Introduction
Schizophrenia is a highly heritable disorder, which effects approximately 1% of the general population (Harrison & Weinberger, Reference Harrison and Weinberger2005). The underlying genetic architecture of schizophrenia has been hard to define, partly because it is a pathophysiologically complex, clinically heterogeneous disorder. Increasingly, however, evidence points to a substantial proportion of trait variation being polygenic; consisting of many common single-nucleotide polymorphisms (SNPs), each of small effect (Purcell et al. Reference Purcell2009; Lee et al. Reference Lee2012; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). At present, the diagnosis of schizophrenia is based solely on the clinical picture, and no objective biological marker exists for predicting which individuals are more likely than others to transition to psychosis. Genetic stratification could provide a means by which individuals who are most likely to transition to psychosis are identified pre-morbidly. Recently, there has been success in predicting whether first-episode patients (FEPs) will develop schizophrenia, as opposed to any other psychoses, through the use of polygenic risk scores for schizophrenia (PGRS-SCZ) (Vassos et al. Reference Vassos2017). The accuracy of these scores is continually improving as the Genome-Wide Association Study (GWAS) discovery sample grows (Vassos et al. Reference Vassos2017). Early detection has important clinical implications, as the duration of untreated psychosis has been shown to be associated with worse clinical outcome (Fraguas et al. Reference Fraguas2014), and early intervention may improve a patient's prognosis, or even prevent the onset of psychotic disorders (Heinssen & Insel, Reference Heinssen and Insel2015).
Schizophrenia is also characterized by a host of structural brain alterations, frequently in frontal and temporal cortex (Lawrie & Abukmeil, Reference Lawrie and Abukmeil1998; Fornito et al. Reference Fornito2012; Haijma et al. Reference Haijma2013). These findings have also been shown in some studies of unaffected relatives and in well individuals who later go on to develop schizophrenia (Boos et al. Reference Boos2007; Fusar-Poli et al. Reference Fusar-Poli2011; Cooper et al. Reference Cooper2014). Brain changes may provide crucial insights into the genetic mechanisms of schizophrenia, increasing our understanding of the disorder's underlying aetiology (Bois et al. Reference Bois2015). In examining the relationship between structural brain alterations and genetic risk, prospective studies of currently unaffected individuals at familial high risk (HR) of developing schizophrenia allow researchers to assess whether brain alterations are specifically associated with the disorder itself, or more accurately characterized as trait or vulnerability markers for schizophrenia (Johnstone et al. Reference Johnstone2005; McIntosh et al. Reference McIntosh2011), whilst avoiding potential confounding influences; such as anti-psychotic medication and effects of adaptation to chronic illness.
Brain structural findings in schizophrenia are widespread and may reflect dysconnectivity between diverse regions, particularly between and within frontal and temporal regions (White & Hilgetag, Reference White and Hilgetag2011). These differences may be revealed in measures of cortical complexity, such as the gyrification index (GI) (Zilles et al. Reference Zilles1988), a measure of cortical folding thought to reflect underlying neuronal connectivity (White & Hilgetag, Reference White and Hilgetag2011). A temporal relationship between neuronal migration and patterns of gyrification has also been shown, with disorders that affect early migration having more alterations in the resulting gyrification patterns (White & Hilgetag, Reference White and Hilgetag2011). Developmental studies of gyral folding demonstrate that gyrification remains relatively stable following birth (Zilles et al. Reference Zilles1988), and is associated with genetic factors (Harris et al. Reference Harris2007; Nanda et al. Reference Nanda2014). As such, patient-control differences may reflect changes that take place early in neurodevelopment.
Studies have shown increased gyrification in the frontal lobes of people with schizophrenia (Falkai et al. Reference Falkai2007; Palaniyappan et al. Reference Palaniyappan2011), their unaffected relatives (Falkai et al. Reference Falkai2007), participants with an at-risk mental state (Sasabayashi et al. Reference Sasabayashi2017) and those at familial HR who later developed schizophrenia (Harris et al. Reference Harris2007). Increases in temporal lobe gyrification have been found in FEPs when compared with controls (Harris et al. Reference Harris2004) as well as in those with an at-risk mental state (Sasabayashi et al. Reference Sasabayashi2017); however, this was not reported within those at familial HR (Harris et al. Reference Harris2007).
