Introduction
According to the Mendelian segregation, a specific allele at a specific chromosomal locus has an equal chance (50:50) to exist in an individual gamete without any selective pressure. Gene-drive dynamics occur when a particular allele is segregated in the way to disrupt the Mendelian segregation. Gene drive systems have capabilities to provide innovative solutions to major environmental and public health problems (Hamilton, Reference Hamilton1967; Curtis, Reference Curtis1968). The majority of developed gene drives are based on naturally occurring selfish genetic elements that work by rising in frequency with each generation without providing any fitness benefit to their hosts, causing non-Mendelian inheritance patterns (Burt, Reference Burt2014). These characteristics of gene drive systems enable the scientist to overcome the genetic and evolutionary drawbacks of some desirable traits by disseminating them to wild-type populations or completely suppressing the population (Alphey et al., Reference Alphey, McKemey, Nimmo, Neira Oviedo, Lacroix, Matzen and Beech2013; Burt, Reference Burt2014). As a result, gene drive systems facilitate the development of innovative strategies for reducing or eliminating the insect-born diseases and even overcome the development of pesticides resistances in a cost-effective and eco-friendly manner (Champer et al., Reference Champer, Buchman and Akbari2016). Furthermore, gene drives might also be used to spread desired traits to the population more quickly than natural selection to protect the endangered species by disseminating pathogen-resistant genes among targeted populations (Champer et al., Reference Champer, Buchman and Akbari2016).
The idea of utilization of gene drive to suppress the vector and pest population emerged in 1960 when naturally occurring male-producing factors were used to suppress the Aedes aegypti (Craig et al., Reference Craig, Hickey and VandeHey1960; Curtis, Reference Curtis1968). Subsequently, a sex-determining chromosomal region in mosquitoes and its potential use for population suppression were defined (Hickey and Craig, Reference Hickey and Craig1966). Later on, Hamilton demonstrated the genes that distribute through the non-Mendelian process might cause population elimination (Hamilton, Reference Hamilton1967) and Curtis explored the prospect of the transformation of malaria mosquito populations by employing self-spreading chromosomal variants (Curtis, Reference Curtis1968). The basic research in molecular biology and genetics continue until the possible, and advanced genetic engineering tools are available. In 1992, an idea and mechanism of utilizing transposable elements were proposed to drive a genetically engineered gene into the mosquito populations (Kidwell and Ribeiro, Reference Kidwell and Ribeiro1992). A substantial stimulus was the discovery of selfish genetic elements that originate in numerous single-cell organisms, called homing-endonuclease genes (HEGs) (Burt, Reference Burt2003). Subsequently, homing endonuclease-based gene drive (HEGD) was developed with HEGs and used to suppress the population of malaria vectors (Stoddard, Reference Stoddard2005). Population biologists and geneticists continued to study how to use different selfish genetic elements as the systematic source for the development of a gene-drive system in mosquitoes (James, Reference James2005; Adelman et al., Reference Adelman, Jasinskiene, Onal, Juhn, Ashikyan, Salampessy, MacCauley and James2007; Windbichler et al., Reference Windbichler, Menichelli, Papathanos, Thyme, Li, Ulge, Hovde, Baker, Monnat and Burt2011). Then, the CRISPR/Cas9-based endonuclease system in bacteria was discovered and modified to produce an artificial drive system (Jinek et al., Reference Jinek, Chylinski, Fonfara, Hauer, Doudna and Charpentier2012; Rode et al., Reference Rode, Estoup, Bourguet, Courtier-Orgogozo and Débarre2019). CRISPR/Cas9-based gene drive (CCGD) was developed and used in Drosophila melanogaster (Gantz and Bier, Reference Gantz and Bier2015), Anopheles stephensi (Gantz et al., Reference Gantz, Jasinskiene, Tatarenkova, Fazekas, Macias, Bier and James2015), Anopheles gambiae (Kyrou et al., Reference Kyrou, Hammond, Galizi, Kranjc, Burt, Beaghton, Nolan and Crisanti2018), Aedes aegypti (Basu et al., Reference Basu, Aryan, Overcash, Samuel, Anderson, Dahlem, Myles and Adelman2015; Williams et al., Reference Williams, Franz, Reid and Olson2020) and Tribolium castaneum (Drury et al., Reference Drury, Dapper, Siniard, Zentner and Wade2017) to suppress and replace the wild-type population.
Nowadays, there is great interest in using of gene-drive technologies to suppress the pest populations (James, Reference James2005). A systemic approach requires for designing effective and safe gene drive systems that involve in the establishment of this system by discovering naturally occurring or synthetic genetic mechanisms for gene spread (Letourneau and Burrows, Reference Letourneau and Burrows2001; James Reference James2005). The most essential and critical criterion is that the gene-drive system must be strong enough to transfer the desirable effector genes to the next population (Boete and Koella, Reference Boete and Koella2003). Four gene-drive systems, i.e. meiotic drive (MD), under-dominance (UD), HEGD and CCGD have been successfully developed and used to control the pest populations. The genetic alteration in gene-drive systems would continue in the future to manage the wild-type population of harmful pests (Godfray et al., Reference Godfray, North and Burt2017; Asad et al., Reference Asad, Munir, Xu, Li, Jiang, Chu and Yang2020). The vital purpose of this review is to discuss different types of gene drives and their potential applications to control the pest and disease vector populations.
Gene-drive systems in insects
Meiotic drive (MD)
MD takes place when a specific heterozygous locus segregates at a higher rate than the typical Mendelian ratio by disabling or disturbing the homologous chromosome (Benedict, Reference Benedict2014). The sex chromosome-based MD states the inadequate transmission of X and Y chromosomes from individuals of heterogametic sex, resulting in a bias sex ratio between the offspring of the population (Helleu et al., Reference Helleu, Gérard and Montchamp-Moreau2015). A lot of articles and reviews have discussed the development and detailed history of MD. We are focusing on X and Y MDs that have been successfully developed in insects during the past few years.
