Introduction
Traditional agroecosystems developed by Native American farmers, including those in the Upper Midwest of the USA, represent accumulated experience of the interaction with the environment and, for centuries, served as laboratories for crop germplasm evolution, dynamic conservation and sustainable utilization (Altieri and Merrick, Reference Altieri and Merrick1986). Ever since its introduction to North America 3000 years ago and due to subsequent multiple introductions into the Upper Midwest (Moeller and Schaal, Reference Moeller and Schaal1999), valuable genetic resources of maize landraces have been developed and incorporated into self-sustained agroecosystems and therefore contributed to the conservation of large genetic diversity of the crop (Flint-Garcia et al., Reference Flint-Garcia, Bondar and Scott2009).
Identification of maize landraces has been carried out by Native American farmers on the basis of characters that show the greatest range of variation and perceptual salience (Boster, Reference Boster1995). The large variation in the multifactorial and seemingly non-adaptive kernel colour trait displayed by Native American maize landraces is an evidence of recurring selection for perceptual distinctiveness. Selection for kernel colour traits allowed Native American farmers to distinguish between and maintain large diversity within maize landraces for various traditional uses (Gibson, Reference Gibson2009).
Traditional and subsistence farmers in the Old and the New World use perceptual distinctiveness traits to indicate an adaptation to certain agroecological and edaphic conditions (Chambers et al., Reference Chambers, Brish, Grote and Gepts2007), particular nutritional or medicinal qualities, or an association with specific rituals and beliefs (Gibson, Reference Gibson2009). Local names of maize landraces, in this and other studies (Benz et al., Reference Benz, Perales and Brush2007), reflect a long-established tradition of referring to a landrace by the most prominent colour of its kernels (e.g. Purple Mountain and Pink Lady), and occasionally by its origin (e.g. Bear Island) or traditional use (e.g. Dakota Black Popcorn). However, farmers might not be consistent in naming and describing landraces, and a particular landrace may assume different names at different localities.
Traditional maize-based farming systems may not be closed or isolated enough to prevent gene flow. Therefore, each maize landrace, maintained by Native American farmers as an open-pollinated population, may represent a collection of highly heterozygous and heterogeneous populations, and its diversity, including kernel colour diversity between or within populations and landraces, may directly reflect a region's agroecological conditions (Gibson, Reference Gibson2009) and cultural diversity (Nabhan, Reference Nabhan1985). Kernel colour may have been a trait critical for maintaining landrace integrity and was used by Native American farmers as a ‘bar-code’ for landrace identification (Wood and Lenné, Reference Wood and Lenné1997), and in order to propagate distinctive phenotypes in isolation (Benz et al., Reference Benz, Perales and Brush2007). Over time, however, inbreeding or farmer selection for distinctive kernel colour(s) may have contributed to repeated and prolonged genetic bottlenecks in maize (Benz et al., Reference Benz, Perales and Brush2007). A comparative study was designed to quantify and partition diversity in colour variation, and to explore its association as a perceptual distinctiveness trait with kernel shape (i.e. morphometrics) and nutritional traits in kernels of a germplasm collection of maize landraces which was assembled from Native American sources. The long-term objective was to assess the persistence of traditional landraces in Northern MN as a region of rapid agricultural and socio-economic change.
Materials and methods
Landrace germplasm
Eleven Native American maize landraces were used in a comparative study under typical field conditions of the corn-growing region in the Upper Midwest. Seven of the landraces (Supplementary Table S1, available online only at http://journals.cambridge.org) were donated by the White Earth Land Recovery Project (47°20′22.50″N 95°43′41.50″W; Winona LaDuke, personal communications). These landraces were identified as: Bear Island Flint (BIF); Dakota Black Popcorn (DBP); Hochunk corn (HCC); Manitoba Flint corn (MFC); Pink Lady corn (PLC); Purple Mountain Purple (PMP); Seneca Blue Bear Dance Flint (SBD). Three landraces donated by a private farmer, based on kernel coloration patterns, were designated as: dark-grey (JDG); purple-black (JPB); vertical red stripe (JVR). A fourth landrace was obtained from a local market (JAG). The last four landraces were grown for three consecutive years (2007–2009) under organic management prior to conducting the field experiment. For the purpose of this study, the first set of landraces will be referred to as population [1] and the second set as population [2] based on their origin and management practices prior to conducting the experiment. Sixteen accessions from six landraces in population [1] and 11 accessions from the four landraces in population [2] were used in this study. One accession from a seventh landrace (DBP) in population [1] was included in the statistical analyses where appropriate. Kernels from landrace accessions grown in the farmer's field at the White Earth Reservation, and from those grown in a common-garden experiment in 2008–2009 were used in field experiments in 2010 and 2011.
Field experiment
Field experiments in a completely randomized design were designed to carry out two field experiments to characterize 28 accessions in 11 landraces of Native American maize during two growing seasons (2010 and 2011) at the Swan Lake Research Farm (45°41′04.48″N 95°47′52.86″W), Morris, MN, USA. Sowing date, seeding rate and management practices for maize typical of the Upper Midwest were used in both years, except weed control which was carried out by hand. Ears were harvested manually from each plant upon reaching full maturity and labelled for future kernel sampling and analyses.
Kernel measurements
Kernel dry weight (KWT) was determined on randomly sampled kernels from ears harvested at full maturity in 2010 and 2011. Ten kernels per accession from each landrace were taken at random for further physical and chemical analyses. Digital imagery using a Nikon D70S camera with 1504 × 1000 pixel resolution captured physical measurements (kernel area, perimeter, width, length, major axis and minor axis) on a total of 590 individual kernels from all landraces. For each digital image, the selected kernels and a scale of 1 cm in length were manually positioned on a platform to ensure that the kernels were totally separated and their embryos were facing up. The number of kernels in each image was verified by the number of objects generated by the ImageJ software program (Ferreira and Rasband, 2011).