It is important to note that decreases in gyrification, within these regions, have also been reported within schizophrenia patients (Sallet et al. Reference Sallet2003; White et al. Reference White2003; Bonnici et al. Reference Bonnici2007; McIntosh et al. Reference McIntosh2009; Palaniyappan et al. Reference Palaniyappan2011) but not in those at HR. It may be that these differential results are due to the developmental stage (Bonnici et al. Reference Bonnici2007) and duration of the illness (McIntosh et al. Reference McIntosh2009; Mancini-Marïe et al. Reference Mancini-Marïe2015). Overall, it appears that the link between gyrification and schizophrenia is not completely clear. However, whilst the evidence is limited, the pattern of increased gyrification in relation to both frontal and temporal regions has been previously reported within those with, and at risk of, developing schizophrenia. Nevertheless, further investigation into the association between gyrification and schizophrenia, particularly in relation to familial HR samples, is required.
Despite previously mixed findings, gyrification has been identified as a potential endophenotype for schizophrenia (White & Gottesman, Reference White and Gottesman2012) and has been linked to genetic loading for the disorder. Increased PGRS-SCZ was associated with decreased gyrification in fronto-parietal regions in two independent populations (Liu et al. Reference Liu2016), but further investigation is needed to determine if PGRS-SCZ can be linked to gyrification within a schizophrenia patient or HR sample.
We have therefore constructed a PGRS-SCZ, obtaining marker weights from the most recent Psychiatric Genomics Consortium-2 (PGC2) GWAS data (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) in participants from the Edinburgh High Risk Study (EHRS). We then tested for an association between PGRS-SCZ and the transition to frank schizophrenia in those at familial HR for the disorder. We hypothesized that there would be a greater burden of common genetic risk variants in those at HR who subsequently made the transition, compared with those at HR who remained well and controls. We also assessed whether there was an association between PGRS-SCZ and frontal and temporal indices of gyrification within the HR cohort, as well as their subsequent clinical outcomes, with the expectation that PGRS-SCZ would be associated with altered gyrification.
Methods and materials
Participants
The present analysis was conducted on data from a prospective longitudinal cohort study of people at high familial risk of schizophrenia, conducted over 10 years (Johnstone et al. Reference Johnstone2005). Participants came from families multiply affected with schizophrenia. The recruitment and clinical assessment process for the EHRS have been described in detail elsewhere (Hodges et al. Reference Hodges1999; Johnstone et al. Reference Johnstone2000, Reference Johnstone2005). In summary, at baseline/recruitment HR individuals aged 16–25 years, with no personal history of psychiatric disorder, were contacted throughout Scotland based on the criteria that they had at least two first- and/or second-degree relatives with a diagnosis of schizophrenia. Healthy controls (HC) without personal or family history of major psychiatric disorder were recruited from the same social and geographical networks as the HR subjects in order to minimize potential confounding environmental influences. The groups were similar in sex distribution, paternal social class and education, with the vast majority of HC and HR individuals being either in full-time employment or education at entry into the study; however, genotyping was not conducted until the end of the study.
Informed consent was obtained from all participants, as approved by the Psychiatry and Clinical Psychology sub-committee of the Multi-Centre Research Ethics Committee for Scotland. All applications for continuation and amendment to this study have been filed appropriately with the Scotland Research Ethics Committee, and conformed to the Declaration of Helsinki.
