and Parro2018; Fernandez-Martinez et al., Reference Fernandez-Martinez, Severino, Moreno-Paz, Gallardo-Carreno, Blanco, Warren-Rhodes, Garcia-Villadangos, Ruiz-Bermejo, Barberan, Wettergreen, Cabrol and Parro2019). As for the classes of PLFAs, the proportions of normal saturated, hydroxy and polyunsaturated fatty acids were higher in BBS-1, while cyclopropyl and monounsaturated fatty acids were higher in Yungay. These results indicated that general bacteria, eukaryotes and hydroxy PLFAs increased, but Gram-negative bacteria became less dominant (Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019).
Various PCA results of desert microbial PLFAs have been previously demonstrated to be convenient to clearly separate regions of different environmental characteristics (Liu et al., Reference Liu, Li, Xing, Zhao and Pan2013; Li et al., Reference Li, Zhang, Hu, Chen, Zhu, Shen and Fang2017; Lu et al., Reference Lu, Ma, Chen, Yangzom and Jiang2018; Bao et al., Reference Bao, Gao, Wang, Yang, Ren and Zhao2020; Zuo et al., Reference Zuo, He, He, Li, Xue, Li and Wang2020). According to PCA, (1) the dominance of fatty acid hydroxylation was associated with viability (Fig. 3b): hydroxylation-driven alteration of membrane PLFAs might suggest the rapid degradation rate of dead cells after the unprecedented rainfall (Gutteridge, Reference Gutteridge1982; Green and Scow, Reference Green and Scow2000; Wilhelm et al., Reference Wilhelm, Davila, Eigenbrode, Parenteau, Jahnke, Liu, Summons, Wray, Stamos, O'reilly and Williams2017), which decreased the percentage of non-viable cells and concomitantly increased viability. (2) The link between C16:0, C18:0, C16:1ω9 and the two environmental factors conductivity and nitrate implied that the general bacteria and Proteobacteria rich in these even-numbered PLFAs were halophilic and had nitrate metabolisms. (3) Other saturated and unsaturated PLFAs contributed more to the biomass and isotopic biomarkers (Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019). Moreover, C16:0, C18:0 and C16:1ω9 showed preferences to acidic pH; C17:0, C19:0, C20:0, 3-OH C14:0, C18:1ω9, C18:21ω9,12 and cyC19:0 showed preferences to alkaline pH; and C14:0, C15:0, a-C15:0, i-C16:0 and 2-OH C12:0 did not associate with soil pH values. (4) Higher organic C and N isotopic ratios (δ 13C-TOC and δ 15N-TON) in TZ-5 and TZ-6 implied more active microbial community because of the transformation of C and N via biological activity, such as the incorporation of atmospheric CO2, denitrification and organic N loss (Carlson and Ingraham, Reference Carlson and Ingraham1983; Marsh et al., Reference Marsh, Sims and Mulvaney2005; Hare et al., Reference Hare, Loftus, Jeffrey and Ramsey2018; Knief et al., Reference Knief, Bol, Amelung, Kusch, Frindte, Eckmeier, Jaeschke, Dunai, Fuentes and Mörchen2019).
The microbial concentrations converted from PLFAs were on the same level as the viable cell counts determined by trypan blue assay (Table S2) of low precision, and were consistent with the cell numbers determined by other studies (Connon et al., Reference Connon, Lester, Shafaat, Obenhuber and Ponce2007; Lester et al., Reference Lester, Satomi and Ponce2007; Shirey, Reference Shirey2013). By analysing the relationships between PLFAs and TOC as well as microbial P (Shen et al., Reference Shen, Zerkle, Stueken and Claire2019, Reference Shen, Smith, Claire and Zerkle2020b), we can understand the origin and degradation of organics in the extremely dry soils. Within the hyperarid core, all the concentrations of PLFAs, TOC and microbial P shared an identical trend: PONR site had the highest biomass, MES had the lowest (Azua-Bustos et al., Reference Azua-Bustos, Caro-Lara and Vicuna2015) and Yungay was in-between (Maier et al., Reference Maier, Drees, Neilson, Henderson, Quade and Betancourt2004; Kuhlman et al., Reference Kuhlman, Venkat, La Duc, Kuhlman and Mckay2008; Azua-Bustos et al., Reference Azua-Bustos, Urrejola and Vicuna2012). PLFAs and TOC both showed a steep leap from site TZ-4 to TZ-5 (Fig. 4a). The ratios of PLFA to TOC in the hyperarid core were more than twice of the ratios in the south transition zone (Fig. 4a and Table 3), indicating PLFAs degraded slower at increasing aridity (Ewing et al., Reference Ewing, Michalski, Thiemens, Quinn, Macalady, Kohl, Wankel, Kendall, Mckay and Amundson2007; Wilhelm et al., Reference Wilhelm, Davila, Eigenbrode, Parenteau, Jahnke, Liu, Summons, Wray, Stamos, O'reilly and Williams2017; Jones et al., Reference Jones, Olivera-Ardid, Klumpp, Knief, Huil, Lehndorff and Bol2018). Although the degradation of PLFAs was dependent on ambient temperatures (Ranneklev and Baath, Reference Ranneklev and Baath2003), the mean annual temperatures within all Atacama sampling sites were 17.8 ± 1.3°C (Table 1) (Shen et al., Reference Shen, Smith, Claire and Zerkle2020b). Thus, the impacts of variable temperatures should be insignificant, and the aridity was the primary regulator of PLFA degradation.