Chemical analyses
Kernel samples were dried at 45°C in a forced air oven until no further reduction in weight occurred. Kernels were ground and placed through a 1 mm screen (Thomas Scientific, Swedesboro, NJ, USA). Then, one subsample was used to determine C and N and another to determine micro- and macronutrients. C and N were determined on kernel samples as the percentage of dry weight using a LECO FP-428 analyser (LECO, St Joseph, MI, USA). Then, the C:N ratio was calculated for each subsample. Per cent nitrogen values were used to estimate the protein content as N% × 6.25. Determination of micro- (Al, Cu, Fe, Mn and Zn) and macronutrients (Ca, K, Mg, P and S) in maize kernels followed the procedure outlined in the US-EPA 5051 method. The procedure was adapted using the Mars Xpress microwave system for the CEM (CEM Corp., Matthews, NC, USA) sample preparation note XprAg-1. The procedure used 55 ml Teflon cubes in a 40-unit carousel. A 0.5 g sample weight was digested with 6.5 mm nitric acid (70% trace material analysis) using a 15 min ramp programme set to a power maximum of 1200 W and held for 15 min. The samples were allowed to cool to room temperature, transferred to 50 ml volumetric flasks and taken to volume with Milli-Q water (Millipore Corp., Billerica, MA, USA). Chemical analyses were completed using the Varian Vista-Pro charged-coupled device (Varian, Inc., Palo Alto, CA, USA) simultaneous inductively coupled plasma-optical emission spectroscopy instrument. MNUSDA-STD 1A and MNUSDA-STD 2A were prepared and used in the analyses as elemental standards (Inorganic Ventures, Lakewood, NJ, USA).
Kernel colour analysis
The three-dimensional red–green–blue colour space of each kernel captured in digital images was used to calculate their L*a*b* quantitative colour space descriptors (referred to hereafter as ‘colour traits’). L* indicates darkness–lightness, a* indicates redness–greenness and b* indicates yellowness–blueness descriptors. The mode of each colour trait was used in the statistical analyses (Darrigues et al., Reference Darrigues, Hall, van der Knapp and Francis2008).
Statistical analyses
Secondary statistics were calculated from the six morphometric measurements on individual kernels to quantify the shape of each kernel (Ferreira and Rasband, 2011; http://JmageJ.nih.gov/ij/docs/guide; accessed 27 October 2012) as follows: circularity (CIR) was estimated as 4π × [kernel area/(perimeter)2]; Feret's diameter (FDM) as the longest distance (cm) between any two points along the boundary of the kernel; aspect ratio (ASR) as the major axis/minor axis; roundness (RND) as 4 × [kernel area/π × (major axis)2]; solidity (SLD) as (kernel area/kernel convex area); density (DEN) as [kernel weight/(length × width × minor axis)] (g/cm3). Secondary statistics were used in the subsequent statistical analyses.
Principal components analysis (PCA) was used on the standardized means of all traits as a linear dimensionality reduction technique to identify orthogonal directions of maximum variance in the original dataset at the prediction (R 2) and validation (Q 2) stages of model building. Landraces were included in model building as categorical variables. Loadings of traits and landraces on the first two principal components (PCs) were used to interpret the results of PCA. A mixed model was employed to perform variance components analysis (Payne et al., Reference Payne, Harding, Murray, Soutar, Baired, Welham, Kane, Gilmore, Thompson, Webster and Wilson2007). The procedure estimates the effects of fixed (years, populations and their interaction) and random (accessions within landraces, landraces within populations and year × accessions within landrace) factors on each trait. KWT was used as a covariate in the mixed model. The variance component attributed to year × landraces within populations was not significant for all traits and was removed from the final mixed model.
Canonical discriminant analysis using a matrix of all traits, with landraces as a classification factor, was conducted to identify which trait combination can be used to discriminate between landraces at a multivariate level. The first two canonical discriminant roots, accounting for the largest amount of variation, were used to develop two-dimensional plots of landraces and traits. Per cent correct classification of each landrace and the standardized coefficients for each trait on the first two canonical discriminant roots are reported. Least squares means (LS means) were calculated for all traits and landraces, and mean separation was performed on LS means using Duncan's multiple range test (DMRT; P= 0.05). The mode of each colour descriptor was used in all statistical analyses as a more reliable indicator of kernel colour than the mean (Darrigues et al., Reference Darrigues, Hall, van der Knapp and Francis2008). Per cent pairwise significant differences (%PSD) between LS means were calculated and reported to illustrate the magnitude of significant differences between accessions within each landrace. Partial least squares regression (PLS) was employed to predict the protein content, the C:N ratio and the micro- and macronutrient contents (quality traits) as functions of colour traits. Prediction and validation models were developed using the non-linear iterative partial least squares (NIPALS) module in a multivariate software program (The Unscrambler software, version 10.1.; CAMO, Oslo, Norway). A cross-tabulation of non-zero frequencies of quality traits × colour traits and landraces × colour traits was subjected to log-linear analysis to test for a significant association between them. The variance accounted for by the validation model (Q 2), the residual mean squares error (RMSE) and the colour traits(s) with a significant regression coefficient (β) are reported. Finally, a separate and two-way joint hierarchical clustering of standardized secondary kernel shape descriptors, colour traits and quality traits (i.e. protein content, C:N ratio and micro- and macronutrients), in addition to landraces, was performed using Euclidean distance and the unweighted pair group method with the arithmetic average (UPGMA) linkage clustering procedure. The joint colour-coded matrix of traits and landraces was used to identify landraces with preferred levels of single or multiple trait combinations (Cañas et al., Reference Cañas, Amiour, Quillere and Hirel2011). Statistical analyses were carried out using relevant modules in GenStat – Version 10 (Payne et al., Reference Payne, Harding, Murray, Soutar, Baired, Welham, Kane, Gilmore, Thompson, Webster and Wilson2007).
Results
Total variation
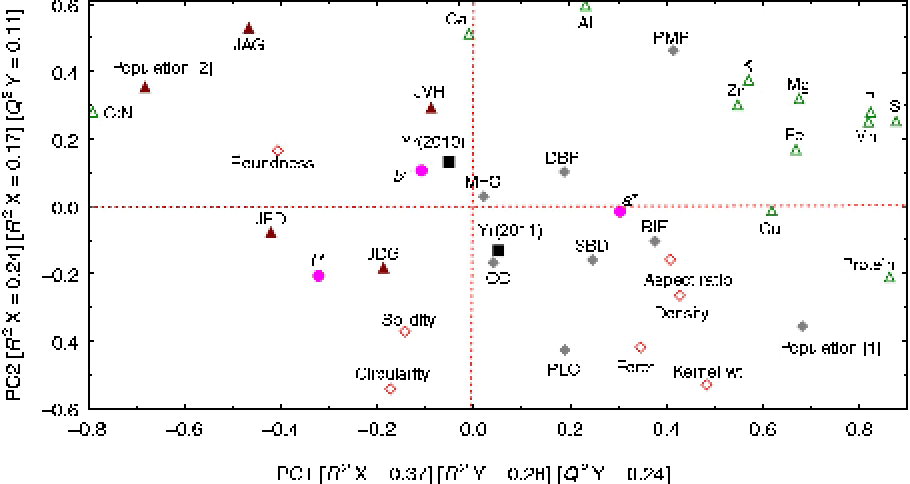
Calibration (R 2) and validation (Q 2) variances explained by the first (PC1; 0.37 and 0.28, respectively) and second (PC2; 0.24 and 0.17, respectively) PCs separated landraces and their kernel shape, quality and colour traits into four groups (Fig. 1). Loadings (i.e. correlation coefficients of traits or landraces with each PC) of landraces and traits on both PCs separated landraces according to their source (i.e. population). Landraces in population [1] have larger values for kernel weight, protein and micro- and macronutrients contents, and lower C:N ratios than landraces in population [2]. Kernel shape traits were separated along PC1, with larger values of kernel weight, DEN and ASR being the characteristics of landraces in population [1], and larger values of kernel RND, SLD and CIR being the characteristics of landraces in population [2].

