Published online by Cambridge University Press: 05 October 2004
Phylogenetic relationships among Trypanosoma rangeli isolates from man, wild mammals and triatomine bugs from widespread geographical origin were inferred by comparison of the small subunit of ribosomal gene sequences. The phylogenetic trees indicated that the subgenus Herpetosoma is polyphyletic and strongly supported division of this group into two monophyletic lineages, one made up of T. rangeli, T. rangeli-like and allied species and other consisting of T. lewisi and related taxa. Based on phylogenetic analysis, morphology, behaviour in vertebrate and invertebrate hosts and epidemiology we propose: a) the validation of Herpetosoma as a taxon comprised only for species of group lewisi and the maintenance of T. lewisi as the type-species of this subgenus; b) the classification of T. rangeli, T. rangeli-like and allied species into a ‘T. rangeli-clade’ more closely related to Schizotrypanum than to T. lewisi or T. brucei. The phylogenetic tree disclosed at least 4 groups within the clade T. rangeli, all confirmed by polymorphism of the internal transcribed spacer, thus conferring for the first time phylogenetic support to groups of T. rangeli and corroborating the high complexity of this taxon. Grouping was independent of their mammalian host-species and geographical origin, indicating that other factors are determining this segregation.
Understanding the phylogenetic relationships among trypanosomes from man, domestic and wild mammals is a crucial problem in the evolutionary history of American trypanosomiasis. Trypanosoma rangeli and Trypanosoma cruzi are the only trypanosomes infecting man in Latin America and sharing vertebrate and invertebrate hosts. Vectors of T. rangeli are triatomine bugs, mainly of the genus Rhodnius. Like T. cruzi, T. rangeli also lacks mammalian host specificity and has been reported infecting a variety of mammal species. The animals found naturally infected with T. rangeli belong to 5 distinct mammalian orders, which include human and non-human primates, Xenarthra, Didelphimorphia, Carnivora and Rodentia (Hoare, 1972; Añez, 1982; D'Alessandro & Saraiva, 1992, 1999; Vallejo et al. 2003).
Human infection by T. rangeli is highly prevalent in Central and Northwest South America, where this species is sympatric with T. cruzi (D'Alessandro & Saraiva, 1999; Vallejo et al. 2003). In contrast, only three confirmed human cases were reported in Brazil, all in the Amazon Region (Coura et al. 1996), where the presence of triatomines and sylvatic mammals infected with T. rangeli are common (Deane et al. 1972; Miles et al. 1983; Ziccardi et al. 2000; Maia da Silva et al. 2004). Outside Amazonia, T. rangeli was only reported in sylvatic mammals and triatomines of Southern and Southeast Brazilian regions (Steindel et al. 1991; Ramirez et al. 1998).
Despite sharing hosts, T. rangeli and T. cruzi are remarkably different with respect to their behaviour in both invertebrates and vertebrates. In contrast to T. cruzi, T. rangeli may be harmful or even lethal to its vector, inducing failures to moult, body abnormalities and reduced ability (or even the inability) of the insect to feed (Añez, 1984; D'Alessandro & Saraiva, 1992, 1999). In contrast to T. cruzi, T. rangeli is harmless to its mammalian hosts. The parasitaemia is generally very low and may persist for a long time (Hoare, 1972; D'Alessandro & Saraiva, 1992, 1999). The infection produced by T. rangeli is poorly understood and nothing definitive is known about the mode and the site of reproduction. Dividing blood trypomastigotes and intracellular tissue forms, such as amastigotes typical of Schizotrypanum species, have not been confirmed in vertebrate hosts of T. rangeli (Añez, 1981, 1982; D'Alessandro & Saraiva, 1992, 1999; Eger-Mangrich et al. 2001).