The two basic types, X and Y chromosome-based MD systems have been used in mosquito and Drosophila. In Drosophila, the X chromosome-based MD is associated with abnormal behavior of the Y chromosome during meiosis II, which can lead to failure of sister chromatid segregation (Keais et al., Reference Keais, Hanson, Gowen and Perlman2017). The X chromosome-based MD can be very strong, often leading to 100% female progeny or near to it (Helleu et al., Reference Helleu, Gérard and Montchamp-Moreau2015).
Y-drive is also linked with the breakage of X chromosome during anaphase I, which leads to the failure of crossover. The segregation distortion occurs during spermatogenesis and leads to Y chromatid non-disjunction at meiosis II or breakage of X chromosome for a failure of spermatogenesis. During meiosis, the non-random segregation of chromosomes occurs through the spermatogenesis of heterogametic male flies of Trichosia pubescens, although in a female-only single gamete is formed during meiosis. The development of only a single gamete is due to monocentric and monopolar spindles during the first meiotic division. In this case, non-random segregation of genetic material is due to the deactivation of paternal chromosomal kinetochores that are incapable of holding the microtubules of the spindle. (Rutkowska and Badyaev, Reference Rutkowska and Badyaev2008).
The potential for the MD is probably high in all sexual organisms, which can alter the natural populations by using the synthetic MD (Burt, Reference Burt2003). The sex-chromosome drive should cause either local population collapse or the preference of suppressor alleles, and therefore, it is difficult to use. However, it will be self-limiting and continuous-release, similar to that required with the sterile insect technique (Alphey et al., Reference Alphey, McKemey, Nimmo, Neira Oviedo, Lacroix, Matzen and Beech2013).
Under-dominance (UD)
Underdominance (UD), also known as heterozygote inferiority, occurs when heterozygotes (or their progeny) have lower fitness than parental homozygotes (Hartl et al., Reference Hartl, Clark and Clark1997). There are two basic types of engineer UD, two-locus UD and single-locus UD (fig. 1) (Dhole et al., Reference Dhole, Vella, Lloyd and Gould2018).
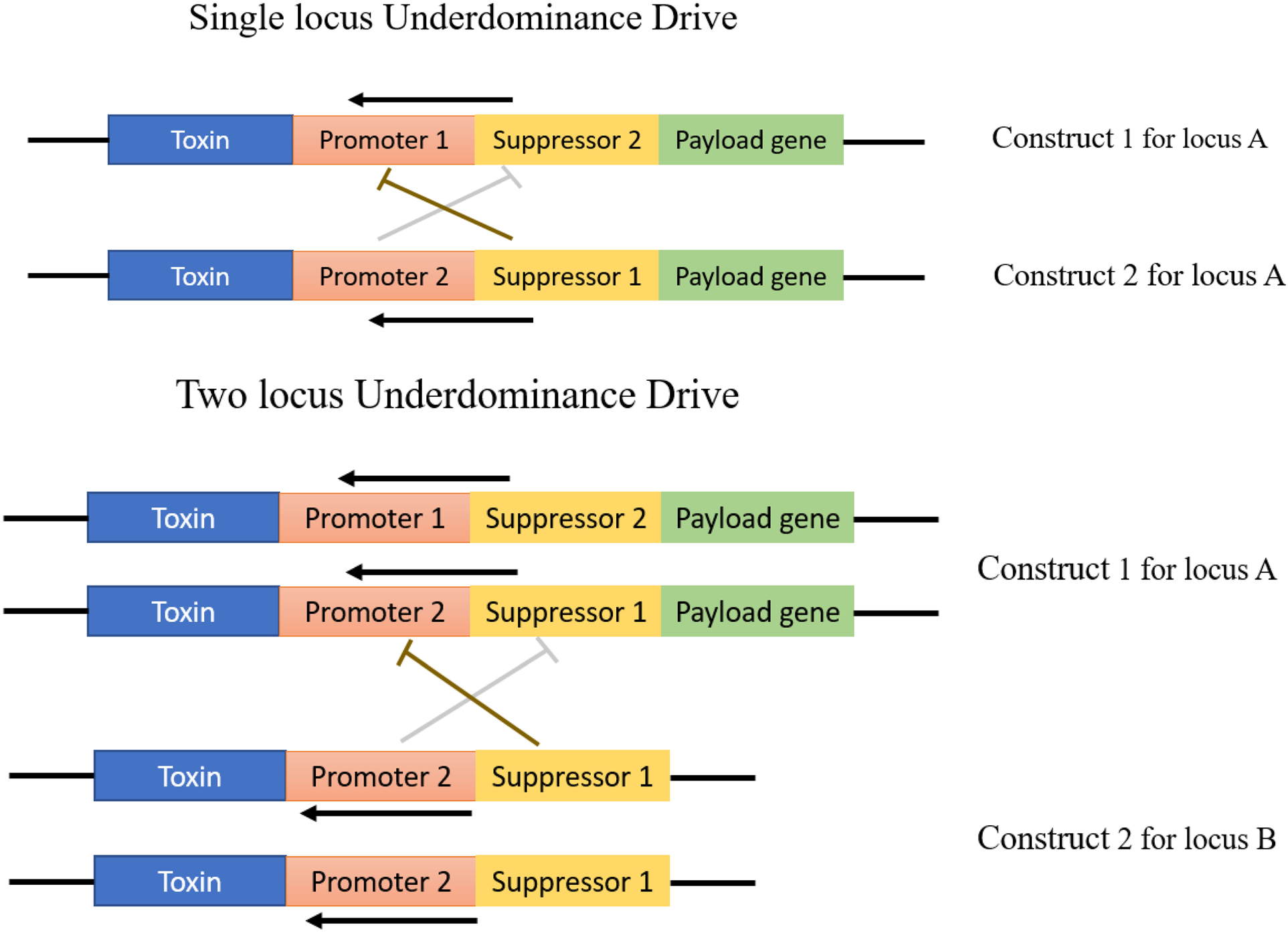
Figure 1. Representation of under-dominance drive. The upper two constructs of under-dominance drive for single locus A and lower two constructs under-dominance for two locus of A and B. Each construct contains a promoter, a suppressor for each promoter, a toxin and payload gene.