Fig. 1 (colour online) Calibration (R 2) and validation (Q 2) variance explained by the first (PC1) and second (PC2) PCs derived from the 11 Native American maize landraces and their kernel shape, nutrients and colour space descriptors.
Colour traits were separated along both PCs, with larger values of L* and b* and smaller (more negative) values of a* being associated with landraces in populations [2] and [1], respectively. L* and b* were also associated with a larger C:N ratio, while a* was associated with higher protein and nutrient contents; however, protein and nutrients had negative and positive loadings on PC2, respectively. Micro- and macronutrients displayed different loadings on both PCs. Ca and Al had small and large loadings on PC1 and PC2, respectively. The remaining nutrients were separated into three groups with increasing loadings on PC1 and decreasing loadings on PC2.
Variance components
LS means and their coefficients of variation (CV%) of kernel shape, quality and colour traits and the results of a mixed model (variance components analyses; Supplementary Table S1, available online only at http://journals.cambridge.org) suggested that the total variance of these landraces is characterized by a complex multivariate structure. The total variation in different traits was shaped by several factors and their interactions. Traits can be grouped into three categories based on their level of variation. The kernel shape descriptors and two quality traits (i.e. protein and C:N ratio) displayed smaller CV% when compared with kernel weight and most micro- and macronutrients; whereas the colour traits and the nutrients Al, Ca, and Cu had the largest variation.
Kernel weight, as a covariate in the variance components analyses, had a significant effect on most traits, except CIR, SLD, K, Mg, S and colour traits. Main fixed factors in the mixed model, but not their interaction, had significant effects on most traits. Significant differences (P< 0.05) were found for kernel weight, L*, protein, C:N and Mn due to annual variation, and significant differences (P< 0.05) between populations were found for most kernel shape traits, none of the colour traits, and only for a few quality traits (protein, C:N, Mn and S). Statistically, all traits (except a marginally significant value for P; P= 0.06) were stable over both years as indicated by a non-significant (P>0.05) population × year interaction.
All three random factors (i.e. accessions within landrace, landraces within population and year × accession within landrace) in the mixed model (Supplementary Table S1, available online only at http://journals.cambridge.org) explained variable portions of total variation, ranging from small (all kernel shape traits, except CIR), to intermediate (colour traits) to large (most quality traits). Differences between accessions within landraces explained significant portions of variation in all traits; differences between landraces within populations explained significant portions of variation in all traits, except kernel weight, solidness, DEN and b*, and the interaction due to year × accessions within landraces explained significant portions of variation in all traits, except Feret's dimension, ASR, RND, a* and Mn. Although colour traits displayed large variation (expressed as CV), and were independent of kernel weight (i.e. the covariate), they showed stability over years, populations and their interaction (except the significant effect of year on L*), and exhibited significant variation due to all three random factors except the non-significant effects of landraces within populations on b* and year × accessions within landraces on a* (Supplementary Table S1, available online only at http://journals.cambridge.org).
Discrimination between landraces
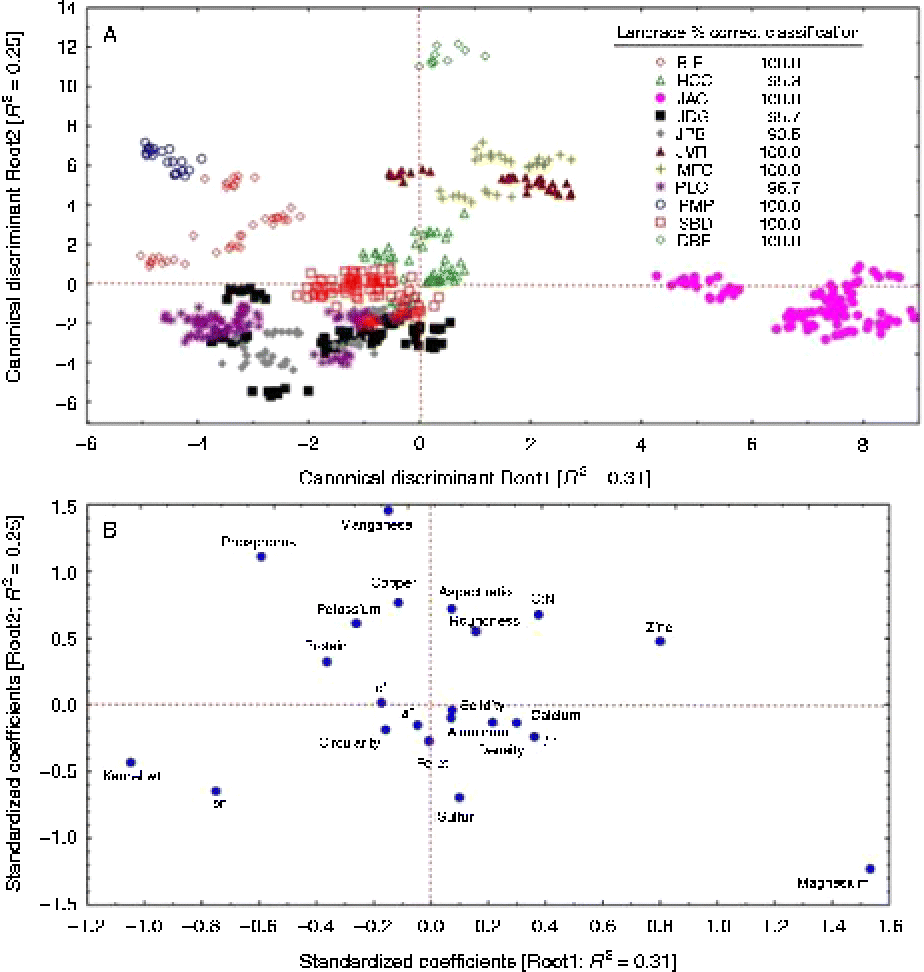
Two discriminant functions (i.e. roots) explained 0.56 of the total variation and discriminated between landraces with a large (97.8%) average per cent correct classification (Fig. 2(a)). Seven of the landraces were 100% correctly classified and the remaining four showed a few ( < 10%) misclassifications. All traits (Fig. 2(b)) contributed to this correct classification as indicated by their standardized coefficients. Five nutrients (Cu, Mn, K, P and Fe, in increasing order) had negative standardized coefficients on Root1, and the remaining five nutrients (Al, S, Ca, Zn and Mg, in increasing order) had positive values on Root1. Similarly, five nutrients (Mn, P, Cu, K and Zn, in increasing order) had positive, and the remaining five nutrients (Al, Ca, Fe, S and Mg, in increasing order) had negative standardized coefficients on Root2. The colour traits and the kernel shape descriptors had relatively smaller standardized coefficients. The colour traits were separated along Root1, with L* having positive and both a* and b* having negative and relatively smaller standardized coefficients than L*.