The presence and absence of psychotic and other symptoms was established by Present State Examinations at 18-month intervals on up to five occasions, which was the main clinical assessment used for the study (Wing et al. Reference Wing, Cooper and Sartorius1974). During the course of the study, 21 HR individuals developed International Classification of Diseases, 10th Edition (ICD-10) schizophrenia, HR[ill], 19 of whom had full clinical assessments and 17 had a usable structural magnetic resonance imaging (MRI) scan before disorder onset. Only a subset of these (n = 9) were genotyped and are included in the current analysis. Those in the HR[ill] group were formally diagnosed after an average of 929 days (s.d. = 138) and were not offered re-scanning or re-assessment once the diagnosis had been made. In contrast, some individuals had psychotic symptoms at one or more assessments of the study, but were never ill enough to meet criteria for diagnosis, as they either had only one key symptom, or their symptoms were too transient or mild to satisfy diagnostic criteria – these individuals were designated HR[symp] (n = 60). The remaining HR subjects, who never had any psychotic symptoms throughout the course of the study were designated HR[well] (n = 66). None of the HR individuals were on any form of anti-psychotic medication at the time of scanning. Participants for the HC group (n = 36) were recruited from the social network of the HR individuals and had no personal or family history of psychotic illness, but could have a family history of other psychiatric illness. HCs were otherwise as similar to the HR participants as possible. Numbers of participants who had both structural MRI and genotyped data amounted to 46% of the initial sample and were designated to sample groups as follows: HC (n = 16), HR[well] (n = 31), HR[symp] (n = 28), HR[ill] (n = 9). More detailed demographic information is presented in Table 1.
Table 1. Demographic information for healthy controls and the high-risk sub-groups

* p⩽0.05.
Significant effect of age was due to the HR[well] cohort being significantly older than the HR[ill] group.
All demographics were measured at the time of scan.
Handedness was measured using the Annett Hand Preference Questionnaire.
Genotyping and derivation of PGRS
Information on the genotyping process can be found in online Supplementary Data.
Polygenic profile scores were generated using imputed genotype data. Imputation was performed in accordance with the ENIGMA2 1000 genomes protocol (ENIGMA2 Genetics Support Team, 2012). SNPs with an imputation R 2 quality score of >0.3 were retained for further analysis resulting in 6 145 246 SNPs. All subsequent analyses were performed in PLINK (Purcell et al. Reference Purcell2007). Further QC criteria were applied to imputed data. Individuals with missingness >2% were excluded, as were SNPs with a genotype call rate of <98%, Hardy–Weinberg equilibrium p value <1 × 10−6, a minor allele frequency of <5%, or those that were strand ambiguous. Clump-based linkage disequilibrium pruning (r 2 0.2, 300 kb window) was performed to create a SNP set in approximate linkage equilibrium. Marker weights (logarithm of the odds ratio) and p value association statistics for SNPs were derived from the most recent PGC GWAS of schizophrenia (9.8 million autosomal SNPs) (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). PGRS were generated at five p value thresholds by multiplying the number of copies of the reference allele by its marker weight for each SNP, and summing across all SNPs, as described in Purcell et al. (Reference Purcell2009).
Imaging parameters
Structural brain images were acquired using a 42 SPE Siemens (Erlangen, Germany) Magnetom scanner operating at 1.0 T. The sequence was a three-dimensional (3D) magnetization prepared rapid acquisition gradient echo sequence consisting of a 180° inversion pulse followed by a fast low-angle shot collection (flip angle 12°, repetition time 10 ms, echo time 4 ms, inversion time 200 ms, relaxation delay time 500 ms, field of view 250 mm), giving 128 contiguous slices with a thickness of 1.88 mm. The sequence was selected in order to obtain optimal grey and white matter contrast.