In a two-allele UD system, one allele is generally fixed in large interbreeding insect populations, whereas the other allele is lost; this bi-stable switch, like the action of UD tends to make it a gene-drive mechanism (Dhole et al., Reference Dhole, Vella, Lloyd and Gould2018). The initial frequency of the two alleles is the primary determinant for which one to lose. Hence, for two alleles, the heterozygote X/x (X and x are two alleles) has lower fitness than the parental homozygote X/X or x/x. These homozygous individuals can be reproductively separated from each other at extreme UD conditions when heterozygote individuals will be 100% lethal (Eppstein et al., Reference Eppstein, Payne and Goodnight2009). The model of two-locus UD represents two constructs that are positioned at independently segregating loci (fig. 1) (Dhole et al., Reference Dhole, Vella, Lloyd and Gould2018). In single-locus UD, both constructs can insert a single locus (Marshall and Hay, Reference Marshall and Hay2012). The model of one-locus UD contains two engineered constructs, and each composes of four different linked elements of a coding gene for a lethal toxin, a suppressor specific to the promoter of another construct, a promoter to drive the toxin gene and a payload gene (the desired gene to be spread into population) (fig. 1) (Dhole et al., Reference Dhole, Vella, Lloyd and Gould2018).
The mechanism to transform the insect population can be demonstrated by a transgenic construct based on UD. It presumes that wild-type (W+/W+) and under-dominant transgenic homozygotes (Ud/Ud) have an equal chance of fitness. By releasing (Ud)/(Ud) homozygotes into a target wild-type population, the (Ud) allele frequency might be increased above a 0.5 threshold frequency and then (Ud) alleles will probably eradicate the wild-type alleles for following generations. The eradication of these wild-type alleles continues even after the stopping release of transgenic under-dominant alleles (Davis et al., Reference Davis, Bax and Grewe2001; Reeves et al., Reference Reeves, Bryk, Altrock, Denton and Reed2014).
In the engineer UD system, it is presumed that even one copy of the heterozygous suppressor at one locus is enough to suppress all other copies of promoters on other constructs. So, only those individuals will survive who have one copy of the transgenic allele or no transgenic allele (Marshall and Akbari, Reference Marshall, Akbari and Adelman2016; Edgington and Alphey, Reference Edgington and Alphey2017). The single-locus UD requires a high release of a population containing drive payload, making it difficult to engineer. In contrast, the two-locus UD system requires a low release of a population containing drive payload (Dhole et al., Reference Dhole, Vella, Lloyd and Gould2018).
Homing endonuclease-based gene drive (HEGD)
The discovery of HEGD was initially stimulated by the finding of selfish genetic elements that originated in numerous single-cell organisms, named HEGs, and has been used to suppress the population of malaria vectors and other diseases related to human (Stoddard, Reference Stoddard2005).
These HEG-encoded proteins can recognize and cut a 20- to 30-bp DNA sequence (Stoddard, Reference Stoddard2005). However, once the HEGs come across into contact with a chromosome harboring the recognition sequence, then induce the double-strand DNA break (DSB), which is frequently repaired by homology-directed repair (HDR) and non-homologous end joining (NHEJ) (fig. 2). In the HDR, the HEG uses itself as a repairing template for this process, and as a consequence, the HEG element copies itself into its corresponding allele (Stoddard, Reference Stoddard2011). Under these dynamics, the HEG frequency can be rapidly increased in a population (Windbichler et al., Reference Windbichler, Menichelli, Papathanos, Thyme, Li, Ulge, Hovde, Baker, Monnat and Burt2011; Hammond et al., Reference Hammond, Galizi, Kyrou, Simoni, Siniscalchi, Katsanos, Gribble, Baker, Marois and Russell2016). The bias inheritance of HEG is through cutting the homologous chromosome and stimulating the copy of itself during the repair of the break. Various efforts have revolved about the prospect of using this gene-drive system to block the transmission of malaria by targeting mosquitoes (Galizi et al., Reference Galizi, Doyle, Menichelli, Bernardini, Deredec, Burt, Stoddard, Windbichler and Crisanti2014). If this procedure takes place in the germline, the ratio of gametes that comprise the HEGD will be higher than 0.5 and it has been proved to be over 0.9 in a few cases (Burt, Reference Burt2003).