Fig. 2 (colour online) Biplots, per cent correct classification, variance (R 2) accounted for by the (a) first two canonical discriminant functions and (b) standardized coefficients of kernel shape, nutrient and colour space descriptors of the 11 Native American maize landraces.
Quantitative variation between and within landraces
Descriptive statistics (LS means and CV%) and mean separation between landraces (DMRT = 0.05) and between accessions within landraces (%PSD) for kernel shape, kernel colour and quality traits are presented in Supplementary Table S2 (available online only at http://journals.cambridge.org). Most differences between landraces, both between and within populations, were highly significant (P< 0.05). Populations differed with respect to the level and extent of significant differences between landraces. There were no significant differences between landraces within population [1] in two traits (protein content and C:N ratio), while there were no significant differences between landraces within population [2] in four traits (CIR, ASR, RND and b*).
When averaged over accessions in each landrace, kernel weight ranged from 21.7 mg (JAG) to 33.2 mg (BIF and PLC), CIR ranged from 0.58 (PMP) to 0.82 (BIF and JDG), FDM ranged from 0.94 (JVR) to 1.10 cm (PLC), ASR ranged from 1.12 (JDG) to 1.29 (PMP), RND ranged from 0.78 (PMP) to 0.90 (JDG), solidness displayed a very narrow range (0.97–0.98) and DEN ranged from 1.15 (JAG) to 1.51 g/cm3 (BIF). Similarly, when colour traits averaged over accessions within landraces, L* ranged from 23.3 (BIF) to 78.0 (MFC), a* ranged from − 1.7 (JVR) to 20.1 (PMP) and b* ranged from 10.4 (BIF) to 20.9 (JVR).
Quality traits (including protein content, C:N ratio and micro- and macronutrients) displayed a wide range of values. The protein content ranged from 9.4 (JAG) to 13.9% (BIF). Although there were no significant differences in the C:N ratio between landraces in population [1], its value ranged from 20.9 (BIF) to 31.3 (JAG). Micronutrient (Al, Cu, Fe, Mn and Zn) contents (μg/g) ranged from 0.52 (MFC) to 3.3 (JAG) for Al, from 0.4 (MFC) to 2.3 (BIF) for Cu, from 21.5 (JAG) to 48.3 (PMP) for Fe, from 5.1 (JPB) to 14.0 (PMP) for Mn and from 15.4 (JPB) to 32.8 (PMP) for Zn. Macronutrients (Ca, K, Mg, P and S) contents (μg/g) ranged from 23.6 (SBD) to 46.1 (JVR) for Ca, from 3318 (HCC) to 4363 (PMP) for K, from 1126 (JPB) to 1625 (PMP) for Mg, from 2929 (JPB) to 4136 (PMP) for P and from 946 (JPB) to 1465 (BIF) for S.
A matrix of 220 trait–landrace combinations, 132 and 88 of which belong to populations [1] and [2], respectively, is presented in Supplementary Table S2 (available online only at http://journals.cambridge.org). There were no significant differences between accessions within landraces in 35% of all 220 combinations, whereas, in 42 and 23% of these combinations, there were significant differences between all (or 100% of accessions) or at least between two accessions within a particular landrace (0.0>%PSD < 100 of all accessions), respectively. There were no significant differences between accessions within landraces in 38 and 30% of trait–landrace combinations in populations [1] and [2], respectively. The respective values for ‘all accessions within landrace’ were 45 and 38%, and for ‘at least two accessions within landrace’ were 17 and 32%.
In population [1], landraces with two accessions each (BIF, MFC, PMP and SBD) displayed less %PSD between accessions within landraces compared with landraces with three (HCC) or five (PLC) accessions. Two landraces (JPB and JVR) in population [2], each with two accessions, displayed contrasting (36 and 73%, respectively) %PSD values, whereas landraces with three (JDG) or four (JAG) accessions displayed %PSD values comparable with those in population [1]. The kernel shape, colour and nutrient (including protein content and C:N ratio) groups of traits displayed increasing (51, 67 and 74%, respectively) %PSD values. On average, the %PSD values for kernel shape traits in populations [1] and [2] were 48 and 57%, respectively; the respective values for colour traits were 67 and 67% and for quality traits were 71 and 79%.
Validation of PLS models
Per cent significant variances (β* indicates a significant regression coefficient; P< 0.05) explained by L*a*b* and RMSE in the PLS validation models for kernel nutrient composition are presented in Supplementary Table S3 (available online only at http://journals.cambridge.org). Each of L*a*b* and L*b* explained 37% of the total variance in the whole germplasm, L*a* explained 14% and the remaining 12% were explained by single colour traits. There were 15 significant pairwise differences between landraces (33%PSD). These significant differences were found between a group of landraces with average explained variance >50% (MFC, 61.2%; JVR, 59.3%, BIF, 56.8%; JPR, 51.0%) and a second group composed of the remaining landraces with < 50% average variance (HCC, 43.7%; PMP, 40.3%; PLC, 28.8%; JAG, 24.3%; SBD, 22.0%). Variance and RMSE estimates were independent of each other for each trait, except for Fe (r= − 0.92; P< 0.01) and K (r= − 0.64; P< 0.05). Colour traits explained significant portions of the total variance in protein content and C:N ratio of all landraces. The largest (64.5%) and smallest (19.2%) significant variances in protein were explained by L*a*b* and L*b*, respectively; however, these estimates were not necessarily associated, respectively, with the smallest and largest RMSE values. The combined L*b* comprised 50% of all colour traits in jointly explaining significant variances in both protein content and C:N ratio.
