INTRODUCTION
Mesocestoides corti is an endoparasitic platyhelminth used as experimental model to study the class Cestoda (Markoski et al. Reference Markoski, Bizarro, Farias, Espinoza, Galanti, Zaha and Ferreira2003), which also harbours helminths of medical and veterinary importance, such as those from the genera Echinococcus and Taenia. Despite of several studies in the field of cestode biology, little is known about the molecular mechanisms underpinning developmental processes of these organisms, such as the strobilation process. This process comprises cestode development from the larval stage to adult and includes proglottization (generation of serially repeated reproductive organs) and total or partial segmentation (Olson et al. Reference Olson, Littlewood, Bray and Mariaux2001).
Several studies have been conducted to characterize genes and proteins involved in cestode development, but the current knowledge is still limited to allow the definition of developmental pathways involved in strobilation and other typical cestode developmental processes (Olson et al. Reference Olson, Zarowiecki, Kiss and Brehm2012). However, difficulties to obtain parasite samples and safety issues regarding manipulation of pathogenic forms have imposed limitations for molecular studies. Mesocestoides corti, which is easily cultivated and is regarded as non-infective for humans, has allowed us to circumvent some of these experimental limitations, especially those concerning the availability of biological material and safety (Markoski et al. Reference Markoski, Bizarro, Farias, Espinoza, Galanti, Zaha and Ferreira2003). Moreover, the in vitro M. corti culture system allows monitoring of the whole strobilation process under controlled experimental conditions (Markoski et al. Reference Markoski, Bizarro, Farias, Espinoza, Galanti, Zaha and Ferreira2003, Reference Markoski, Trindade, Cabrera, Laschuk, Galanti, Zaha, Nader and Ferreira2006).
In a previous study conducted by our group, subtracted cDNA libraries enriched in sequences differentially expressed in M. corti larvae (tetrathyridia) or strobilated adult worms were used to provide the first report on differential gene expression between the larval and adult stages of this organism (Bizarro et al. Reference Bizarro, Bengtson, Ricachenevsky, Zaha, Sogayar and Ferreira2005). We also performed a proteomic analysis of strobilating tetrathyridia in order to identify proteins differentially represented in early strobilation events (Laschuk et al. Reference Laschuk, Monteiro, Vidal, Pinto, Duran, Cerveñanski, Zaha and Ferreira2011). These studies revealed several genes and proteins with apparent differential expression between tetrathyridia and adult worms or upon strobilation induction, including a putative M. corti orthologue of SET/TAF-Iβ proteins (McSET/TAF).
SET/TAF-Iβ proteins form a subfamily of the nucleosome-assembly protein (NAP) family. They consist of an N-terminal NAP domain, which is conserved among NAP-family members, and a C-terminal acidic tail, implicated in histone binding and chromatin modification (Akey and Luger, Reference Akey and Luger2003). SET/TAF-Iβ orthologues of different species have been described as inhibitors of PP2A protein (Li et al. Reference Li, Makkinje and Damuni1996) and as part of inhibitor of acetyltransferases (INHAT) complexes (Kim et al. Reference Kim, Kim, Son, Chae, Oh, Kim, Pak and Seo2012). As chromatin regulators, they have been implicated in several important cellular processes, including DNA replication (Nagata et al. Reference Nagata, Kawase, Handa, Yano, Yamasaki, Ishimi, Okuda, Kikuchi and Matsumoto1995), transcriptional co-activation (Kato et al. Reference Kato, Miyaji-Yamaguchi, Okuwaki and Nagata2007), cell differentiation (Kim et al. Reference Kim, Kim, Kim, Lee and Seo2010) and apoptosis induction (Fan et al. Reference Fan, Beresford, Oh, Zhang and Lieberman2003).
In this work, the full length McSET/TAF encoding gene (McSET/TAF), retrieved from the M. corti unannotated genome draft, was analysed regarding its exon–intron structure. The McSET/TAF deduced amino acid sequence was also analysed and the protein was established as a true member of the SET/TAF-Iβ subfamily of NAP proteins. We also investigated the expression of McSET/TAF transcripts and of the McSET/TAF protein in tetrathyridia and in three time points after strobilation induction, demonstrating the progressive increase of McSET/TAF expression during strobilar development and providing the first evidence for the involvement of a protein from the SET/TAF-Iβ family in the regulation of cestode development.
MATERIALS AND METHODS
Parasite material
Mesocestoides corti tetrathyridia were maintained by intraperitoneal infection of 3-month-old female Balb/c mice. Parasites collected from mice were used for in vitro cultures in modified RPMI medium (Gibco), as previously described (Markoski et al. Reference Markoski, Bizarro, Farias, Espinoza, Galanti, Zaha and Ferreira2003). Briefly, tetrathyridia were collected from euthanized mice by peritoneal aspiration and washed three times in the culture medium prior to culture. Approximately 5000 tetrathyridia were collected from each infected mouse and cultures were carried out into six-well plates, with about 50 tetrathyridia in 3 mL of medium per well, at 37 °C and 5% CO2 for 2 days prior to strobilation induction. All experimental procedures for in vivo maintenance of M. corti tetrathyridia in mice hosts were approved by the Ethical Committee of the Universidade Federal do Rio Grande do Sul (UFRGS) (Project no. 21625).