FreeSurfer analysis
Cortical reconstructions were generated using the surface-based stream of the software FreeSurfer, version 5.3.0 (http://surfer.nmr.mgh.harvard.edu/fswiki/recon-all/). Briefly, this processing includes motion correction and averaging of T1-weighted images, removal of non-brain tissue using a hybrid watershed/surface deformation procedure, automated Talairach transformation, intensity normalization (Ségonne et al. Reference Ségonne, Pacheco and Fischl2007), tessellation of the grey matter–white matter boundary, automated topology correction (Sled et al. Reference Sled, Zijdenbos and Evans1998; Fischl et al. Reference Fischl, Liu and Dale2001) and surface deformation to place the grey/white and grey/cerebrospinal fluid borders optimally (Fischl et al. Reference Fischl, Sereno and Dale1999). This method uses both intensity and continuity information from the entire 3D MR volume in segmentation and deformation procedures to produce representations of cortical thickness and surface area (Fischl & Dale, Reference Fischl and Dale2000). The maps are created using spatial intensity gradients across tissue classes and are therefore not simply reliant on absolute signal intensity. Reconstructed data sets were visually inspected for accuracy, and all segmentation errors were corrected by a trained, group-blinded rater (CB).
Gyrification indices
Local gyrification indices (LGI) were obtained with the method of Schaer et al. (Reference Schaer2008), which operates on the images reconstructed through the FreeSurfer pipeline described above. Schaer et al’s method is a vertex-wise extension of Zilles’ original 2D GI, which is a measure that gives an estimate of the inner folded contour compared with the outer perimeter of the cortex (Zilles et al. Reference Zilles1988). A detailed description of Schaer et al’s method can be found in online Supplementary Data.
Statistical analysis
All statistical analyses were computed with R version 3.2.2 (http://www.r-project.org). Analyses of covariance (ANCOVAs) were used to implement mixed-model linear analyses. A genetic relationship matrix (GRM) was created using autosomal SNPs that passed QC (Gorjanc et al. Reference Gorjanc, Henderson, Kinghorn and Percy2007). The ASReml-R (www.vsni.co.uk/software/asreml) software package was used to implement mixed linear model analyses with an inverse relationship matrix derived from the GRM fit as a random effect. These analyses were performed to determine whether genetic relatedness within the sample impacted on the associations reported, a method described previously (McIntosh et al. Reference McIntosh2016).
To account for potential confounding due to population stratification, multi-dimensional scaling (MDS) components were created following the ENIGMA 1000 genomes protocol (ENIGMA2 Genetics Support Team, 2012), using the software package, PLINK (Purcell et al. Reference Purcell2007). Four MDS components were included in subsequent analyses models within this study, consistent with previous publications (McIntosh et al. Reference McIntosh2013; Whalley et al. Reference Whalley2015). Results presented here primarily refer to PGRS-SCZ at the p ⩽ 0.1 threshold, as this threshold was shown to explain the most phenotypic variance in the discovery cohort (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Results at the four other thresholds (p ⩽ 0.01, 0.05, 0.5 and 1) are presented in online Supplementary Data. All analyses are subject to a statistical significance level of p < 0.05.
Group comparisons in mean PGRS
Gender, age (mean centred), MDS components 1–4 and group status (HR[well], HR[symp], HR[ill] and HC) were entered as fixed effects in the ANCOVA models, with family structure fitted as a random effect in subsequent ASReml-R models. Family structure was inferred using genomic data from 61 268 SNPs across the genome and an inverse G matrix created. PGRS-SCZ was entered as the dependent variable in both models.
For the ASReml-R models, Wald's conditional F-test was used to calculate p values for fixed effects.
All ANCOVA and ASReml-R models were corrected for multiple comparisons using false discovery rate (FDR) correction, with a rate of p = 0.05 (Genovese et al. Reference Genovese, Lazar and Nichols2002), with pairwise comparisons adjusted using Tukey from the R package ‘lsmeans’.
PGRS and frontal and temporal gyrification
The covariates were entered as in the above analyses, with the addition of global gyrification (left/right) as a covariate, to assess localized, as opposed to global changes in gyrification. Frontal/temporal (left/right) gyrification was entered as the dependent variable, with PGRS-SCZ as the predictor variable of interest. These models were conducted for controls, the HR cohort as a whole, and then with the HR group split by subsequent clinical outcome. For the latter model, a group by PGRS-SCZ interaction was used as the predictor of interest with further analyses of individual sub-groups, FDR corrected, to explore significant relationships.