Most (86%) of the PLS validation models accounted for significant (β*) portions of variances in micro- and macronutrients, being explained by the variation in colour traits; the remaining 15% of the models (6% of PLS validation models in population [1] and 9% in population [2]) were not significant. The PLS validation models for quality traits indicated that L*a*b* explained 19 and 14% of the variation in micro- and macronutrients, respectively; the respective variances accounted for by L*a* were 9 and 4%, and by L*b* were 8 and 19%. A few PLS validation models included only one colour descriptor. The L* descriptor accounted for significant variances in C:N ratio in MFC, in two micronutrients (Al and Mn) in BIF, SBD and JDG, and one macronutrient (S) in HCC and JDG. The variation in a* accounted for significant variances in protein in MFC, Fe in JVR and Mg in BIF, while b* accounted for a significant variance in P in JAG and both a*b* accounted for significant variances in Cu and Mg in SBD and PLC, respectively.
A log-linear model, used to test the non-zero, two-way frequency tables for ‘quality traits × kernel colour traits’ and ‘landraces × colour traits’, resulted in a maximum likelihood ratio χ2 statistics of 33.6 (P= 0.14) and 18.4 (P= 0.61), respectively. Only L*a*b*, L*a* and L*b*, which accounted for 74% of all combinations of colour traits and quality traits (Supplementary Table S3, available online only at http://journals.cambridge.org), were used in constructing the log-linear models to avoid the use of structural zeros. The results suggested that the two-way associations between ‘kernel colour traits’ and each of ‘quality traits’ and ‘landraces’ fit the specified model in each case.
Joint landrace–trait clustering
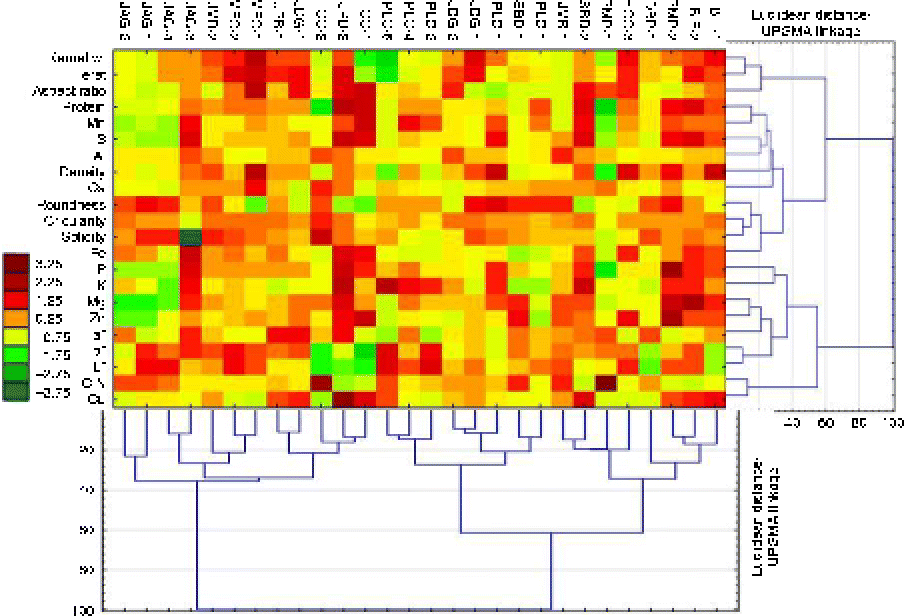
A separate hierarchical and joint clustering of landrace accessions and kernel traits resulted, at 50% of maximum Euclidean distance, in separating accessions and kernel traits into three and four sub-clusters, respectively (Fig. 3). The level of variation displayed by the standardized values of all traits (mean zero and variance 1.0) was large as indicated by the colour-coded scale (a maximum of 3.25 SD above average to a minimum of 3.75 SD below average; Fig. 3). It is possible to delineate sections in Fig. 3 of the above- or below-average values of single or multiple kernel shape, quality or colour traits and their association with each other or with specific colour patterns in certain landrace accessions. High-DEN kernels in some landraces (e.g. BIF-1, HCC-3 and MFC-1) were heavier than average, and with an elliptical shape based on their ASRs, regardless of their colour pattern(s).

Fig. 3 (colour online) Joint and hierarchical clustering of kernel shape, nutrient and colour space descriptors measured on kernels of the 28 accessions from the 11 Native American maize landraces expressed as standard deviation units from a zero mean and unit variance for each trait using Euclidean distance and a UPGMA linkage clustering procedure.
The clearly different L*a*b* colour patterns for the landraces BIF-1 and BIF-2, for example, were associated with different levels of protein and other ‘quality traits.’ Four landrace accessions (BIF-2, PMP-2, SBD-2 and HCC-1) in population [1], and one (JPB-1) in population [2] had above-average protein contents; their colour patterns (L*, a*, or b* ± SD) can be quantified as follows: BIF-2 (L* +0.75, a* − 0.75, b* +0.75); PMP-2 (L* +0.75, a* − 0.75, b* +0.25); SBD-2 (L* − 0.75, a* +0.25, b* +0.25); HCC-1 (L* − 1.75, a* +0.25, b* − 0.75); JPB-2 (L* − 1.75, a* +1.25, b* − 0.75). These landrace accessions were also characterized by above-average contents of nutrients, except Al and Ca, and a low C:N ratio. Each one of these landrace accessions had large (>mean+2.25 SD) content(s) of one or more nutrients as follows: BIF-2 had a large M content, PMP-2 had large P and Zn contents, SBD-2 had large Cu, K, Mn and S contents, HCC-1 had large Mn and Zn contents and JPB-2 had large Cu, K and S contents.