Strobilar induction
Tetrathyridia strobilation was performed as described (Markoski et al. Reference Markoski, Bizarro, Farias, Espinoza, Galanti, Zaha and Ferreira2003). Parasites were pre-incubated in modified RPMI media supplemented with 20% fetal bovine serum (FBS, Cultilab, Brazil) containing 0·662% (w/v) trypsin (Sigma), equivalent to 105 Na-benzoyl-l-arginine ethyl ester (BAEE) units ml−1, at 39°C for 24 h. After 24 h induction, the parasites were incubated with 3 ml per well of the fresh modified RPMI media supplemented with 20% fetal bovine serum (FBS, Cultilab, Brazil) and maintained at 39°C under 5% CO2 for at least 7 days. Fully strobilated parasites were selected in stereo microscope. All cultures were performed in quadruplicates and strobilation efficiency upon induction was 70–80%. In control cultures (not induced by trypsin treatment) strobilation rate was under 60%.
Four developmental stages of M. corti were selected during strobilar induction for preparing RNA and protein extracts: bona fide tetrathyridia (TT), tetrathyridia 24 h post strobilation induction (24 h-PI), strobilating worms 72 h post-induction (72 h-PI) and fully strobilated worms (SW).
Protein extracts
All samples (TT, 24 h-PI, 72 h-PI and SW) were washed ten times with 40 mm Tris–HCl buffer (pH 7·2) and homogenized using a glass tissue grinder in an ice bath. The parasite lysate was clarified by centrifugation at 4 °C (15 000 g for 30 min). The protein content of the soluble supernatant was estimated using a Qubit™ quantitation fluorometer (Invitrogen) and qualitatively assessed by SDS–PAGE. Aliquots were stored at −20 °C until use.
McSET/TAF gene prediction, amino acid sequence analysis and phylogeny
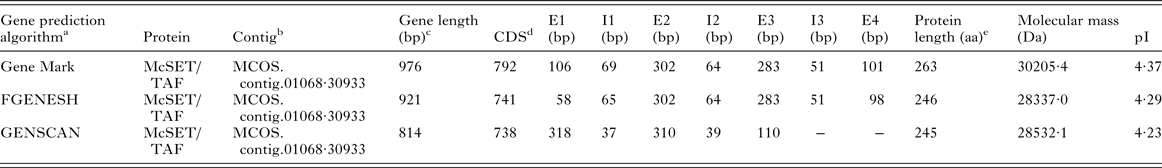
A previously isolated partial McSET/TAF cDNA (Bizarro et al. Reference Bizarro, Bengtson, Ricachenevsky, Zaha, Sogayar and Ferreira2005) was used as a query against the M. corti genomic contigs deposited at the Sanger Institute (ftp://ftp.sanger.ac.uk/pub/project/pathogens/HGI/) using the BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST/). The McSET/TAF gene structure was predicted using the GeneMark (Besemer and Borodovsky, Reference Besemer and Borodovsky2005), FGENESH (Salamov and Solovyev, Reference Salamov and Solovyev2000) and GENESCAN algorithms (Burge and Karlin, Reference Burge and Karlin1998). The amino acid sequence deduced from the full-length coding region of the McSET/TAF gene (corresponding to the McSET/TAF protein) was analysed using Pfam Ver. 27·0 (http://pfam.xfam.org/), and ProtParam tool (www.expasy.org) in order to determine protein domains and molecular masses/isoelectric point, respectively. Phylogeny analysis was performed using SET/TAF-Iβ orthologous proteins recovered from the GeneDB (http://www.genedb.org) (Logan-Klumpler et al. Reference Logan-Klumpler, De Silva, Boehme, Rogers, Velarde, McQuillan, Carver, Aslett, Olsen, Subramanian, Phan, Farris, Mitra, Ramasamy, Wang, Tivey, Jackson, Houston, Parkhill, Holden, Harb, Brunk, Myler, Roos, Carrington, Smith, Hertz-Fowler and Berriman2012) and NCBI (http://www.ncbi.nlm.nih.gov/) databases using the BLAST tool. Protein sequences were aligned using ClustalW2 and visualized using GeneDoc ver. 2·6·002. Phylogenetic trees were constructed by the Neighbour-Joining method (Saitou and Nei, Reference Saitou and Nei1987) and all evolutionary analyses were conducted using the MEGA 5·2 tool (Tamura et al. Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011). The bootstrap consensus tree inferred from 10 000 replicates was taken to represent the evolutionary history of the analysed taxa. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved the full-length McSET/TAF deduced amino acid sequence and orthologous proteins from Danio rerio (NCBI no. NP_958876·1), Drosophila melanogaster (NCBI no. NT_033777·2), Echinococcus granulosus (GeneDB no. EgrG_000465500·1), Echinococcus multilocularis (GeneDB no. EmuJ_000465500·1), Homo sapiens (NCBI no. NP_003002·2), Mus musculus (NCBI no. NP_001191804·1), Schistosoma japonicum (GeneDB no. Sjp_0002330·1), Schistosoma mansoni (GeneDB no. Smp_155060·2) and Taenia solium (GeneDB no. TsM_000881900·1). All positions containing gaps and missing data were eliminated. There were a total of 232 positions in the final dataset.