Mean gyrification
Comparisons of mean gyrification utilized the means of left and right, frontal and temporal gyrifications within the sample.
Results
Demographics
Statistical analysis of demographic variables revealed a significant difference in age between groups, with pairwise comparisons suggesting this was due to the HR[well] cohort being significantly older than the HR[ill] group (p = 0.044). However, all statistical models controlled for age within this study.
No other significant differences were found for any other demographics of the sample (Table 1).
Group comparisons in mean polygenic scores
Mean PGRS-SCZ for each group, at all five thresholds, are reported in online Supplementary Data, Table S1.
The main effect of group on PGRS-SCZ was significant at the p ⩽ 0.1 threshold (F 3,74 = 2.91, p = 0.040). Differences at other thresholds can be seen in online Supplementary Data, Table S1.
Subsequent pairwise comparisons, adjusted using Tukey, revealed that this was because the HR[ill] cohort had significantly increased PGRS-SCZ compared with controls (p = 0.037). These results are illustrated in Fig. 1. No other contrasts reached significance (online Supplementary Data, Table S2).

Fig. 1. Boxplot illustrating differences in mean PGRS-SCZ by group, where mean polygenic loading for the HR[ill] group (n = 9) was significantly different to the HC group (n = 16). Y-axis represents PGRS-SCZ. X-axis are the four groups: HR+ = HR[symp], HR =HR[well], ILL = HR[ill] and CON = healthy controls. Two outliers are highlighted in this figure; however, these individuals were <3 standard deviations away from the mean and the decision was therefore made to include them within the analysis.
Importantly, this main effect was also significant when adjusted for relatedness in ASReml-R (F 3,74 = 3.22, p = 0.028).
Associations between polygenic scores and frontal and temporal gyrification
Frontal lobe
For the HR cohort, as a whole, there was a significant effect of PGRS-SCZ on left frontal (F 1,59 = 7.37, p = 0.010) and right frontal gyrifications (F 1,59 = 5.38, p = 0.024), where an increase in genetic loading was associated with bilateral hypergyrification. These findings survived FDR correction (left frontal: p corrected = 0.035, right frontal: p corrected = 0.048) and are illustrated in Figs 2a and b. These results were also apparent when controlling for relatedness: left frontal gyrification (F 1,58.9 = 7.12, p = 0.010, p corrected = 0.039), right frontal gyrification (F 1,59 = 5.38, p = 0.024, p corrected = 0.048). No effect was found for HC (online Supplementary Data, Table S3).