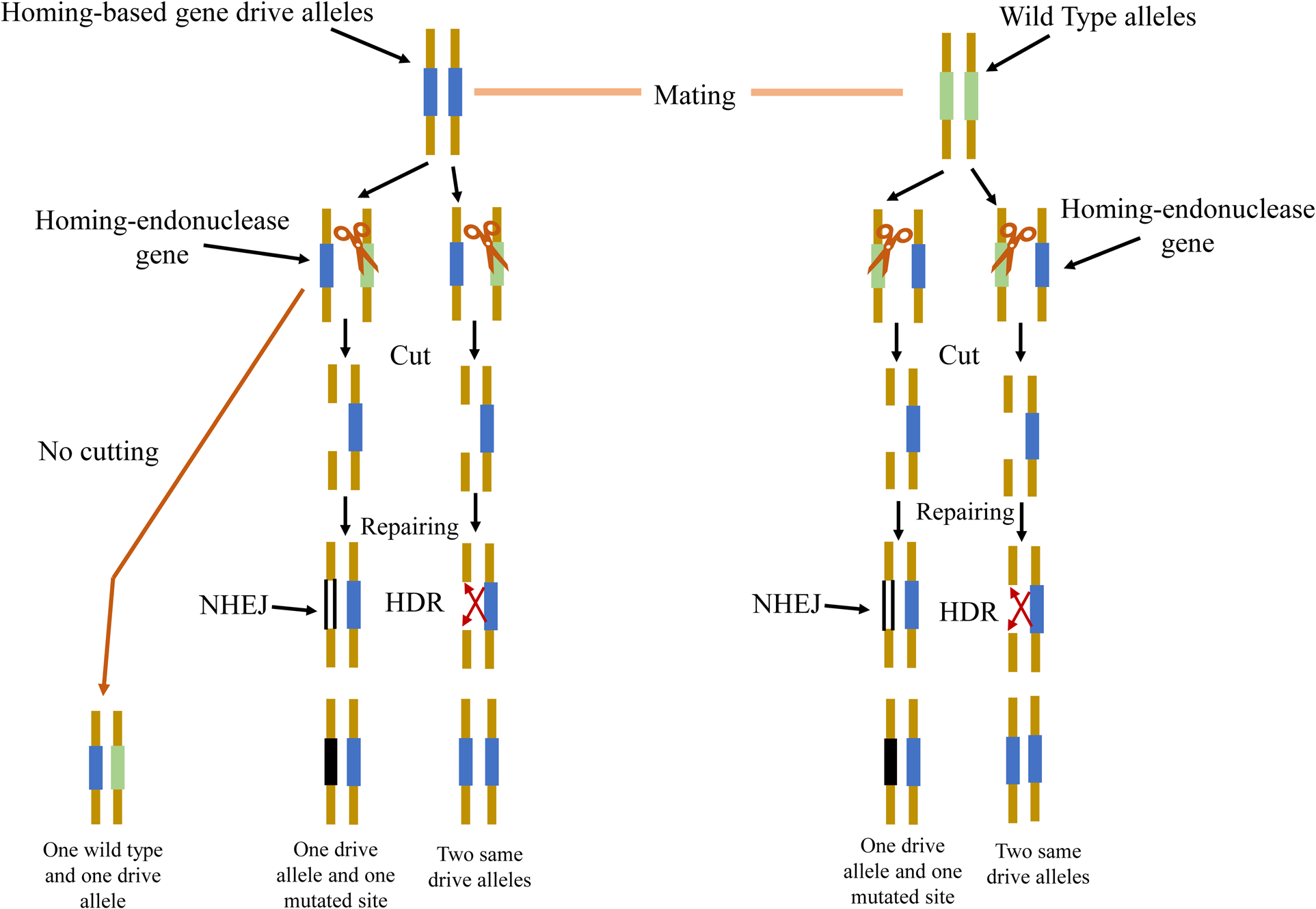
Figure 2. Representation of HEG-based gene drive. When the endonuclease-based gene drive organism mates with a wild-type organism, the endonuclease cutting system will be activated and cut the wild-type chromosome. If DNA break is repaired by using homologous recombination, then it will use drive chromosome containing the desired fragment as a template, copying drives into the wild-type chromosome (Bottom right). If endonuclease fails to cleave the sequence, no repairing occurs (bottom left). If homologous recombination fails to copy the template, the break will be repaired by non-homologous end-joining (bottom central).
Therefore, HEGD can be quickly spread to a population. Once a large number of individuals in the population are heterozygous, the population will collapse as heterozygote pairings will create non-viable or sterile offspring (Champer et al., Reference Champer, Buchman and Akbari2016). It is hard to redesign the HEG for a new target sequence (Chan et al., Reference Chan, Takeuchi, Jarjour, Huen, Stoddard and Russell2013b).
CRISPR/Cas9-based gene drive (CCGD)
More recently, the development of the CRISPR/Cas9 system has simplified the engineering nucleases to cut the specific genomic sequence (Fu et al., Reference Fu, Sander, Reyon, Cascio and Joung2014). Cas9 was initially identified in the bacterial innate-immunity system and discovered as a novel class of sequence-specific endonuclease, whose target specificity was determined by its guide RNA (gRNA) (Kondo and Ueda, Reference Kondo and Ueda2013; Mali et al., Reference Mali, Yang, Esvelt, Aach, Guell, DiCarlo, Norville and Church2013). Cas9 can be easily redesigned to target a new sequence merely by replacing the specificity of the gRNA, which is only a 20-bp sequence (fig. 3). The only limitation of this system is that the Cas9 will cleave the sequence, which is flanked by a protospacer-adjacent motif (PAM) at the 3′ end (Jinek et al., Reference Jinek, Chylinski, Fonfara, Hauer, Doudna and Charpentier2012).