RT-qPCR
Total RNA was extracted from M. corti TT, 24 h-PI, 72 h-PI and SW samples (with ~150 parasites each) using TRIzol® Reagent (Invitrogen) following the manufacturer's instructions. Isolated RNA was quantified by Qubit® 2·0 Fluorometer (Qubit® RNA Assay Kit), treated with RNase-free DNaseI (Fermentas) and used for reverse transcription using M-MuLV Reverse Transcriptase (Fermentas) with Oligo(dT)18 Primer (Fermentas). RT-qPCRs were performed in a 7500 Real-Time PCR System (Applied Biosystems) in a 96-well microtiter plate format. Each reaction was prepared with 10 μL of cDNA (diluted 1:100), SYBR Green (1X) (Invitrogen), 5 pmol of gene-specific forward and reverse primers, 5 mm dNTPs, 0·24 U Platinum Taq DNA Polymerase (Invitrogen) and RNase/DNase free water for a final volume of 20 μL. The applied cycling conditions were as follows: 5 min DNA polymerase activation at 95 °C, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 10 s, and elongation at 72 °C for 15 s. Negative control reactions (without RT enzyme) were carried out to exclude the possibility of amplification due to the presence of contaminating DNA. All samples and corresponding negative controls were amplified in triplicates.
Based on previous studies (Bizarro et al. Reference Bizarro, Bengtson, Ricachenevsky, Zaha, Sogayar and Ferreira2005; Koziol et al. Reference Koziol, Costábile, Domínguez, Iriarte, Alvite, Kun and Castillo2011), the M. corti orthologues of the receptor-mediated endocytosis-8 (ARME-8), chorea-acanthocytosis (CHOREIN), receptor-mediated endocytosis-8 (CRME-8), lipopolysaccharide (LPS)-responsive and beige-like anchor (LRBA), eyelid/OSA brahma complex gene (OSA), poly(A)-binding protein (PAPB), suvar3-complex (SBF1), programmed cell death 4 (PDCD4), splicing factor, arginine/serine-rich 6 (SR6) and tropomyosin (TROPO) genes were selected as potential references genes for M. corti RT-qPCR data normalization. The primers used for the amplification of sequences from McSET/TAF and selected reference genes are shown in the Appendix (Table A1). The geNorm software (Vandesompele et al. Reference Vandesompele, De Preter, Pattyn, Poppe, Van Roy, De Paepe and Speleman2002) was used for determination of reference genes and normalization of real-time PCR expression data, based on the expression levels of target genes in the TT stage. The calculations of the relative expression of each gene were made by the 2−ΔΔCt method (Livak and Schmittgen, Reference Livak and Schmittgen2001). Statistical analyses were performed using the Duncan's test at 5% significance, using SPSS 20 software (IBM Corp).
McSET/TAF partial cDNA cloning
Total RNA from M. corti TT stage was extracted and used for cDNA synthesis as described above. The cDNA synthesis was followed by PCR amplification with High Fidelity DNA Polymerase (Fermentas) using gene-specific primers for McSET/TAF (5′-ATCTTGGAGAGCGTGGACT-3 and 5′-GCCAGACGTAATATCTTCG-3′). The primers were designed according to the previously published partial McSET/TAF cDNA sequence (Bizarro et al. Reference Bizarro, Bengtson, Ricachenevsky, Zaha, Sogayar and Ferreira2005), which codes for 93 amino acids near the N-terminal end of McSET/TAF (corresponding to amino acids 40–132 of the protein) (GenBank Acc: KJ472143). The amplified coding sequence (McSET/TAF 42–132 ), with 273 pb, codes for McSET/TAF amino acids 42–132 (McSET/TAF42–132). In the first PCR amplification, 24 nt recombination tags FrecI (5′-TATTTTCAGGGAGAATTCCCGGGT-3′) and RrecI (5′-GCGAGGCAGATCGTCAGTCAGTCA-3′), matching the cloning vector, were added to the ends of the 5′ gene-specific primers. Secondary PCR amplification was performed using the primary reaction as template and the primers FrecII (5′-TGGTTCCGCGTGGATCTGAAAACCTGTATTTTCAGGGAGAATTCCCGGGT-3′) and RrecII (5′-GGTTTTCACCGTCATCACCGAAACGCGCGAGGCAGATCGTCAGTCAGTCA-3′). The two rounds of PCR amplification resulted in a McSET/TAF 42–132 amplicon tagged with 50 bp matching pGEX-TEV in its 5′ and 3′ ends.
The final amplification product was then cloned into the pGEX-TEV plasmid, a version of pGEX-4-T1 (GE Healthcare) modified in our laboratory to include the sequence coding for a cleavage site for tobacco etch virus (TEV) protease into the vector polycloning site (data not show). Cloning was performed by in vivo homologous recombination (Parrish et al. Reference Parrish, Limjindaporn, Hines, Liu, Liu and Finley2004), with minor modifications. In frame cloning of the McSET/TAF42–132 coding sequence with the vector encoded glutathione S-transferase (GST) was confirmed by sequencing using the Dyenamic ET Dye Terminator Cycle Sequencing kit (GE Healthcare) in a MEGABACE 1000 Sequencing System.
Expression and purification of recombinant McSET/TAF42–132
Recombinant McSET/TAF42–132 expression in Escherichia coli and purification were performed essentially as described by Moitinho-Silva et al. (Reference Moitinho-Silva, Heineck, Reolon, Paes, Klein, Rebelatto, Schrank, Zaha and Ferreira2012), with some modifications. Briefly, recombinant plasmids carrying the coding sequence for the McSET/TAF42–132 were transformed into BL21-CodonPlus-RIL (Stratagene) and cultures were induced with 0·1 mm IPTG for 3 h at 37 °C. After induction, cells were harvested and lysed, and 0·5% sarkosyl was used for protein solubilization. The GST-tagged recombinant protein was purified by affinity chromatography on Glutathione-Sepharose 4B (GE Healthcare) and cleaved with TEV protease for 16 h at 34 °C to be released from the GST moiety. Eluted protein concentration was measured using a Qubit™ quantitation fluorometer and Quant-iT™ reagents (Invitrogen) and purity was assessed by SDS–PAGE.