The colour patterns characterizing four accessions (JAG-1, 2, 3 and 4; Fig. 3) in population [1] differed from the rest and were associated with below-average protein and nutrient contents, and an above-average C:N ratio. However, three of these accessions (JAG-1, JAG-2 and JAG-4) differed from JAG-3 in a few traits. Colour patterns of these accessions can be quantified as follows: JAG-1 (L* +1.25, a* − 0.75, b* +1.25); JAG-2 (L* − 0.75, a* +0.25, b* − 0.75); JAG-4 (L* +1.75, a* − 0.75, b* +0.25); JAG-3 (L* +0.25, a* +1.25, b* +1.25). Their higher protein and nutrient contents were slightly higher than the other landraces in this sub-cluster. Finally, colour patterns (L* − 1.75, a* − 0.75, b* − 1.75) of two accessions (HCC-1 and HCC-2) selected from the same landrace and linked at a small Euclidean distance (~20.0; Fig. 3) and their association with kernel shape and quality traits were almost identical. Nevertheless, HCC-2 had an above-average C:N ratio (+3.25 SD) when compared with HCC-1 which had a below-average C:N ratio ( − 0.75 SD). This difference in the C:N ratio, directly or indirectly, resulted in major differences between these two landraces in some kernel shape (e.g. kernel weight, CIR, SLD and DEN) and quality (protein and most micro- and macronutrients) traits.
Discussion
The introduction of maize into North America and its continued evolution after domestication led to its tremendous diversification into highly diverse landraces (Nabhan, Reference Nabhan1985). Farmers' selection and management, when combined with macro-environmental heterogeneity (Perales et al., Reference Perales, Brush and Qualset2003; Olson et al., Reference Olson, Morris and Mendez2012) and cultural diversity (Nabhan, Reference Nabhan1985) in Native American communities, may explain why distinct and diverse landraces occur in geographic proximity and under seemingly similar environments.
Arguably, perceptual distinctiveness of kernel shape and colour traits is the single most important tool that enabled each landrace to be distinguished in traditional complex cropping systems (Gibson, Reference Gibson2009). The current maize germplasm collection, although not exhaustive, came from a relatively small geographical region in the Upper Midwest of the USA; it exhibited large levels of variation in several kernel shape, quality and colour traits. Selection may have been relaxed for a number of interrelated traits at the community level and hence the large level of variation between landraces within the community (Perales et al., Reference Perales, Brush and Qualset2003). It was possible to assess the adaptive nature of the large genetic variation in this germplasm at three hierarchical levels: populations; landraces within populations; accessions within landraces, in addition to their interaction with the environment. Native American farmers typically mediate the evolution of maize landraces by exchanging and mixing seed lots, by imposing selection on landrace populations through management practices and by choosing ears and kernels with desirable shape, nutritional and colour traits (Cleveland et al., Reference Cleveland, Soleri and Smith1994).
Germplasm diversity
A century after warning (Baur, 1914; cf. van de Wouw et al., Reference van de Wouw, Kik, van Hintum, van Treuen and Visser2009) of the consequences of the disappearance of traditional landraces for the future of plant breeding, subsistence farmers, including Native American farmers, continue to cultivate and sustainably conserve and use a highly diverse germplasm pool of landraces (Bellon, Reference Bellon1996). Diverse maize landraces, both within and between populations in this and other Native American communities (Chambers et al., Reference Chambers, Brish, Grote and Gepts2007; Miller, Reference Miller2010), provide a variety of diets, forge social ties and fulfil certain rituals. Additionally, landrace diversity continues to serve as an insurance to cope with emerging economic, social and environmental changes (Shiferaw et al., Reference Shiferaw, Prasanna, Hellin and Bänziger2011; Prasanna, Reference Prasanna2012). Maize landrace diversity may become more critical to maintain maize production under future climate change scenarios (Ureta et al., Reference Ureta, Martinez-Meyer, Peralis and Alvarez-Buylla2012). Maize landraces display a clear spatial structure, corresponding to isolation-by-distance locally and to clinal variation regionally (van Etten, Reference van Etten2006), and are likely to respond differently to alternative future climate scenarios (Ureta et al., Reference Ureta, Martinez-Meyer, Peralis and Alvarez-Buylla2012). It is therefore prudent to understand factors determining the level of diversity and distribution of landraces in order to have a better insight of how they may respond to climate change.
The Native American community of the White Earth Reservation is actively involved in restoring traditional food crops, including maize landraces that fit their socio-economic, cultural and environmental conditions. In spite of a gradual decline in the number of Native American farmers (Nabhan, Reference Nabhan1985; USDA, 2009), and a decline in the number and diversity of maize landraces on Native American farms, a resurgence of native food production offers opportunities to restore, conserve and sustainably use these and other crop landraces (Winona LaDuke, personal communications). The number of maize landraces cultivated on the White Earth Reservation (7), or managed by a single farmer (3) in this study, falls within the reported range of 1 (Soleri and Smith, Reference Soleri and Smith1995), as few as 1–7 (Louette et al., Reference Louette, Charrier and Berthaud1997) and as many as 17 landraces managed by a Native American household (Gibson, Reference Gibson2009), each with different kernel colour(s). The number of maize landraces that can be maintained and managed by a farmer (Boster, Reference Boster1995) or a community (Perales et al., Reference Perales, Brush and Qualset2003; Gibson, Reference Gibson2009) is usually constrained by the ability to observe and remember perceptual differences between landraces. Obviously, conscious selection resulted in the increased variation of those traits considered as valuable for landrace identification (Boster, Reference Boster1995). This model of perceptual selection and identification is generally accepted (Boster, Reference Boster1995; Bellon, Reference Bellon1996; Chambers et al., Reference Chambers, Brish, Grote and Gepts2007), although with some reluctance (Benz et al., Reference Benz, Perales and Brush2007), as a necessary condition for landrace maintenance and is not random. Some researchers have contended that for a crop with significant gene flow such as maize, a reduction in the number of landraces may not necessarily affect diversity much at a regional scale (van de Wouw et al., Reference van de Wouw, Kik, van Hintum, van Treuen and Visser2009). Maize seed is exchanged frequently in this (www.protectseed.com) and other Native American communities (Zimmerer, Reference Zimmerer1991), thus potentially adding more spatial and temporal diversity. A relatively small validation (0.45) variance explained by two PCs suggested that germplasm collection is highly diverse when compared with a much larger maize germplasm collection (Jaradat and Goldstein, Reference Jaradat and Goldstein2013). Seven PCs were necessary to extract from the 22 traits in this study in order to account for 0.76 of total validation variation (data not presented).