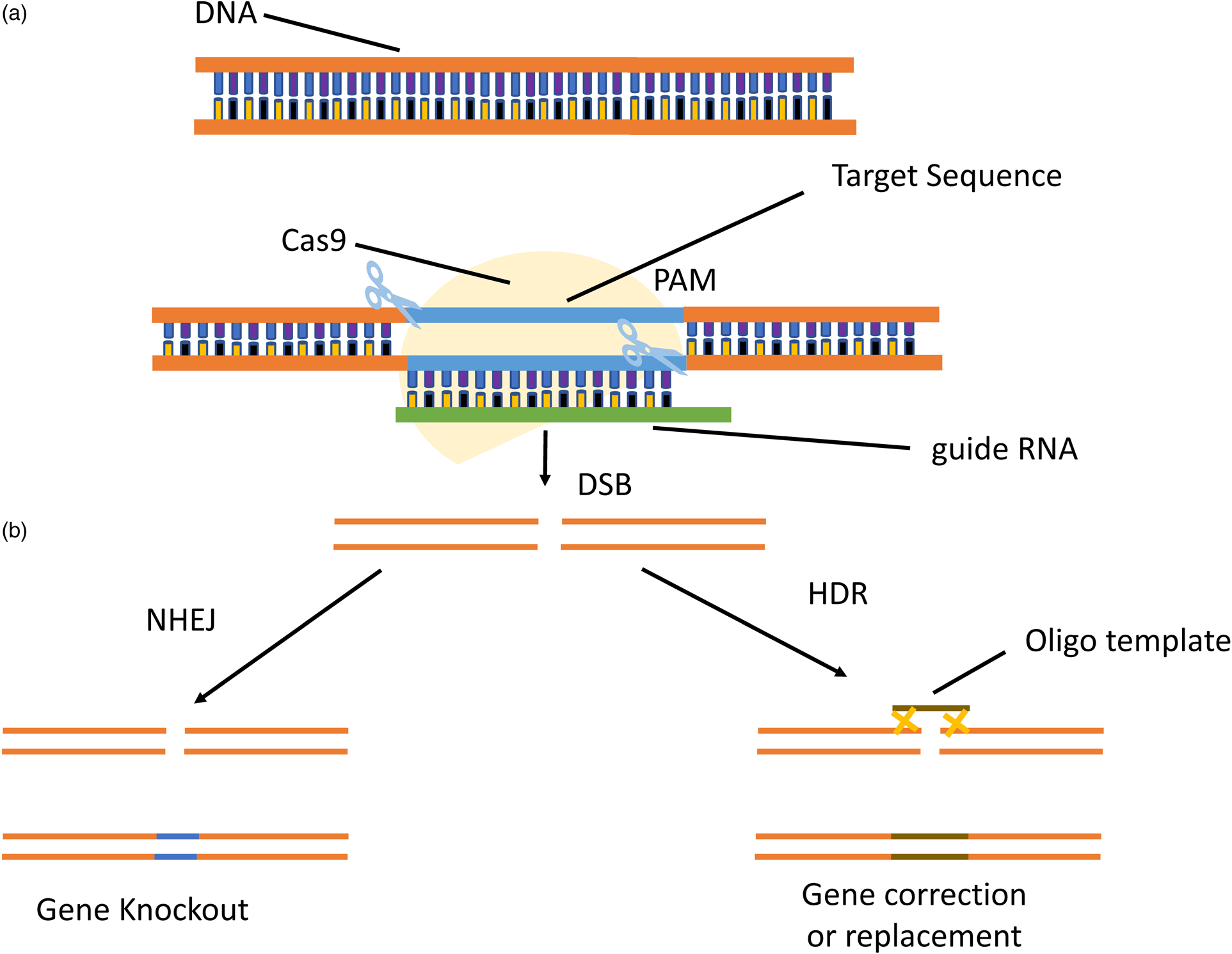
Figure 3. RNA guided CRISPR/Cas9 process. (a) The gRNA and Cas9 protein are delivered into the target cell. The gRNA directs Cas9 to bind target DNA. Cas9 will play a role as an endonuclease and cause a double-strand break (DSB), (b) DSB may be repaired by two processes: one is NHEJ resulting in gene knockout and the other is HDR resulting in gene correction and replacement.
In CRISPR/Cas9-based gene-drive system, a CRISPR/Cas9 construct contains at least a coding sequence of the Cas9 endonuclease and a 20-bp gRNA that binds with the target site of the host genome. RNA-guided Cas9 causes a break of double-stranded DNA in the wild-type allele at heterozygous state (same as homing endonucleases), which might be repaired through homology-directed repair (HDR) (fig. 4). Developing this procedure in the germline will drive this Cas9 homologous element into the wild population (Drury et al., Reference Drury, Dapper, Siniard, Zentner and Wade2017).