Antiserum production and antibody purification
For specific polyclonal antiserum production, a rabbit was immunized by subcutaneous injection using 150 μg of recombinant protein in complete Freund's adjuvant (Sigma). Immunization was followed by three boosters of 150 μg of protein in incomplete Freund's adjuvant (Sigma) every two weeks. Blood sample was collected one week after the last immunization and the serum was separated by centrifugation. IgG antibody purification from non-immune control serum and from polyclonal serum from the immunized animal was carried out in HiTrap Protein G HP columns (GE Healthcare), according to the manufacturer's protocol. All experimental procedures for polyclonal antiserum production in rabbits were approved by the Ethical Committee of the UFRGS (Project no. 21625).
Immunoblots
Twenty micrograms of protein extracts from M. corti developmental stages TT, 24 h-PI, 72 h-PI and SW were resolved by 12% SDS–PAGE and transferred to polyvinylidene fluoride membranes (PVDF) (Hybond™-ECL™, GE Healthcare) as described by Monteiro et al. (Reference Monteiro, de Carvalho, Zaha and Ferreira2010). Rabbit polyclonal antiserum anti-McSET/TAF42–132 (diluted 1:20 000,v/v) was used as primary antibody, and horseradish peroxidase (HRP)-labelled anti-rabbit IgG (ECL™, GE Healthcare) was used as secondary antibody (1:9000 v/v dilution). Antigen–antibody complexes were detected using ECL detection reagent (GE Healthcare) and imaged using a VersaDoc imaging system (Bio-Rad). All experiments were performed in duplicate, with protein samples corresponding to biological replicates (from independent parasite cultures). The ImageJ software (NIH, Maryland, USA) was used for expression relative quantifications.
Immunohistochemistry
Samples of each M. corti developmental stage (TT, 24 h-PI, 72 h-PI and SW) were fixed, dehydrated and embedded in paraffin as described by Koziol et al. (Reference Koziol, Domínguez, Marín, Kun and Castillo2010). Sections 8 μm thick were mounted, rehydrated and blocked with 1% bovine serum albumin, 0·05% Tween in PBS for 1 h at 37 °C. Processed sections were then incubated in a humid chamber for 1 h at 37 °C with purified anti-McSET/TAF42–132 antibodies, diluted 1:200 (v/v) in blocking solution, followed by six washes for 20 min each with PBS. Sections were then incubated with goat anti-rabbit IgG conjugated with Alexa Fluor 488 (Invitrogen), diluted 1:500 (v/v) in blocking solution, in a humid chamber for 1 h at 37 °C. After three additional washes in PBS, sections were incubated with 50 μm DAPI for 20 min at 37 °C, mounted with Fluoromount (Sigma) and observed under a confocal microscope (Olympus FluoView™ 1000), available at the Centro de Microscopia Eletrônica of the UFRGS (CME-UFRGS). Images were digitally captured and processed using the software Olympus Fluoview-F1000.
RESULTS
McSET/TAF gene prediction, amino acid sequence analysis and phylogeny
In order to determine the full-length coding sequence of the M. corti SET/TAF-Iβ orthologous gene and protein (McSET/TAF and McSET/TAF, respectively), we performed a blastN analysis using the previously available McSET/TAF partial cDNA sequence (Bizarro et al. Reference Bizarro, Bengtson, Ricachenevsky, Zaha, Sogayar and Ferreira2005) as query against the M. corti draft genome sequence available at Sanger Institute (ftp://ftp.sanger.ac.uk/pub/project/pathogens/HGI/). A single contig (MCOS.contig.01068·30933 contig), containing only one identifiable gene with homology to the query sequence (e-value of 5·2e–57), was retrieved, indicating that McSET/TAF is encoded by a single copy gene in the M. corti genome. No evidence of paralogue sequences were found in the performed searches.
The MCOS.contig.01068·30933 contig sequence was analysed with three gene prediction algorithms, and the predicted exon–intron structures for the identified McSET/TAF gene are shown in the Appendix (Table A2). The longest predicted coding DNA sequence (that predicted by the GeneMark algorithm), which also resulted in the highest similarity with flatworm orthologues, was used for subsequent in silico analyses. Based on that, the McSET/TAF gene is composed of four exons and three introns, with a coding region of 792 bp encoding a 263 aa-long protein (Fig. 1A). The analysis of the deduced amino acid sequence revealed the presence of both a NAP domain and a C-terminal acidic tail (Fig. 1B), typical of NAPs of the SET/TAF-Iβ subfamily.

Fig. 1. Structure of McSET/TAF gene, deduced amino acid sequence of the encoded protein and phylogenetic relationships. (A) Exon–intron structure as predicted in silico. Numbered grey bars represent exons, lines represent introns, and the cloned region, coding for McSET/TAF amino acids 42–132 (McSET/TAF42–132), is indicated (*); (B) McSET/TAF deduced amino acid sequence and predicted domains, which grey-shadowed amino acids belong to the NAP domain and the C-terminal acidic tail is boxed. The cloned and expressed segment of the protein is underlined; (C) Phylogenetic analysis of McSET/TAF inferred using the Neighbour-Joining method. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The percentages of replicate trees in which the associated proteins clustered together in the bootstrap test (10 000 replicates) are shown next to the branches. The phylogenetic tree was generated with the full-length McSET/TAF deduced amino acid sequence and orthologous proteins from D. rerio (Dr), D. melanogaster (Dm), E. granulosus (Eg), E. multilocularis (Em), H. sapiens (Hs), M. musculus (Mm), S. japonicum (Sj), S. mansoni (Sm) and T. solium (Ts).