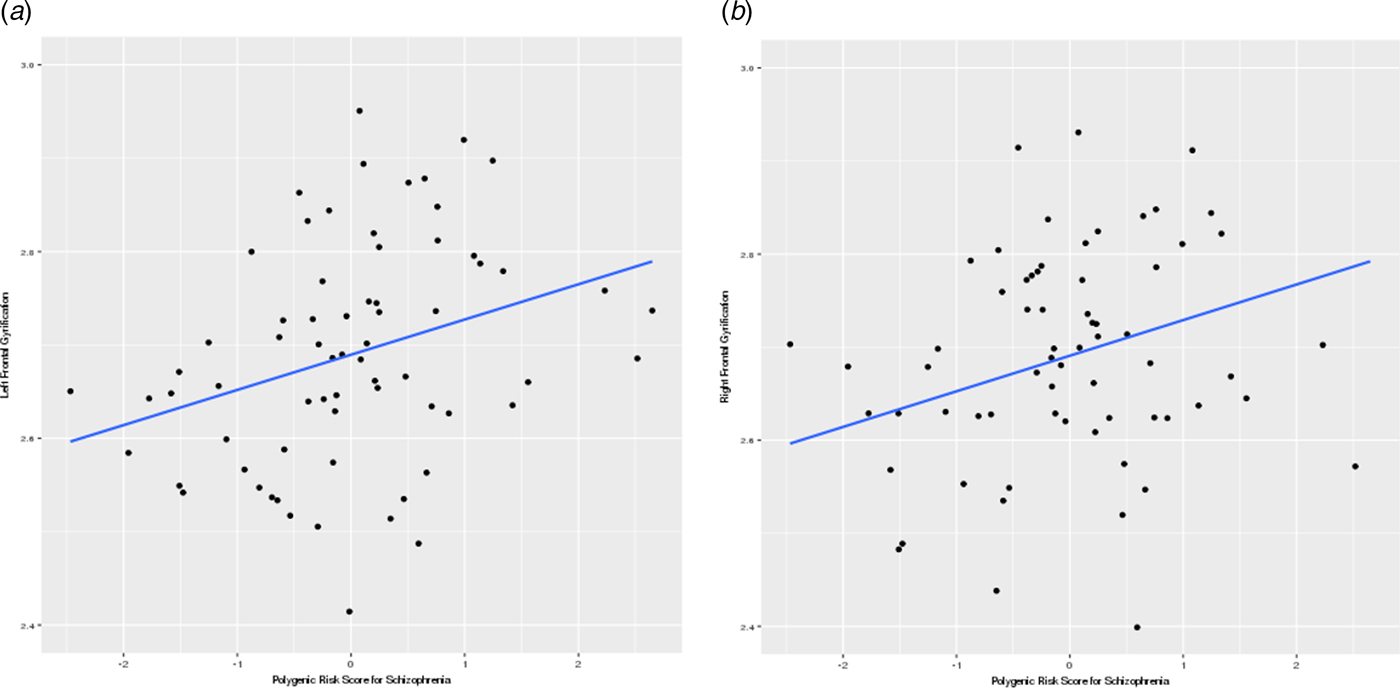
Fig. 2. Scatter plots showing a positive relationship between PGRS-SCZ and frontal gyrification in the (a) left and (b) right hemisphere. Y-axis represents (a) left and (b) right frontal gyrifications. X-axis represents PGRS-SCZ.
Analysis of the differences between the HR sub-groups revealed a significant interaction of group and PGRS-SCZ on the right frontal lobe (F 2,56 = 4.78, p = 0.012), which survived FDR correction (p correted = 0.050).
Further exploration of the HR sub-groups individually revealed a significant positive relationship with polygenic loading and right frontal gyrification for the HR[symp] group (F 1,20 = 10.63, p = 0.004, p correted = 0.012). Only a trend-level effect, in the same direction, was found for the HR[ill] group, although a large effect size (η 2 = 0.98) suggests that, had this cohort included a larger number of participants, this result may have reached significance (Table 2). No effect was found within the HR[well] group (online Supplementary Data, Table S4).
Table 2. Significance and effect size values for the effect of polygenic risk for schizophrenia on the left and right frontal lobes, within the high-risk sub-groups

Effect size = partial η 2.
There was also a positive effect of PGRS-SCZ within the left frontal lobe for the HR[symp] sub-group (F 1,20 = 6.21, p = 0.022), but this did not survive FDR correction (p corrected = 0.065). No significant effects were found for the remaining HR sub-groups in this region (Table 2).
Temporal lobe
No associations between PGRS-SCZ and temporal gyrification reached significance in the temporal lobe for either the HR cohort, HR cohort split by subsequent clinical outcome or in controls (online Supplementary Data, Tables S3 and S4).
Group comparisons in mean gyrification
Models comparing mean gyrification between groups failed to reach significance within any of the areas of interest (online Supplementary Data, Table S5). However, inspection of the means suggested that there was a pattern of incremental increase in gyrification for participants at HR who went on to develop symptoms and transition into schizophrenia (Table 3).