Traditionally, T. rangeli is classified in the subgenus Herpetosoma of Stercoraria together with T. lewisi, which is the type species of this subgenus. Two sections without taxonomic status were created by Hoare (1972) to separate mammalian trypanosomes whose development occurs exclusively in the gut of vectors and which are transmitted by faeces (Stercoraria) from trypanosomes transmitted by inoculation of metacyclic forms in the saliva of tsetse fleas (Salivaria). The life-cycle of T. rangeli in the triatomine vectors has been reported to have features of both the stercorarian and the salivarian trypanosome life-cycles. Although T. rangeli develops in the insect's gut, trypanosomes in faecal material are not metacyclic forms (Tobie, 1964; Añez, 1983a,b) and faecal contamination is not the usual mechanism of transmission (D'Alessandro & Saraiva, 1999). After initial development in the gut, the flagellates penetrate through the gut wall, a route peculiar to this species, and develop in the haemolymph and salivary glands. The triatomine-bugs transmit T. rangeli by inoculation of saliva containing metacyclic forms during feeding or probing (Tobie, 1970; Añez, 1983a,b; Hecker, Schwarzenbach & Rudin, 1990).
On account of this unusual combination of transmission routes and life-cycle features, producing infective forms in both the posterior and anterior stations of the vector, the systematic position of T. rangeli has been a subject of controversy. Hoare (1972) considered T. rangeli an aberrant species of the subgenus Herpetosoma. Añez (1982) proposed removing T. rangeli from the subgenus Herpetosoma, creating the subgenus Tejeraia within Salivaria. Initial molecular analyses only served to increase the controversy surrounding the evolutionary relationships of T. rangeli because data from a few isolates about sequences of β-tubulin (Amorim, Momen & Traub-Cseko, 1993) and cysteine-protease (Tanaka et al. 1994) genes, and karyotyping (Henriksson et al. 1996) also suggested that T. rangeli is more related to T. brucei than to T. cruzi. However, D'Alessandro & Saraiva (1999) supported its maintenance within Stercoraria as a Herpetosoma species while Stevens & Gibson (1999), in a broad phylogenetic analysis of SSU rRNA sequences, positioned 5 isolates of T. rangeli within Stercoraria and more close to Schizotrypanum spp. than to all other trypanosomes, thus indicating that the taxonomic position of T. rangeli and the taxonomy of the whole Herpetosoma must to be reviewed (Stevens & Gibson, 1999; Stevens et al. 1999a, 2001).
Polymorphism among T. rangeli populations was shown by: zymodemes (Miles et al. 1983; Steindel et al. 1994), DNA fingerprinting (Macedo et al. 1993), mini-exon gene sequences (Grisard, Campbell & Romanha, 1999), karyotyping (Henriksson et al. 1996) and random amplification of polymorphic DNA (RAPD) (Steindel et al. 1994). According to these studies the genetic polymorphism among isolates from different geographical origins permitted their partition into 2 groups. We recently identified at least 4 distinct genetic lineages among a large number of isolates by RAPD analysis, thus revealing that the complexity of T. rangeli is higher than previously described (Maia da Silva et al. 2004). While the taxonomic position of T. rangeli from man and triatomines is now relatively well established, trypanosomes from sylvatic mammals classified as T. rangeli, T. rangeli-like and allied species were under-represented in the previous studies and the little available data suggested that reclassification of some isolates may be prudent (Stevens & Gibson, 1999; Stevens et al. 1999a,b, 2001; Maia da Silva et al. 2004). Due to the genetic variability within T. rangeli and considering that, besides the biased selection for human and insect isolates, previous studies included few isolates representative of all distinct genetic groups of T. rangeli, we do not have sufficient data yet to define the evolutionary relationships among T. rangeli isolates and allied species. Study of several isolates from distinct host species and geographical origin is required to evaluate the segregation of T. rangeli into lineages, their evolutionary relationships and thus, their taxonomic status. With these points in mind, we investigated the phylogenetic relationships based on polymorphism in SSU and ITS rDNA sequences of 34 isolates of T. rangeli from man, triatomine bugs, non-human primates and other sylvatic mammals (opossum, anteater, sloth, rodent) from a widespread geographical region.
In this study we used 34 isolates of T. rangeli and allied species from distinct hosts and geographical origin, embracing widespread geographical origins from Central and South Americas. From Brazil, we analysed isolates from 5 States from northern (Amazonia Region) and southern (Santa Catarina State) regions (Fig. 1, Table 1). Isolation, culturing and classification of all new isolates were done as previously described (Maia da Silva et al. 2004). Trypanosomes used in this study were tested using 2 T. rangeli-specific PCR assays (Vargas et al. 2000; Maia da Silva et al. 2004). Other trypanosome species used in this study were cultivated in LIT medium at 28 °C (Table 1). Trypanosomes are cryopreserved in the Trypanosomatid Culture Collection (TCC) of the Department of Parasitology of the University of São Paulo, SP, Brazil.