Multiple sequence alignments of McSET/TAF and its orthologues from a wide range of eukaryotic species, including other helminths and model organisms, are shown in the Appendix (Fig. A1). Regarding other Platyhelminthes, the M. corti McSET/TAF amino acid sequence displayed 68% identity and 76% similarity with the orthologous sequences from the cestodes E. granulosus and E. multilocularis, 71% identity and 79% similarity with T. solium sequence, 47% identity and 63% similarity with the trematode S. japonicum and 48% identity and 63% similarity with S. mansoni. Comparisons with D. melanogaster sequence showed identity/similarity levels of 38%/56%. With mammalian sequences, levels of identity/similarity were 40/57% and 39/57% for mouse and human orthologues, respectively. With D. rerio, the levels of identity/similarity were 41/57%.
The McSET/TAF deduced amino acid sequence and the previously aligned orthologous sequences were also used to infer a phylogenetic tree (Fig. 1C). In this tree, McSET/TAF appears in a ‘more primitive’ branch, along with orthologues from other helminths, while orthologues from D. melanogaster and vertebrates formed another major branch. Taken together, our phylogenetic analysis, the overall identities and similarities with other eukaryotic SET/TAF-Iβ proteins, and the identification of typical NAP domain and acidic tail clearly point out that McSET/TAF can be considered a true SET/TAF-Iβ orthologue.
RT-qPCR standardization and assessment of McSET/TAF mRNA levels during strobilation
For RT-qPCR standardization, 10 M. corti genes (listed in Table A1, Appendix) were evaluated as potential normalizers for M. corti transcriptional data. Based on the expression stability (M) values obtained in TT, 24 h-PI, 72 h-PI and SW samples (Fig. A2, Appendix), five genes were selected as the more stable ones (with the lowest M values), namely PDCD4 (M = 0·5), PAPB (M = 0·53), SR6 (M = 0·57), CHOREIN (M = 0·72) and TROPO (M = 0·8). Using the established normalizing genes and standardized RT-qPCR conditions, we then assessed the McSET/TAF mRNA levels in TT, 24 h-PI, 72 h-PI and SW. The RT-qPCR results, summarized in Fig. 2A, revealed that the McSET/TAF mRNA was expressed in all tested parasite stages, with significantly different levels only between TT and SW, and higher expression level in the SW stage.
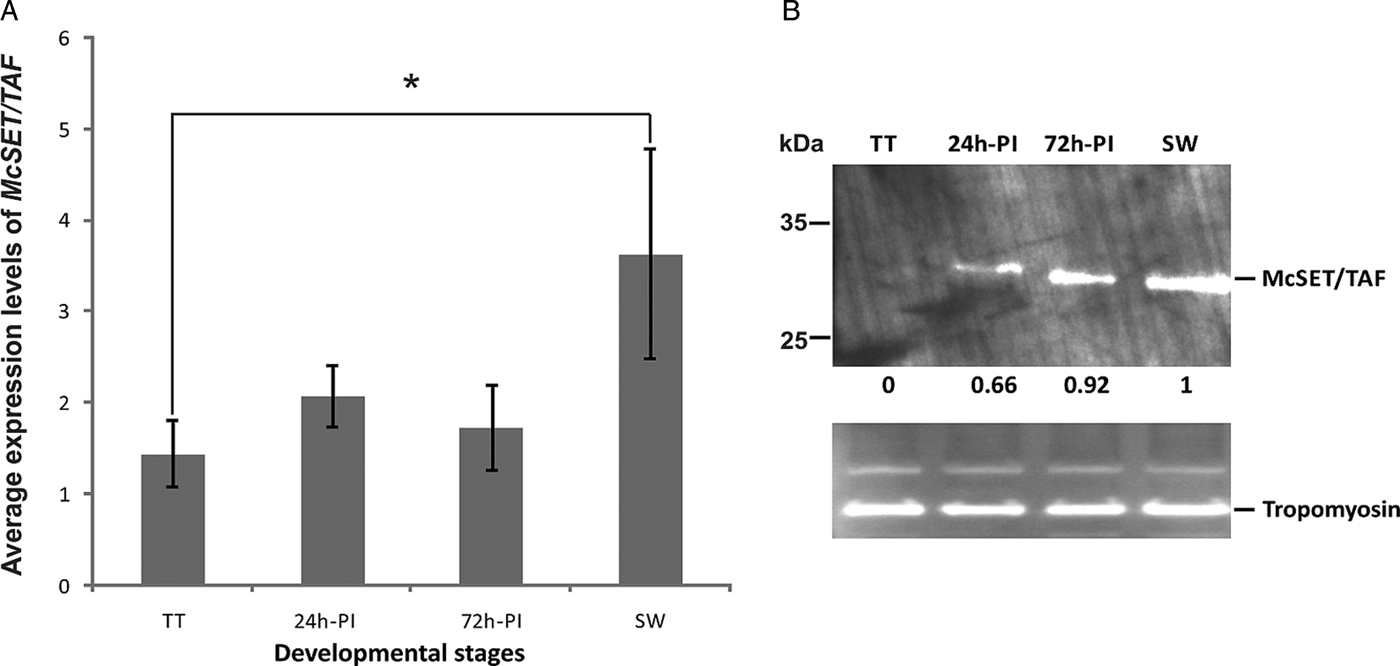
Fig. 2. McSET/TAF gene and protein expression levels in different M. corti developmental stages. (A) The levels of McSET/TAF gene transcripts were normalized to the expression level of PDCD4, PAPB, SR6, CHOREIN and TROPO gene transcripts. Normalized data were submitted to analysis of variance and differences with >5% significance by Duncan test are indicated (*). The bars indicate standard deviation between replicates; (B) McSET/TAF protein levels assessed by immunoblot using anti-McSET/TAF purified IgG. All lanes were loaded with 20 μg of each protein extract and relative McSET/TAF expression levels were normalized against the M. corti tropomyosin band for each sample. In the comparisons between McSET/TAF bands from different samples, the SW band was used as reference, and the relative expression values for all samples are indicated. Expression data were normalized and compared using the IMAGE J software.