Diversity in kernel shape descriptors
Kernel shape descriptors were based on morphological traits that are shaped by the interaction of genotypes with the environment that can result in different phenotypic expressions of the same genotype (Chambers et al., Reference Chambers, Brish, Grote and Gepts2007). A long-term outcome is the creation of multiple types that can be identified as landraces (Mercer et al., Reference Mercer, Martínez-Vásquez and Perales2008); preferences for local seed may have pushed these landraces apart (Perales et al., Reference Perales, Brush and Qualset2003). Kernel shape descriptors were temporally stable; they displayed, as a group, the smallest levels of variation (CV, 5–16%) and were statistically dependent on kernel weight, except CIR and SLD (Supplementary Table S1, available online only at http://journals.cambridge.org). Both traits had negative loadings on PC1 (Fig. 1) and, along with RND, formed a sub-cluster that was separated from kernel weight at 60% Euclidean distance (Fig. 3). Although there were significant differences between populations, most of their variations were due to differences between accessions within landraces (Supplementary Table S1, available online only at http://journals.cambridge.org), followed by differences between landraces (Supplementary Table S2, available online only at http://journals.cambridge.org). CIR was the only trait where the year × accessions within landrace interaction was significant (Supplementary Table S1, available online only at http://journals.cambridge.org); however, with a small variance (4.4%). The remaining traits, with a non-significant genotype × environment interaction (Ortiz et al., Reference Ortiz, Sevilla, Alvarado and Crossa2008), are among the best descriptors of kernel shape.
Unlike Peruvian highland maize races (Ortiz et al., Reference Ortiz, Sevilla, Alvarado and Crossa2008) where the variance component between races was larger and more important than the variance components for accessions within races for most internal ear traits, the variance for accessions within landraces in this study was larger than, almost equal to or smaller than the variance for landraces within populations for 13, 6 and 3 traits, respectively (Supplementary Table S1, available online only at http://journals.cambridge.org). Therefore, accessions within landraces and landraces within populations can be considered as rich sources of kernel shape, quality and colour traits or trait combinations (van Etten, Reference van Etten2006).
Diversity in kernel colour traits
Maize landraces are well known for their kernel colours and patterns, including uniform black, blue, grey-blue, pink, red, white and yellow, or with streaks (Gibson, Reference Gibson2009); all of which have been documented in the current germplasm collection. Colour streaking appeared on light-coloured kernels (large L* values, e.g. MFC-2 and PLC-3; Fig. 3), with the streaks often being orange or red and may indicate the presence of transposable elements or jumping genes (Wood and Lenné, Reference Wood and Lenné1997). The colour traits (Fig. 1) were associated with different traits and different landraces; these associations confirmed earlier findings (Floyed et al., Reference Floyed, Rooney and Bockholt1995; Jaradat and Goldstein, Reference Jaradat and Goldstein2013) and were supported by the results of rigorous statistical testing as indicated by the per cent variance in quality traits explained by different combinations of kernel colour traits (Supplementary Table S3, available online only at http://journals.cambridge.org).
Kernel colour variability in the current germplasm collection, as quantified by the magnitude and variation of colour traits, is an indicator of how complex, and culturally and geographically widespread, this perceptual distinctiveness trait is. Kernel colour, a perceptually distinctive, but generally considered as a non-adaptive trait (Gibson, Reference Gibson2009), dominated maize classification by Native American farmers (Moeller and Schaal, Reference Moeller and Schaal1999) and may serve as a critical trait to maintain landrace integrity (Wood and Lenné, Reference Wood and Lenné1997). However, it is not universally used as a significant trait for distinguishing between different landraces (Benz et al., Reference Benz, Perales and Brush2007), and may have been used by farmers to eliminate off-type coloration patterns in their fields (Zimmerer, 1997). Additionally, Zimmerer (Reference Zimmerer1991) ruled out repeated selection for kernel colour as a perceptual distinctiveness trait in maize because ‘it would create unmanageable number of cultivars within a few years, and may not be associated with agro-ecologically significant or favorable traits’. Nevertheless, kernel colour traits in this germplasm displayed the largest variation (CV 48, 70 and 159%, for L*, b* and a*, respectively; Supplementary Table S1, available online only at http://journals.cambridge.org), and had significant standardized coefficients and contributed to a large (93.5–100%) correct classification of landraces in the multivariate discriminant analysis (Fig. 2(a) and (b)). On average, total variance in kernel colour traits explained by random factors (47.8%) was larger than that of kernel shape traits (15.08%) or micro- and macronutrients (31.5%). Compared with a larger maize germplasm collection (Jaradat and Goldstein, Reference Jaradat and Goldstein2013) of different genetic backgrounds (CV L*= 3.2, a*= 16.07 and b*= 10.9%), the current Native American maize landraces displayed a larger variation in all the three colour traits (CV for L*= 48.0, a*= 191.0 and b*= 69.0%; Supplementary Table S1, available online only at http://journals.cambridge.org). Additionally, kernel colour traits were temporally stable and independent of kernel weight (covariate) at the landrace and population levels, and exhibited larger diversity between accessions within landraces when compared with the variation between landraces within populations (Supplementary Table S1, available online only at http://journals.cambridge.org). The pattern of variance partitioning may suggest the presence of different structures of variation, depending on the traits examined (Bellon, Reference Bellon1996).
Quality traits
Maize kernel composition is important for human nutrition (Flint-Garcia et al., Reference Flint-Garcia, Bondar and Scott2009), especially for Native American communities where it comprised, until recently, a major source of energy, protein and micro- and macronutrients (Arvanitoyyannis and Vlachos, Reference Arvanitoyyannis and Vlachos2009). Detailed information on landrace-specific composition can be used in breeding and improvement programmes to enhance the nutrient content of more commonly used landraces (Burlingame et al., Reference Burlingame, Charrondiere and Mouille2009). In addition to their inherently high protein content, Native American maize landraces proved to be rich sources of macro- and micronutrients when compared with commercial maize varieties (Arvanitoyyannis and Vlachos, Reference Arvanitoyyannis and Vlachos2009) and maize germplasm developed for protein quality (Jaradat and Goldstein, Reference Jaradat and Goldstein2013).
Protein and C:N ratio
Protein content (12.09 ± 1.89) and C:N ratio (24.55 ± 4.47) are two important antagonistic quality parameters in maize kernels. They exhibited a negative relationship on both PCs (Fig. 1), discriminated between landraces with almost equal negative and positive standardized coefficients, respectively (Fig. 2(b)), were dependent on kernel weight and responded to fixed and random factors in almost the same quantitative manner (Supplementary Table S1, available online only at http://journals.cambridge.org). Qualitatively, protein and C:N were positively and negatively associated with micro- and macronutrients, respectively (Fig. 1). A high protein content was a characteristic of landraces in population [1] having a larger kernel weight and kernel DEN; while a high C:N ratio was a characteristic of landraces in population [2] having a smaller kernel weight and DEN.