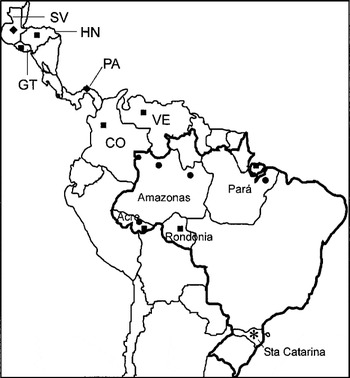
Fig. 1. Geographical origin of Trypanosoma rangeli isolates and allied species employed in this study. Groups of T. rangeli defined in this study: A ([squf ]), B ([bull ]), C ([lozf ]) and D (*).
DNA of trypanosomes obtained by the phenol-chloroform extraction method was used as template for PCR amplification of a 900 bp DNA fragment containing partial SSU rRNA sequence using primers 609F and 706R (Fig. 2). Amplifications were done in reactions of 50 μl containing 100 ng of DNA, 2.5 U of Taq DNA polymerase, 0·2 mM each dNTP and 200 pM of primers. The reactions were cycled 30 times as follows: 1 min at 94 °C, 2 min at 48 °C and 1 min at 72 °C. Amplification products of 3–5 separated PCR reactions were purified by Spin-X (Costar), pooled and automatically sequenced by primer walking in both directions. For restriction analysis, amplified DNA was digested with Rsa I (Biolabs) enzyme, analysed on 2·0% agarose gels and stained with ethidium bromide. Polymorphism of restriction enzyme sites on the aligned SSU rRNA sequences of T. rangeli isolates was investigated using the Clone program (CLONE MANAGER version 3.11; www.scied.com/ses_cm6.htm).

Fig. 2. Schematic diagram of the ribosomal gene showing the annealing sites for oligonucleotides (in box) used for PCR amplifications and DNA sequencing.
DNA fragments corresponding to ITS1, 5.8S and ITS2 sequences of the ribosomal gene were amplified using primers IR1 and IR2 (Cupolillo et al. 1995). For exclusive amplification of ITS1 sequences the primers IR1 and 5.8R (Fig. 2) were used as described by Cupolillo et al. (1995). Amplified products were electrophoresed on 2% agarose gels and transferred to nylon membranes (Hybond-N, Amersham Pharmacia). Membranes were hybridized with amplified fragment generated from DNA of T. rangeli isolates, labelled by random primed synthesis with [α-32P] dCTP (Ready to Go kit, Amersham Pharmacia). Southern blots were pre-hybridized for 1 h and hybridized, at 43 °C, for 16–18 h in 2×SSC, 2% SDS, 4·0 mM sodium pyrophosphate and 40 mg/ml of salmon sperm DNA. Membranes were washed 3 times for 15 min each in 0·1×SSC, 2% SDS and 4 mMsodium pyrophosphate at 55 °C. Amplified ITS1 sequences of selected isolates were separated in 2% agarose gels, stained with ethidium bromide, excised from the gels, purified by Spin-X (Costar) and cloned (pGEM kit, Amersham Pharmacia). Sequences of 2–3 clones from each isolate were determined by automated sequencing.
SSU rRNA (V7-V8) sequences from 27 isolates of T. rangeli and allied species, from 3 new isolates of T. cruzi from Amazonia (2 from monkeys and 1 from man), and from T. blanchardi and T. rabinowitschae were determined in this study and aligned with sequences from GeneBank (Table 1). Alignments were made manually using as guide the general alignment in the rRNA database (http://rrna.uia.ac.be/). Phylogenies were inferred using both Maximum Parsimony (MP) and Maximum-Likelihood (ML) analysis. MP analysis was done as before (Stevens et al. 1999b). ML parameters were optimized using the hierarchical likelihood test in Modeltest 3-06 (Posada & Crandall, 1998) and estimates using TreePuzzle 5.0 (Strimmer & Von Haeseler, 1996). Distance matrix was calculated using Neighbour Joining (Saitou & Nei, 1987) with parameters from the Tamura & Nei substitution model. Similarity matrix was performed using Point Replacer v2.0 (http://www.geocities.com/alvesjmp/software.html). Bootstrap analyses with 100 replicates were performed using PAUP 4.0b10. The trees were rooted using Trypanoplasma borreli as outgroup for Trypanosoma.