Temporal and spatial expression pattern of McSET/TAF during M. corti development
The McSET/TAF protein expression in TT, 24 h-PI, 72 h-PI and SW stages was assessed both by immunoblot and immunofluorescence using polyclonal antibodies generated in rabbit against the recombinant polypeptide McSET/TAF42–132. In immunoblot experiments (Fig. 2B), the anti-McSET/TAF42–132 antibodies detected a band corresponding to a ~30 kDa protein, in accordance with the expected molecular mass of McSET/TAF (see Appendix, Table A2). A faint McSET/TAF band was detected in the TT protein extract, and, in the other analysed developmental stages, the McSET/TAF band showed a gradual increase in its intensity, from 24 h-PI to SW. Apparent small variations in McSET/TAF size between the 24 h-PI, 72 h-PI, and SW extracts could be noticed.
By immunofluorescence (Fig. 3), McSET/TAF expression showed a pattern essentially in line with that observed in the immunoblot experiment described above. McSET/TAF expression was also very weak in the TT stage, and, upon strobilation induction, it increased. Actual differences in McSET/TAF expression between 24 h-PI, 72 h-PI and SW sections are not easy to establish, due to differences in cell densities and distribution in the different developmental stages. The protein showed a uniform pattern of distribution in inner tissues for all samples, as expected for typical SET/TAF-Iβ proteins. In 24 h-PI e 72 h-PI sections, there was a strong staining in the sub-tegumental region, corresponding to sub-tegumental muscle cells or tegumental cells (also known as perinuclear cell bodies of the tegumental syncytium).

Fig. 3. Immunolocalization of McSET/TAF in different M. corti developmental stages. Parasite sections (8 μm) were incubated with purified non-immune IgG (upper panel) and purified anti-McSET/TAF42–132 IgG. Recognition of immune complexes was achieved using Alexa 488-conjugated secondary antibodies. DAPI nuclei staining; antibody staining; merged DAPI and antibody images; and bright field images are shown, from left to right, in the first, second, third and fourth columns, respectively. Scale bars: TT images, 200 μm; 24 h-PI and 72 h-PI, and SW images 50 μm.
DISCUSSION
The characterization of differentially expressed proteins during development of M. corti is important for a better understanding of basic biological aspects of the parasite, such as the molecular mechanisms involved during its development process. To date, only a few cestode genes involved in processes of cell proliferation and differentiation, such as those encoding the protein-kinases Ras, Raf and MPK1 in E. multilocularis (Spiliotis et al. Reference Spiliotis, Tappe, Brückner, Mösch and Brehm2005, Reference Spiliotis, Konrad, Gelmedin, Tappe, Brückner, Mösch and Brehm2006) and the transcription factor Mvlim in M. corti (Lalanne et al. Reference Lalanne, Britos, Ehrlich and Castillo2004). The potential involvement of these genes in developmental processes has been suggested, but further studies are needed to demonstrate it. Recently, the genome sequences of some cestode species were published (Tsai et al. Reference Tsai, Zarowiecki, Holroyd, Garciarrubio, Sanchez-Flores, Brooks, Tracey, Bobes, Fragoso, Sciutto, Aslett, Beasley, Bennett, Cai, Camicia, Clark, Cucher, De Silva, Day, Deplazes, Estrada, Fernández, Holland, Hou, Hu, Huckvale, Hung, Kamenetzky, Keane and Kiss2013; Zheng et al. Reference Zheng, Zhang, Zhang, Zhang, Li, Lu, Zhu, Wang, Huang, Liu, Kang, Chen, Wang, Chen, Yu, Gao, Jin, Gu, Wang, Zhao, Shi, Wen, Lin, Jones, Brejova, Vinar, Zhao, McManus, Chen and Zhou2013), which will facilitate the prospection of developmental genes and proteins for this taxon of parasites.
The M. corti SET/TAF-Iβ related sequence studied here was originally isolated as a partial cDNA enriched in SW libraries (Bizarro et al. Reference Bizarro, Bengtson, Ricachenevsky, Zaha, Sogayar and Ferreira2005). Here, we were able to characterize the whole gene sequence in silico and establish the identity and expression pattern of the encoded protein in undifferentiated larvae and in three stages of the strobilation process. Based on a phylogenetic tree, the McSET/TAF protein appears in a ‘more primitive’ branch, along with orthologues from other helminths, while orthologues from D. melanogaster and vertebrates formed another major branch. This phylogenetic tree, along with the overall identities and similarities of McSET/TAF with other eukaryotic SET/TAF-Iβ proteins and the identification of typical NAP domain and acidic tail clearly point out that McSET/TAF is a member of the SET/TAF-Iβ subfamily of NAP proteins.