Fig. 4. Plots of the concentrations of PLFAs versus (a) total organic carbon (TOC) and (b) microbial phosphorus (P).
Table 3. Mass ratios of PLFAs to TOC and microbial P in each Atacama site

–, not measured.
Concerning the ratios of PLFA to microbial P, they did not change in the hyperarid core, whereas the microbial P in transition zone did not correlate with PLFAs (Fig. 4b). Hence, the membrane PLFA components degraded more rapidly than other phosphorus-containing biomolecules, such as DNA and intracellular calcium phosphate, in the less arid southern sites. The degradation of PLFAs reached a minimum when the mean annual precipitation is lower than 2 mm (Table 1). Moreover, the constant ratios of PLFA/TOC and PLFA/microbial P in the hyperarid Atacama Desert (Table 3) suggested that these microbial organics were derived directly from the native species, with negligible sources from organics in the form of seawater droplets over the Pacific Ocean (Wang et al., Reference Wang, Michalski, Seo and Ge2014; Azua-Bustos et al., Reference Azua-Bustos, Gonzalez-Silva and Corsini2017) transported by wind (Azua-Bustos et al., Reference Azua-Bustos, Gonzalez-Silva, Fernandez-Martinez, Arenas-Fajardo, Fonseca, Martin-Torres, Fernandez-Sampedro, Fairen and Zorzano2019).
By applying PLFA analysis, it should be noted that not all PLFA components are known in every microbial species (Watzinger, Reference Watzinger2015), and most individual PLFAs correspond with multiple microbial taxa and thus have a low taxonomic resolution (White et al., Reference White, Stair and Ringelberg1996; Olsson, Reference Olsson1999; Ruess and Chamberlain, Reference Ruess and Chamberlain2010; Willers, Reference Willers2016). Different researches usually use very different BAME laboratory reference standards for lipid peak identification, which makes conclusions drawn by comparative studies less strong. Besides, the baseline of gas chromatograms of FAMEs can be noisy to pester the peak identification especially for such dry soils of low microbial abundance. Previous studies imply that the living microorganisms may be overestimated because some deceased biomass can remain in these extremely dry soils (Kowalenko and Mckercher, Reference Kowalenko and Mckercher1971; Frostegard and Baath, Reference Frostegard and Baath1996). In spite of these caveats, PLFA analysis is still a valuable and sensitive tool to investigate both structural and adaptational facets of microbial communities in a single experiment.
On Mars, if microbial life ever thrived formerly, these microorganisms can be preserved in the subsurface with minimal degradation in the absence of precipitation for billions of years (Brocks and Banfield, Reference Brocks and Banfield2009). Given abundant chloride, nitrate and sulphate salts on Mars (Altheide et al., Reference Altheide, Chevrier, Nicholson and Denson2009; Hecht et al., Reference Hecht, Kounaves, Quinn, West, Young, Ming, Catling, Clark, Boynton, Hoffman, Deflores, Gospodinova, Kapit and Smith2009; Stern et al., Reference Stern, Sutter, Freissinet, Navarro-Gonzalez, Mckay, Archer, Buch, Brunner, Coll, Eigenbrode, Fairen, Franz, Glavin, Kashyap, Mcadam, Ming, Steele, Szopa, Wray, Martin-Torres, Zorzano, Conrad, Mahaffy and Team2015), membrane lipids (especially even-numbered normal saturated fatty acids) and other microbial organics are expected to be detectable. If we assume the microbial abundance in Martian subsurface soils are about the same as the minimum reported levels in the Atacama Desert, hundreds of cells per gram (Navarro-Gonzalez et al., Reference Navarro-Gonzalez, Rainey, Molina, Bagaley, Hollen, De La Rosa, Small, Quinn, Grunthaner, Caceres, Gomez-Silva and Mckay2003; Bagaley, Reference Bagaley2006), then tens of kilograms of soils might be enough to extract total organic compounds that can be analysed by the Mars Organic Molecule Analyzer equipped with GC-MS and proper separation columns (Goesmann et al., Reference Goesmann, Brinckerhoff, Raulin, Goetz, Danell, Getty, Siljestrom, Missbach, Steininger, Arevalo, Buch, Freissinet, Grubisic, Meierhenrich, Pinnick, Stalport, Szopa, Vago, Lindner, Schulte, Brucato, Glavin, Grand, Li and Van Amerom2017).
Conclusions
The numbers of cells and PLFAs generally increase along the precipitation gradient, but the PLFA compositions do not follow any particular pattern. The degradation of PLFAs is slower as the mean annual precipitation decreases. When the annual precipitation is lower than 2 mm, the PLFA degradation becomes minimal. The biomass in each Atacama Desert site that is not artificially contaminated is predominantly derived from native microbial communities, with negligible foreign organic sources transported from the Pacific Ocean. The abundant cyclopropane structure of membrane PLFAs reflects the starvation and halotolerant responses of Atacama extremophiles to their harsh environments. If potentially extant microbes existed on Mars, cyclopropyl and even-numbered normal saturated phospholipids are expected to be preserved and detectable.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1473550420000294
Acknowledgements
This research was funded by the Natural Environment Research Council (Grant Agreement NEIF BRIS/135/0719) (to J.S.) and the China Scholarship Council (CSC).
Conflict of interest
None.