Table 3. Mean gyrification for each group within the regions of interest, along with their standard deviations

Discussion
To our knowledge, the present study is the first to examine PGRS-SCZ in a prospectively ascertained familial HR cohort, and to show that such scores are associated with transition to schizophrenia. The current results, along with the recent finding that the PGRS-SCZ can be used to predict between FEPs who go on to develop schizophrenia or other psychoses (Vassos et al. Reference Vassos2017), have important implications for our understanding of the aetiology of schizophrenia. It is likely, for example, that genetic and environmental factors, and their interactions, determine who of those at elevated genetic risk go on to get a broader phenotype of psychotic symptoms or the narrower phenotype of schizophrenia. Although PGRS are currently underpowered to be of clinical utility in a predictive capacity, they are sufficiently well-powered to test for association between different phenotypes and the PGRS (Dudbridge, Reference Dudbridge2013). Therefore, using neuroimaging phenotypes in combination with PGRS could become a useful tool in clinical settings in the future. The utility of both measures will increase as collaborative efforts yield ever-larger sample sizes; however, due to the current sample size, the results reported here should be interpreted with caution.
This study is also the first to examine the relationship between PGRS-SCZ and gyrification within a HR sample. We found that within the HR cohort, but not controls, an increased genetic load for schizophrenia was positively and significantly associated with increased bilateral frontal gyrification. As there is a temporal relationship between patterns of neuronal migration and patterns of gyrification, with disorders that affect migration early having more pronounced alterations in the pattern of gyrification, our results suggest that a mechanism by which an increased genetic load for schizophrenia increases frontal gyrification may be involved in the pathogenesis of the disorder. However, the current results require replication in a large data set to provide further evidence for this theory. Nevertheless, our current findings are in keeping with our own and other earlier studies, using less automated measures of GI which found an association with familial risk for schizophrenia and propensity to develop the disorder (Vogeley et al. Reference Vogeley2000; Harris et al. Reference Harris2004, Reference Harris2007).
We also found that increasing PGRS-SCZ was related to right frontal gyrification in the HR[symp] and HR[ill] sub-groups of the HR cohort. Hence, those at HR who developed schizophrenia had a significantly greater proportion of risk alleles for schizophrenia which, for the HR[symp] cohort and, at trend level for the HR[ill] cohort, correlated with gyrification estimates. Although, this finding was only at trend level for the HR[ill] sub-group, the result was also associated with a large effect size and it is suggested that the lack of significance was a result of low numbers within this group. The interaction effect of group and PGRS-SCZ was not found within the left frontal lobe; however, a trend-level effect was found within the HR[symp] group individually.
It is also important to note that, analysis of the HR group as a whole, provided significant effects bilaterally. Thus, it may be that this polygenic risk-influenced gyrification is a ‘trait’ marker of schizophrenia, and provides a putative mechanism by which genetic influences affect subsequent neurodevelopment and increase an individual's risk of developing schizophrenia. Further analysis of cohorts, using a larger sample size, are required to determine if this result is related to the HR status as a whole or can be predicted by subsequent clinical outcome. As schizophrenia is such a complex and heterogeneous disorder, however, it is unlikely to be fully mediated by this one mechanism. It is likely that other genetic and environmental risks, through development and nearer to the time of onset of the disorder, interact with the polygenic risk-influenced gyrification differences which we have found.
Lack of an effect within the temporal lobe was not entirely unexpected. Previous research has linked temporal lobe abnormalities in gyrification to participants with developed schizophrenia (White et al. Reference White2003; Harris et al. Reference Harris2004) rather than those at HR (Harris et al. Reference Harris2007), suggesting that this deficit may be more related to the impact of the disorder rather than its genetic architecture. It would be of interest for future studies to further investigate this by replicating the current result within an independent HR sample as well as to conduct these analyses within a patient population.
Contradictory to our previous findings within this sample (Harris et al. Reference Harris2007), we did not find that gyrification predicted group status within the present study. However, investigation of the means suggests that there is an incremental increase in both the frontal and temporal lobes in those that develop symptoms and subsequently transition to schizophrenia. As outlined in the introduction, it is important to note here that the research into the link between gyrification and schizophrenia has been fairly inconsistent. However, within the current study, it is suggested that the lack of a significant effect may be due to the small sample size; as only a sub-section of the previous sample had genotyped information that could be used in the current analyses. Future studies should further investigate the relationship between transition to schizophrenia and gyrification and, in turn, the association with PGRS-SCZ.