Dynamics of nutrient densities
Factors influencing nutrient densities in maize kernels are numerous and complex (Simic et al., Reference Simic, Sudar, Ledencan, Jambrovic, Zdunic, Brkic and Kovacevic2009). The effect of the genotype × environment interaction on nutrient contents (especially Fe and Zn) is an important aspect of selecting landraces with a stable content of these nutrients (Simic et al., Reference Simic, Sudar, Ledencan, Jambrovic, Zdunic, Brkic and Kovacevic2009; Jaradat and Goldstein, Reference Jaradat and Goldstein2013). The population × year interaction component in the fixed model (Supplementary Table S1, available online only at http://journals.cambridge.org) was non-significant for all nutrients; the marginally significant value (P= 0.06) for P was the only exception. The three random factors in the mixed model (Supplementary Table S1, available online only at http://journals.cambridge.org) explained, on average, 31.6 and 31.3% of the total variance in micro- and macronutrients, respectively. The largest sources of variation for Fe and Zn were landraces within populations and accessions within landraces, respectively.
Non-significant correlations of Fe and Zn with P, and both positive and significant correlations between Fe and Zn have been reported in maize (Menkir, Reference Menkir2008), suggesting that Fe and Zn contents can be improved regardless of the large P content, which could be an indicator of the phytate content. The correlation between P and each of Fe (0.74) and Zn (0.56), and between Fe and Zn (0.81) were significant (P< 0.01) in population [1]; only P was significantly correlated with Zn (0.79) in population [2]. These correlations may have practical breeding and nutritional implications (Simic et al., Reference Simic, Sudar, Ledencan, Jambrovic, Zdunic, Brkic and Kovacevic2009), especially if the kernel phytate content is large (Menkir, Reference Menkir2008). When averaged over landraces, P had larger positive standardized coefficients than Zn on Root1 (Fig. 3(b)) and slightly smaller negative standardized coefficients on Root2 than Fe, while Zn and Fe had opposite standardized coefficients on both roots. These relationships may reflect the dependence of Fe and Zn, but not of P, on kernel weight (Supplementary Table S1, available online only at http://journals.cambridge.org) and the variable contents and complex interrelationships between these nutrients in different landraces (Supplementary Table S2, available online only at http://journals.cambridge.org; Fig. 3(a)).
Kernel colour traits and quality
Unlike modern maize hybrids whose kernels are either white or yellow coloured and predominantly preferred for human food and animal feed, respectively (Floyed et al., Reference Floyed, Rooney and Bockholt1995; Shiferaw et al., Reference Shiferaw, Prasanna, Hellin and Bänziger2011), maize landraces are rich sources of multivariate kernel colours (Kuhnen et al., Reference Kuhnen, Lemos, Capestrini, Ogliari, Dias and Maraschin2011). Kernel colour traits had complex interrelationships with each other and with quality traits, as can be deduced from PCA (Fig. 1) and coefficients of PLS validation regression models (Supplementary Table S3, available online only at http://journals.cambridge.org). Kernel colour traits exhibit significant relationships with other components of the maize kernel, including kernel shape and quality traits (e.g. oil, crude fibre, protein and nutrients, such as Ca, Cu and K) (Kaur et al., Reference Kaur, Singh and Rana2010; Jaradat and Goldstein, Reference Jaradat and Goldstein2013). The positive loading and the association of a* and quality traits on PC1 when coupled with denser kernels (Flint-Garcia et al., Reference Flint-Garcia, Bondar and Scott2009) suggest a potentially larger content of nutrients and protein in denser kernels with a reddish colour (Fig. 1). This deduction is supported by the dissociation on PC1 of a* and quality traits from the C:N ratio, the larger values of which indicate lower kernel quality. Similarly, positive loadings of b* and nutrients on PC2 may indicate a larger carotenoid content (Kuhnen et al., Reference Kuhnen, Lemos, Capestrini, Ogliari, Dias and Maraschin2011).
The large variation in a relatively small germplasm collection of maize landraces for, and interrelationships between, several kernel shape and quality traits, on the one hand, and kernel colour traits, on the other hand, offers many opportunities for quality improvement in this and other Native American maize landraces. Genes can be identified and selected to develop novel kernel colour combinations in landraces or can be incorporated into new commercial varieties to improve food and feed quality of maize products.
Conclusions
Native American farmers identify and manage a wide range of variations in maize landraces largely on the basis of kernel colour; they developed and selected maize landraces for kernel colour traits that allowed them to distinguish between and maintain diverse maize landraces for various traditional uses. Variable selection criteria applied by Native American farmers assured large genetic diversity and maintained between and within maize landraces. A 2-year experiment was conducted to study and quantify physical, chemical and colour characteristics and to identify any potential relationship between colour traits and nutrient content in a collection of Northern flint maize landraces managed by Native American farmers. The long-term objective of this research was to study the dynamics of maize population at a micro-geographical scale in the context of in situ conservation of maize landraces in the White Earth Reservation and elsewhere in the Upper Midwest. The variation in kernel colour was found to be associated with the variation in quality traits, and may be used to select for certain quality attributes in diverse Native American maize landraces. Native American farmers may have selected maize kernels that matched their mental templates of a maize ideotype, and necessarily derived preferential maize landrace names from the most prominent kernel colour and from the landscape that surrounded production or settlement sites. A visual selection procedure, based on quantitative kernel colour estimates, was developed to identify landraces with colour traits that are associated with high levels of quality traits; for example, small L* values were indicative of large Mn and large a*, b* and a*b* values were indicative of large Fe, P and Cu and Mg contents, respectively. Furthermore, this procedure can be used to identify closely related landraces and to maintain a large variation between maize landraces on Native American farms.
Acknowledgements
Thanks to Winona LaDuke and an anonymous Native American farmer who donated the seed of maize landraces for the study. Thanks to Charles Hennen for his assistance in the fieldwork, and Jana Rinke and Jay Hanson for their laboratory work. This research was funded by USDA-ARS Project No. 3645–61600-001-00D, Morris, MN. The use of trade, firm or corporation names in this publication is for information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable. USDA is an equal provider and employer.