The ITS1 sequences determined in this study were aligned with others from GeneBank (Table 1). Alignments of our sequences did not reveal significant polymorphism among sequences from different clones of the same isolates. Phylogenetic analyses were performed as described above for SSU rRNA sequences.
Ten isolates of T. rangeli from man, 2 from triatomines, 13 from non-human primates of Brazilian Amazon, 7 from other wild mammals and 2 from dogs were used in this study. These isolates came from 7 countries, from Guatemala to Brazil (Fig. 1). All new isolates of T. rangeli investigated in this study showed all traditional taxonomic criteria (morphology and behaviour in mice and triatomines) compatible with T. rangeli (Maia da Silva et al. 2004). In addition, all T. rangeli isolates were confirmed by PCR assays developed to detect specifically this species. Both methods employed amplified DNA fragments of identical length for all isolates, independently of their genetic groups (Vargas et al. 2000; Maia da Silva et al. 2004).
Phylogenetic relationships were inferred from comparison of variable regions (V7-V8) of SSU rRNA sequences among 34 isolates of T. rangeli and allied species, including 5 previously published sequences and 27 sequenced in this study. Nineteen sequences of other trypanosome species were also included (5 obtained in this study) (Table 1). Similar strongly supported branching patterns were obtained using both MP (data not shown) and ML (Fig. 3A) methods, as well as using different data sets varying in species, number of samples and aligned sequences (only V7 or V7-V8 for all isolates and V7-V9 for few isolates).
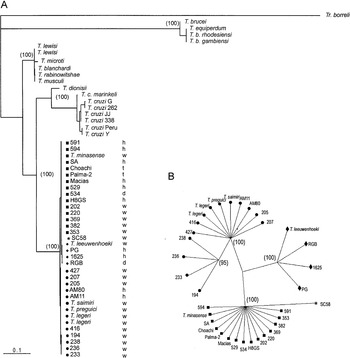
Fig. 3. (A) Phylogenetic tree based on bootstrapped Maximum Likelihood analysis of SSU (V7-V8) rRNA sequences from 52 trypanosomes. (B) Dendrogram of 34 Trypanosoma rangeli isolates and allied species constructed by Maximum Likelihood analysis of V7-V8 SSU rRNA sequences. The numbers in parenthesis refer to the bootstrap values of the clusters in 100 replicates. T. rangeli groups: A ([squf ]), B ([bull ]), C ([lozf ]) and D (*). Source of trypanosomes: h, human; w, wild mammals; t, triatomine bugs; d, dog.
According to the topology of our phylogenetic tree, all isolates of T. rangeli and allied species form a strongly supported monophyletic assemblage (clade T. rangeli), which always clustered together (with 100% of bootstrap support). This clade was characterized by a low inter- and intra-specific sequence divergence (maximum of 2·1% and an average of 0·9%), demonstrating that organisms within this clade are highly genetically related. The closest relative to the T. rangeli clade was T. cruzi (~20% of sequence divergence) and other species of the subgenus Schizotrypanum (T. c. marinkellei and T. dionisii). We also included in the phylogenetic analysis isolates of T. cruzi belonging to the major phylogenetic lineages (T. cruzi I, T. cruzi II and Z3), including new isolates from man and from non-human primates of the Brazilian Amazon (Table 1). Contrasting with the high homogeneity within the clade T. rangeli, significant intra-specific heterogeneity was detected among lineages of T. cruzi (4·5% of sequence divergence) and in the subgenus Schizotrypanum, including T. cruzi, T. c. marinkellei and T. dionisii (7·8% of divergence).