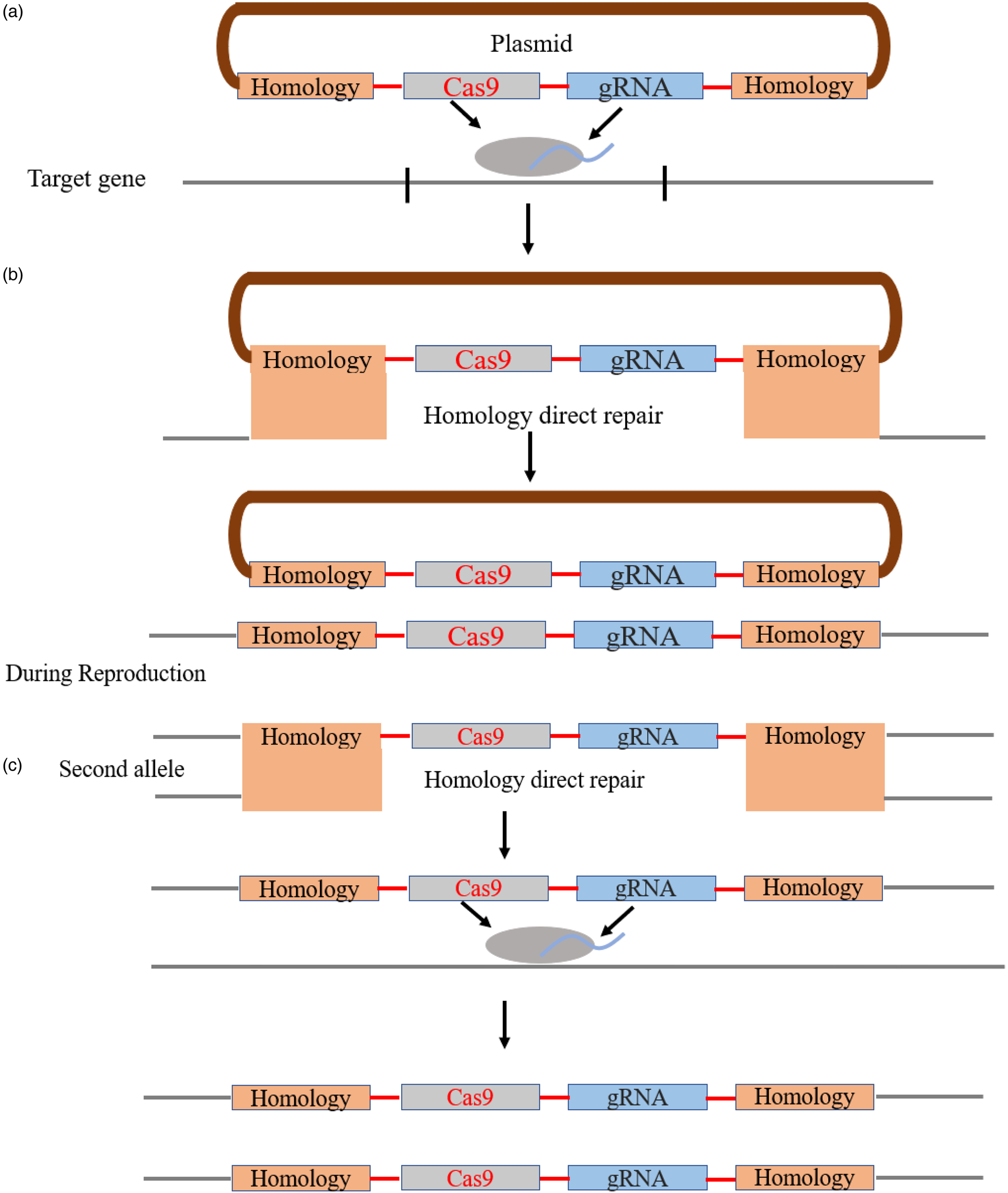
Figure 4. Representation of CRISPR/Cas9 based gene drive system. (a) The plasmid contains gRNA, Cas9 and other payload gene sequences. The gRNA will bind to a specific sequence and Cas9 will cut the double-strand DNA. (b) The gap will be repaired by HDR, and the targeted allele will be modified the same as plasmid containing sections. (c) During reproduction, this mutant allele in the heterozygous parent will be activated and cut the native wild-type allele, which is also repaired by HDR to generate two mutated gametes.
The first necessity for all kinds of endonuclease-based gene drives is to cleave the target sequence. Incomplete and improper cutting was a massive problem in transgenic mosquitoes and fruit flies, and HEG-based gene drive has 72% of cutting efficiency in mosquito and 86% in engineered fruit flies (Windbichler et al., Reference Windbichler, Menichelli, Papathanos, Thyme, Li, Ulge, Hovde, Baker, Monnat and Burt2011; Chan et al., Reference Chan, Huen, Glauert, Whiteway and Russell2013a). In contrast, target cutting rates of RNA-guided Cas9 exceeded up to 85–99% in the male of fruit flies (Kondo and Ueda, Reference Kondo and Ueda2013). Hence, the highly efficient CRISPR/Cas9 system allows researchers to construct a homologous CCGD to transfer any desired trait into the population (KaramiNejadRanjbar et al., Reference KaramiNejadRanjbar, Eckermann, Ahmed, Dippel, Marshall and Wimmer2018). The most exceptional challenging conditions require that the cleave sequence be repaired by HDR instead of non-homologous end joining (NHEJ). The rates of HDR are recognized to vary across the different species, developmental stages (Preston et al., Reference Preston, Flores and Engels2006; Chan et al., Reference Chan, Huen, Glauert, Whiteway and Russell2013a; Asad et al., Reference Asad, Liu, Li, Chen and Yang2022), and different types of cells (Mali et al., Reference Mali, Yang, Esvelt, Aach, Guell, DiCarlo, Norville and Church2013).
The major problem of CRISPR/Cas9 gene drive is the low-frequency rate of HDR. The HDR frequency rate can be increased when Cas9 protein is active at the early development stages, especially in germline tissues (Ren et al., Reference Ren, Sun, Housden, Hu, Roesel, Lin, Liu, Yang, Mao and Sun2013; KaramiNejadRanjbar et al., Reference KaramiNejadRanjbar, Eckermann, Ahmed, Dippel, Marshall and Wimmer2018). The HDR frequency rate increases in silkworms by knocking out the two genes (Ku70 and Ku80) involved in the NHEJ pathway (Ma et al., Reference Ma, Chang, Wang, Liu, Zhang, Lu, Gao, Shi, Zhao and Xia2014; Xu et al., Reference Xu, Xu, Zhan and Huang2019).