The main limitation within this study is the small sample size, particularly in relation to the HR[ill] group, which may impede the generalizability of the results. Nevertheless, the incremental deviation from HC in gyrification estimates across the HR sub-groups (Table 3) along with the case–control differences in PGRS-SCZ (Fig. 1) found within the current study, which are in keeping with previous findings (Harris et al. Reference Harris2007; Falkai et al. Reference Falkai2007; Palaniyappan et al. Reference Palaniyappan2011; Vassos et al. Reference Vassos2017), lend credibility to the results.
Another limitation relates to the parameters of the MRI scans for use with FreeSurfer. The recommended guidelines suggest 1.5–3 T with 1 mm3 isotropic resolution; however, the current MR images were collected at 1 T with a voxel size of 1.8 mm3 (https://surfer.nmr.mgh.harvard.edu/fswiki/). Although we are working with slightly lower spatial resolution parameters than those recommended by FreeSurfer, we have attempted to overcome this potential issue by manually editing all of the data and checking the fidelity of all of the imaging reconstructions. Moreover, by quantifying large-scale shape parameters such as LGI, which is essentially calculated over 25 mm of the cortical surface (Schaer et al. Reference Schaer2008), we are less likely to suffer from voxel-based inaccuracies than, for example, a per-vertex thickness measure that depends on grey–white contrast ratio. Furthermore, the current results rely on a comparison between subjects acquired on the same scanner, and the fact that we only see results in the frontal (and not temporal) regions suggests that the lower field strength does not hamper such comparisons. Thus, despite utilising slightly lower scan resolution than recommended, these results still provide meaningful insight into the relationship between polygenic loading for schizophrenia and frontal gyrification.
In conclusion, we have found that an increased genetic loading for schizophrenia was present in those who developed schizophrenia, compared with those at HR who developed symptoms, remained well or controls. This increased loading was associated with altered bilateral frontal gyrification within the HR cohort, but this association was not present in the control group. Limited evidence for this effect being dependent on subsequent clinical outcome was found within the right frontal lobe; however, small sample sizes impeded our ability to investigate this fully. Understanding the genetic architecture of schizophrenia and polygenic effects on brain structure could have critical implications for our knowledge of the aetiology and pathogenesis of the disorder and thus provide us with a means to provide more targeted interventions for people at an elevated familial risk of developing schizophrenia.
Supplementary Material
The supplementary material for this article can be found at https://doi.org/10.1017/S0033291717003087.
Acknowledgements
The authors would like to thank all the participants and their families for taking part in this study and the radiographers who acquired the MRI scans at the City Hospital in Edinburgh. The authors would also like to extend their thanks to both Lauren Navrady and Simon R. Cox (University of Edinburgh) for additional help with the statistical analyses. Medical Research Council (MRC) [Grant Numbers: G9226254 and G9825423] and the Dr Mortimer and Theresa Sackler Foundation provided financial support.
Declaration of Interest
The author SML has received financial support for research, in the past 3 years, from Roche Abbvie, Sunovion and Janssen, in relation to therapeutic studies of people with schizophrenia. He has also received personal payments for advisory panels and/or educational meetings from Janssen, Forum and Otsuka. AMM is supported by the Health Foundation through a Clinician Scientist Fellowship (Ref: 2268/4295) and by the National Alliance for Research on Schizophrenia and Depression (NARSAD) through an Independent Investigator Award. AMM has previously received financial support from Janssen and Lilly. HCW is supported by a JMAS SIM fellowship from the Royal College of Physicians of Edinburgh and an ESAT College Fellowship from the University of Edinburgh. The authors AMM, HCW and SML have received financial support from Pfizer (formerly Wyeth) in relation to imaging studies of people with schizophrenia and bipolar disorder. ECJ, DGCO, EN, CB, T-KC and LH report no financial conflicts of interest.
Ethical Standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.