T. lewisi and allied species (T. musculi, T. microti, T. blanchardi and T. rabinowitschae) from different host-species and geographical regions were tightly clustered together (100% of bootstrap) into the T. lewisi clade. Species of the T. lewisi clade showed a high intra- and inter-specific sequence similarity (95%), similar to that within the T. rangeli clade. Despite sequence divergence (~20%) separating clades, T. rangeli and T. cruzi showed similarity to the divergence (~20%) between T. rangeli and T. lewisi clades, clade T. lewisi was positioned as an outgroup of the branch containing T. cruzi and T. rangeli in all inferred trees (100% of replicates). Similarly, T. rangeli and T. brucei were separated by a high genetic distance (~35% of sequence divergence) strongly supported (100% of bootstrap) in all inferred trees (Fig. 3A).
Despite the high similarity among V7-V8 rRNA sequences from all members of the clade T. rangeli (99·2% of similarity), phylogenetic analysis based on these sequences segregated these organisms into 4 groups: A, B, C and D (Fig. 2, Table 2). The same branching pattern was observed in all inferred phylogenetic trees. In agreement with tree topology, analysis of the matrix of pairwise differences between SSU rRNA sequences (which provided a measure of absolute genetic differences between taxa) showed significant genetic polymorphism between distinct groups within the clade T. rangeli. According to polymorphism on SSU rRNA sequences, organisms were distributed as follows: Group A, composed of all isolates from Colombia, Venezuela, Honduras and Brazilian isolates from Rondônia and Pará (Marajó Island) States; Group B, formed exclusively by Brazilian isolates from Acre, Amazonas (Manaus and Alto e Médio Rio Negro) and Pará (Belém) States; Group C, constituted by isolates from Panama, El Salvador and Colombia (Fig. 3); Group D, represented in this study exclusively by the isolate SC58, from Santa Catarina State, Southern Brazil, was segregated into a distinct branch, although it clustered close to group A, separated by 1·1% of sequence divergence. Little or no variability was observed within groups: Groups A, C and D were more related to each other (0·7–1·0% of divergence) than to group B (~2·1% of divergence from the other groups). Group B was divided into subclades B and B′, the last composed by isolates from Acre sharing identical sequences with small sequence divergence (0·2%) from the rest of the isolates of Group B (Figs 2, 3).

Aiming to standardize a simple and quick method to type any unknown T. rangeli isolates in groups dispensing SSU rRNA DNA sequencing, PCR-amplified fragments of this gene were digested with Rsa I restriction enzyme. This enzyme was selected after analysis of aligned sequences had detected restriction sites within the amplified fragments capable of generating different fragment patterns according to the groups of isolates. Restriction analysis of SSU rRNA amplified fragments from T. rangeli isolates disclosed three major patterns of DNA fragments in agarose gels, corresponding to the three major groups of T. rangeli (A, B and C) defined in the phylogenetic trees (Fig. 3A). PCR-RFLP patterns generated by digestion with Rsa I enzyme did not distinguish Group D (isolate SC58) from Group A (Fig. 4). Additional analysis with other enzymes can be used for this purpose.

Fig. 4. PCR-RFLP analysis as a tool for quick genotyping of Trypanosoma rangeli. Agarose gel (2%) of the restriction DNA fragments generated by digestion of amplified V7-V8 regions of SSU rRNA with Rsa I enzyme and stained with ethidium bromide.
In order to clarify the grouping patterns within the clade T. rangeli, we have evaluated the polymorphism among T. rangeli isolates using the more divergent sequences of ITS ribosomal gene. For this purpose, we initially evaluated the polymorphism by comparing length and sequence of the PCR-amplified DNA fragments containing the whole ITS (ITS1, 5.8S and ITS2) from DNA of 11 isolates (Fig. 5A). Results showed length differences that permitted us to distribute the isolates into the same groups defined by the tree branching pattern (Fig. 3A). The degrees of conservation of ITS sequences among members of distinct groups of T. rangeli were evaluated by cross-hybridization analysis (Fig. 1B,C). For this purpose, the ITS amplified fragments were analysed by Southern blot hybridization using probes TrA and TrB, which are constituted by the amplified fragments from DNA of T. rangeli SA and AM80, respectively. These isolates were selected because they represent the 2 major groups of isolates from the Brazilian Amazon. Results revealed that each probe cross-hybridized only with organisms belonging to groups from which they were derived, with no cross-hybridization between isolates from groups A and B. Very faint signals, and only when hybridized with probe TrA, were detected for isolates SC58 (Group D) and PG (Group C) (Fig. 5B) and thus are in agreement with tree topology, in which both isolates seemed to be more related to group A than to group B. The polymorphism of whole ITS was assessed for 12 more isolates (Fig. 5D) and, according to the length of the amplified fragments, the 23 isolates examined were distributed into the same groups shown in the dendrogram (Table 2). Thus, this approach provided a quick and simple tool to type T. rangeli in groups.