Application of gene drive systems in insects
Application of meiotic drive for insect pests population suppression
The insect population can be suppressed by targeting female fertility, which has been achieved through MD. The X chromosome-based drive was constructed in the woodland fly; the male was driving X chromosome and produced 80–100% female offspring; most of the male offspring were sterile without Y chromosome (Keais et al., Reference Keais, Hanson, Gowen and Perlman2017). The X-linked protein named heterochromatin protein 1 plays a vital role to develop sex-ratio MD in Drosophila simulans. The heterochromatin protein 1 driver (HP1D2) was constructed and this allelic driver prevents the Y chromatids from segregation during meiosis II to produce the female-biased progeny (Helleu et al., Reference Helleu, Gerard, Dubruille, Ogereau, Homme, Loppin and Montchampmoreau2016). The natural fluctuation of Y-linked sex-ratio suppressors and chromosomal capability to regulate the gene expression was investigated in D. simulans. The Y-linked sex-ratio suppressors induce 63–93% female-based progeny (Branco et al., Reference Branco, Tao, Hartl and Lemos2013). The natural occurrence of Y-chromosome-linked drive (Y-drive) has been identified and exposed to suppress insect populations by enhancing the male population (Lyttle, Reference Lyttle1977; Wood and Newton, Reference Wood and Newton1991). The Y-drive was observed in two mosquito species, Culex pipiens and A. aegypti, with a high ratio of sex bias (Helleu et al., Reference Helleu, Gérard and Montchamp-Moreau2015). The sex-linked MD system has been developed A. aegypti, which is associated with M (a male-determining gene) in chromosome Y, which leads to breakage of homologous chromosome X containing the m allele (Mori et al., Reference Mori, Chadee, Graham and Severson2004). In D. affinis, it has been confirmed that fathers siring all sons do not have a Y chromosome, and no morphological variations are linked with sex-ratio resistance to MD (Unckless et al., Reference Unckless, Larracuente and Clark2015). The persistence of X chromosome in the population of D. neotestacea differs between standard males and X chromosome-based males (Pieper and Dyer, Reference Pieper and Dyer2016). Recently, the tethered gene-drive system has been developed for population suppression, in which a two-locus UD and split homing drive were linked to one part of the split drive. This system shows better population-suppression results than the previous UD systems (Dhole et al., Reference Dhole, Lloyd and Gould2019).
The sex-ratio meiotic-drive system plays a vital role in the process of speciation, sex separation, alteration of linkage pattern disequilibrium, interspecific competition and extinction (Unckless et al., Reference Unckless, Larracuente and Clark2015).
Application of under-dominance for insect pests population suppression
An inherited character of UD causes a majority of males in the progeny of A. aegypti strain and the mass release of male-producing progeny was proposed for the control of A. aegypti (Craig et al., Reference Craig, Hickey and VandeHey1960). A hypothetical model showed that an engineered UD-based gene drive system allows traits to be introduced into pest populations economically and efficiently (Davis et al., Reference Davis, Bax and Grewe2001). The maternal-death under-dominance (UDMEL) gene-drive system was developed to modify and suppress local insect populations. The two maternal toxins individually settle on the different chromosomes. The zygotic antidotes connecting with two maternal toxins individually settling on the different chromosomes are capable of rescuing the maternal death of the other toxin. This model suggested that the spread of transgenic insects can be limited to local environments; a population contained single-locus UDMEL may require the repeated release of wild-type males for population suppression (Akbari et al., Reference Akbari, Matzen, Marshall, Huang, Ward and Hay2013). A UD transgenic construct is modified in D. melanogaster and introduced into the fly population to successfully make the flies refractory to spreading disease into the population (Reeves et al., Reference Reeves, Bryk, Altrock, Denton and Reed2014).
UD gene drive has shown great potential for introducing refractory genes into the pest populations for controlling infective diseases, including malaria and dengue fever. Furthermore, the pest population has been controlled by UD knockdown/rescue system (Magori and Gould, Reference Magori and Gould2006; Akbari et al., Reference Akbari, Matzen, Marshall, Huang, Ward and Hay2013; Reeves et al., Reference Reeves, Bryk, Altrock, Denton and Reed2014; Champer et al., Reference Champer, Buchman and Akbari2016). The UD-based gene-drive population models based on two-locus toxin and antitoxin show that UD could effectively change the neighboring populations (Champer et al., Reference Champer, Zhao, Champer, Liu and Messer2020). The comparison between the two-locus and the one-locus UD systems exhibits that the one-locus UD system requires the massive release of the population compared to the two-locus UD (Dhole et al., Reference Dhole, Vella, Lloyd and Gould2018).
UD requires the mass release of transgenic organisms into the population, which is not economically viable (table 1). Although UD can spread the desired mutation in the wild-type population, it is less stable than other potential gene-drive systems (table 1). In contrast, this system could be well suitable for the replacement of specified target populations, in which further spread is not required (Sinkins and Gould, Reference Sinkins and Gould2006).
Table 1. Comparison of gene-drive techniques

Application of homing endonuclease-based gene drive for insect population suppression
HEGD has been employed to suppress vector and pest populations by homing into a specific target locus or spreading a genetic payload, which leads to recessive lethality or sterility (Deredec et al., Reference Deredec, Burt and Godfray2008). HEGD using I-SceI gene elements has been developed in D. melanogaster with high rates of homing attained at both ectopic and homing sites, suggesting that the HEG-based gene-drive system may be feasible in other dipteran insects (Chan et al., Reference Chan, Naujoks, Huen and Russell2011). The engineered homing endonucleases enhance the targeted mutagenesis and the homing in D. melanogaster (Chan et al., Reference Chan, Takeuchi, Jarjour, Huen, Stoddard and Russell2013b). To produce transgenic A. gambiae, HEG I-SceI was used as a gene-drive component to obtain high rates of site-specific chromosomal cleavage and gene conversion in the male germline (Windbichler et al., Reference Windbichler, Menichelli, Papathanos, Thyme, Li, Ulge, Hovde, Baker, Monnat and Burt2011). The A. gambiae population might be eradicated by targeting the female-fertility gene with HEGs or driving by Y chromosome that transmitted to 75–96% of progeny (Deredec et al., Reference Deredec, Godfray and Burt2011). The combination of Y drive and fertility-targeting HEGD is more convenient to eradicate the disease by eradicating the vector (Deredec et al., Reference Deredec, Godfray and Burt2011). The HEG I-PpoI cleaves the gene responsible for disabling X-carrying spermatozoa on the X chromosome in A. gambiae to produce a male-biased sex ratio of A. gambiae (Windbichler et al., Reference Windbichler, Papathanos, Catteruccia, Ranson, Burt and Crisanti2007).