McSET/TAF gene expression revealed that it is transcribed in all four tested parasite stages, which represent the undifferentiated larval stage (TT) and the early (24 h-PI), intermediate (72 h-PI) and end (SW) stages of strobilation. A significant higher level of expression in SW in comparison with the TT stage was demonstrated. This corroborated the preliminary transcriptomic data reported by Bizarro et al. (Reference Bizarro, Bengtson, Ricachenevsky, Zaha, Sogayar and Ferreira2005) for M. corti, which suggested that the McSET/TAF gene was differentially transcribed between TT and SW stages, with higher expression in the SW stage. Although not significantly different, the McSET/TAF expression was also consistently higher from 24 h-PI to 72 h-PI to SW samples in all experiments performed with independent cultures, indicating a progressive increase in the transcription levels of this gene from TT to SW. During this developmental transition, the variation in McSET/TAF protein size observed in the immunoblot experiments raises the possibility of differential regulation at a post-translational level. This could be due to post-translational modifications, such as phosphorylation, known to occur in SET/TAF-Iβ proteins (Irie et al. Reference Irie, Harada, Araki and Nishimura2012). Other alternatives to explain possible McSET/TAF variants would be the use of alternative translational initiation codons or alternative splicing. SET/TAF-Iβ variants generated by alternative splicing have been described in humans (Nagata et al. Reference Nagata, Kawase, Handa, Yano, Yamasaki, Ishimi, Okuda, Kikuchi and Matsumoto1995), but such post-transcriptional events remain to be investigated in M. corti.
The expression levels of the McSET/TAF gene and the McSET/TAF protein are very low in TT, where lower levels of overall expression are expected, since the parasite is in an undifferentiated stage, in which only vegetative growth and asexual reproduction take place (Markoski et al. Reference Markoski, Bizarro, Farias, Espinoza, Galanti, Zaha and Ferreira2003). In 24 h-PI, 72 h-PI and SW stages, there was an increase in the levels of McSET/TAF, suggesting a greater involvement of this protein in both initial and later strobilar development. Therefore, McSET/TAF expression level increase in parallel with the activation and functioning of processes of growth, differentiation and cell death during strobilation indicates its possible involvement with the cellular machinery that regulates them. Besides, the expression pattern found for SET/TAF-Iβ proteins in different eukaryotic organisms, such as Saccharomyces cerevisiae, D. melanogaster and mouse, is ubiquitous, with the protein being found in both the nucleus and cytoplasm of the cells (Nagata et al. Reference Nagata, Saito, Okuwaki, Kawase, Furuya, Kusano, Hanai, Okuda and Kikuchi1998). This is in line with the ubiquitous and uniform pattern found for McSET/TAF in our immunofluorescence experiments.
SET/TAF-Iβ proteins are known to act both in the nucleus and in the cytoplasm regulating several cellular processes, from DNA replication and gene expression to cell differentiation and apoptosis (Nagata et al. Reference Nagata, Kawase, Handa, Yano, Yamasaki, Ishimi, Okuda, Kikuchi and Matsumoto1995; Fan et al. Reference Fan, Beresford, Oh, Zhang and Lieberman2003; Kim et al. Reference Kim, Kim, Kim, Lee and Seo2010). It is then interesting to speculate the possible functional involvement of McSET/TAF in these and other cellular processes. As a putative histone chaperone of the SET/TAF-Iβ subfamily of NAPs, as inferred from the performed alignment and phylogenetic analyses, McSET/TAF may act as a transcriptional co-activator (Kato et al. Reference Kato, Miyaji-Yamaguchi, Okuwaki and Nagata2007) and, as such, regulate developmental genes important for M. corti strobilation. Furthermore, in immunofluorescence assays, SW sections showed an apparent staining in the tegument surface, which could indicate an ectopic localization of McSET/TAF at this stage, whose tegument can present differences in structure and/or composition (Markoski et al. Reference Markoski, Bizarro, Farias, Espinoza, Galanti, Zaha and Ferreira2003). The presence of typical intracellular proteins in the tegument or among excretion/secretion products of cestodes has been described and associated with potential moonlighting functions (Monteiro et al. Reference Monteiro, de Carvalho, Zaha and Ferreira2010; Lorenzatto et al. Reference Lorenzatto, Monteiro, Paredes, Paludo, da Fonsêca, Galanti, Zaha and Ferreira2012).
Concluding remarks
The McSET/TAF gene codes for a SET/TAF-Iβ protein of the NAP family. Based on a set of five M. corti reference genes standardized as normalizers, it was shown that McSET/TAF transcription level is higher in SW than in TT. It was also shown that the encoded McSET/TAF protein progressively increases during M. corti strobilation, providing the first evidence for the involvement of a protein from the SET/TAF-Iβ family of epigenetic effectors in the regulation of cestode development. McSET/TAF can now be further studied in order to characterize its activities, functions and/or interactions relevant for cestode developmental pathways.
ACKNOWLEDGEMENTS
The authors thank the Centro de Microscopia Eletrônica (UFRGS), for technical support with the confocal microscopy.
FINANCIAL SUPPORT
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (AUX-PE-PARASITOLOGIA 1278/2011), in Brazil. C.B.C was a recipient of a CAPES M.Sc. fellowship. K.M.M. was a recipient of a CAPES postdoctoral fellowship. E.D.S. and J.A.P. were recipients of Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) M.Sc. fellowships. M.C. is a recipient of a CAPES postdoctoral fellowship. A.T. was a recipient of a CAPES PhD fellowship. K.R.L. was a recipient of a CNPq PhD fellowship.