However, the HEGD system has not been tested in other insect species, such as lepidopterans, coleopterans, and orthopterans. Different kinds of homing genes are used in different species, and some of the homing genes work well in specific species (table 1). Therefore, this gene drive works differently in different species, and its efficacy mostly depends upon species and endonuclease genes.
Application of CRISPR/Cas9-based gene drive for insect pest population suppression
The application of a gene-drive system to modify the insect population has been improved by the CRISPR/Cas9 genome editing tool (McFarlane et al., Reference McFarlane, Whitelaw and Lillico2018). A CCGD targeting three genes (AGAP011377, AGAP007280, and AGAP005958) has been constructed in A. gambiae to cause phenotypic recessive female-sterility with a high transmission rate of 99.4% to the progeny (Hammond et al., Reference Hammond, Galizi, Kyrou, Simoni, Siniscalchi, Katsanos, Gribble, Baker, Marois and Russell2016). Another highly effective CCGD adopted from mutagenesis chain reaction (MCR) in A. stephens resulted in a high frequency to copy a 17-kb construct through HDR at target locus with site-specific integration manner, and the autonomous gene-drive components were introduced approximately 99% into progeny by outcrossing wild-type mosquitoes with transgenic lines (Gantz et al., Reference Gantz, Jasinskiene, Tatarenkova, Fazekas, Macias, Bier and James2015). An MCR developed in D. melanogaster targeting yellow gene of D. melanogaster causes the conversion of heterozygous to homozygous flies and the cross of wild-type individuals with transgenic individuals resulted in 95–100% mutation in progeny (Gantz and Bier, Reference Gantz and Bier2015). A CCGD construct targeting the intron 4 and boundary of exon 5 of dsx gene resulted in blockage of female dsx function without disturbing the fertility and development of the male with the efficiency of 100% within 7–10 generations to cause the total population collapse of cage mosquito (Kyrou et al., Reference Kyrou, Hammond, Galizi, Kranjc, Burt, Beaghton, Nolan and Crisanti2018). A CRISPR/Cas9 gene drive targeting the transformer (tra) gene exhibits the high efficiency in sex conversion of the females into males in Ceratitis capitata (KaramiNejadRanjbar et al., Reference KaramiNejadRanjbar, Eckermann, Ahmed, Dippel, Marshall and Wimmer2018). This gene-drive system causes all-male population collapse and it may be used to control other harmful pests.
The CCGD can fulfill the basic requirements of a potential gene drive (table 1). Hence due to the successful construction of CRISPR/Cas9 gene drive in mosquitos and D. melanogaster, it may be constructed in other insect species such as major crop pests to modify or remove the pest population from the field. The major drawback in the application of CRISPR/Cas9 gene drive is a low rate of HDR because gene drive ultimately depends upon HDR (table 1).
Conclusions and future perspectives
Regardless of the possible benefits of various gene-drive systems, these systems may remain under the study, because it is unclear which type of gene-drive system is best suitable to overcome their engineering limitations and regulatory situation. The above-mentioned gene-drive systems can potentially be used to suppress the insect pests population, but these systems have some limitations, which decreases the feasibility. We have discussed the four types of gene-drive systems, including the benefits and drawbacks in the application for insect pest population suppression, which will be helpful in the future to construct a potential gene-drive system to control the pest population. Recently, the CRISPR/Cas9-based gene-drive system in mosquitoes and Drosphilla has been successfully developed for population replacement, and it might be used for other crop pests in the future. The limitation is low HDR efficiency, which is partially due to the formation of resistant alleles. This limitation can be overcome by discovering an effective germline-specific promoter to drive the Cas9 expression effectively in germ cells. Considering the high drive activity of CRISPR/Cas9-based gene-drive system, future genetic and molecular work will be further proceeded to improve system efficiency and broaden their application to modify the wild-type population.
Gene drives show great promise as potentially new solutions to complex biological challenges, but they may also create risks. Payload genes may have unexpected effects, and certain gene drives may result in extinction. A gene drive might potentially have unintended consequences, such as spreading outside the specified geographical region or transferring over into another species. Concerns have also been expressed about the possibility of employing gene drives to cause economic damage. Considering that each of the systems discussed here is vulnerable to the development of drive resistance to variable degrees, more research into molecular designs will be required to prevent the creation of drive resistance. All these gene drive systems discussed above have been successfully constructed and evaluated under laboratory conditions to suppress the wild pest population. Due to certain limitations, these gene drive systems have not been tested under field conditions. It is necessary to conduct safe and controlled field experiments to achieve the numerous potential advantages of gene drive technology in a safe, ethical, and responsible manner. The genetic alteration in gene-drive systems will continue in the future to increase the feasibility of these gene-drive systems so that the wild-type population of harmful pests will be effectively managed under field conditions.
Financial support
This work was financially supported by Fujian Science and Technology Major Project (2018NZ0002).
Conflict of interest
The authors declare that there is no conflict of interest.