APPENDIX

Fig. A1. Alignment of SET/TAF deduced amino acid sequences from M. corti, selected helminths and model organisms. For each alignment, amino acids which are conserved in all the aligned sequences are printed in white on a black background; amino acids conserved in at least five of the aligned sequences are printed in black on gray background; and amino acids conserved in at least eight of the aligned sequences are printed in white on gray background. The following sequences were used: M. corti (Mc) McSET/TAF amino acid sequence deduced from contig MCOS.contig.01068·30933; Danio rerio (Dr) NCBI no. NP_958876·1; Drosophila melanogaster (Dm) NCBI no. NT_033777·2; Echinococcus granulosus (Eg) GeneDB no. EgrG_000465500·1; Echinococcus multilocularis (Em) GeneDB no. EmuJ_000465500·1; Homo sapiens (Hs) NCBI no. NP_003002·2; Mus musculus (Mm) NCBI no. NP_001191804·1; Schistosoma japonicum (Sj) GeneDB no. Sjp_0002330·1; Schistosoma mansoni (Sm) GeneDB no. Smp_155060·2; and Taenia solium (Ts) GeneDB no. TsM_000881900·1.
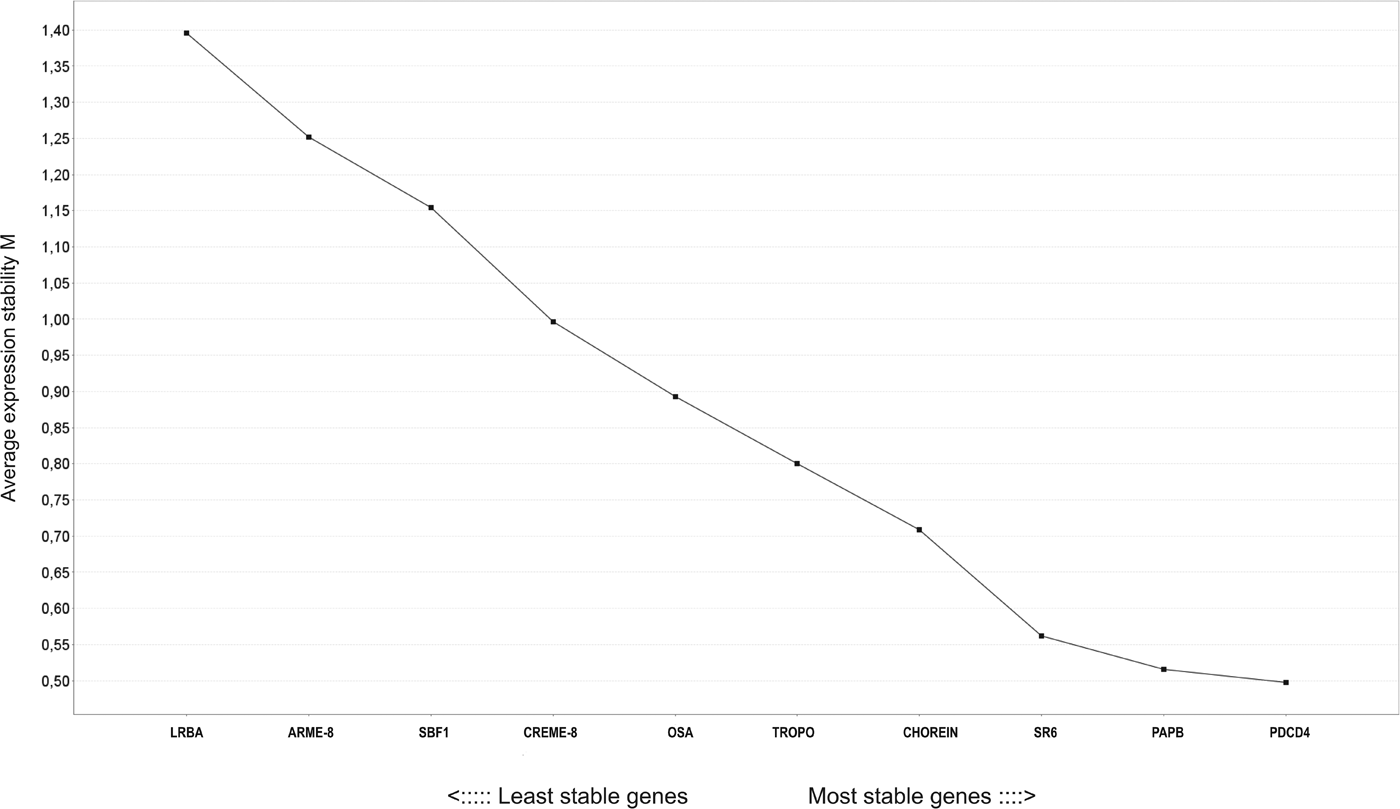
Fig. A2. Analysis of expression stability of potential M. corti reference genes by geNorm program. M values obtained for each of the genes analysed are indicated on the figure. Among the ten genes analysed, those with the lowest M values are PDCD4 (M = 0·5), PAPB (M = 0·53), SR6 (M = 0·57), CHOREIN (M = 0·72) and TROPO (M = 0·8). The LRBA gene (M = 1·40) was considered the less stable.
Table A1. Primer sequences used in RT-qPCR experiments, with corresponding Tm and amplicon sizes

Table A2. Structure and characteristics of McSET/TAF gene and encoded protein predicted by different algorithms