Fig. 5. Agarose gel (2%) showing amplified fragments of whole ITS (ITS1+5.8S+ITS2) from Trypanosoma rangeli isolates and allied species stained with ethidium bromide (A, D). Southern blotting of the gel in (A) hybridized with probes constituted by amplified ITS sequences of T. rangeli isolates: ITS-TrA from SA (B) or ITS-TrB from AM80 (C).
Despite significant length and sequence polymorphism of ribosomal sequences among T. rangeli isolates, results showed higher conservation among isolates of the same groups than between organisms belonging to different groups. Corroborating these data, analysis of T. rangeli ITS sequences showed that both ITS1 and ITS2 are highly divergent among isolates of this species, whereas the 5.8S gene was identical. In order to compare sequences presenting different evolutionary rates, we compared data from the highly conserved SSU with that based on the more variable ITS1 sequences. Length polymorphism of amplified ITS1 from 19 isolates permitted the same grouping originated by whole ITS, despite small heterogeneity within the group B (Fig. 6A, Table 2). For comparative analysis, sequences of isolates of Group B (T. saimiri and AM80), Group C (PG and 1625) and Group D (SC58) were aligned with sequences of T. rangeli from Genebank (isolates Macias, Choachi, H9, Palma-2 and AM11), aiming to obtain a good set of sequences containing members of all major groups of T. rangeli (Fig. 3; Table 2). The genetic relatedness among the isolates assessed through a matrix of pairwise divergence showed high sequence divergence between groups A and B of T. rangeli. Contrasting with the low divergence among SSU rRNA sequences (1·2%) these groups were separated by 39·4% of ITS1 sequence divergence. The topology of the dendrogram based on ITS1 sequences disclosed a branching pattern in concordance with clustering on an SSU rRNA-based tree (Fig. 6B).
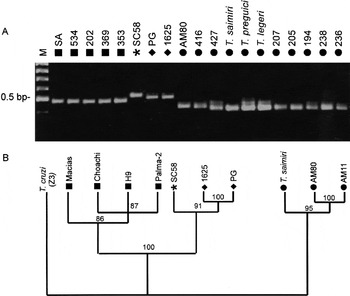
Fig. 6. (A) Agarose gel (2%) showing amplified fragments of ITS1 from Trypanosoma rangeli isolates and allied species stained with ethidium bromide. (B) Dendrogram representing the genetic relatedness among 7 T. rangeli isolates and T. saimiri constructed by maximum likelihood analysis of ITS1 rRNA sequences. The numbers in parenthesis refers to the bootstrap values of the clusters in 100 replicates. T. rangeli groups: A ([squf ]), B ([bull ]), C ([lozf ]) and D (*).
The genus Trypanosoma seems to be among the most diverse lineage within the Trypanosomatidae family and its monophyly has been supported by several studies (Haag, O'h Uigin & Overath, 1998; Hannaert, Opperdoes & Michels, 1998; Stevens & Gibson, 1999; Stevens et al. 1999b, 2001). Molecular phylogenetic studies supported the division of mammalian trypanosomes into 2 major groups, which are represented by stercorarian or salivarian trypanosomes, separated by a very large genetic distance and constituted by highly divergent subgenera. Since the classification of Trypanosoma proposed by Hoare (1972), the taxonomy of this genus has remained remarkably stable despite a lot of data pointing towards the need for an urgent revision. Nowadays, of the three subgenera of Stercoraria, only Schizotrypanum appears to be a monophyletic group, whereas Herpetosoma and Megatrypanum were shown to be polyphyletic in studies based on SSU rRNA (Stevens & Gibson, 1999; Stevens et al. 1999, 2001). In fact, Hoare (1964) had already pointed out that not all stercorarian trypanosomes could be fitted with certainty into subgenera, especially in Megatrypanum and Herpetosoma. Phylogenetic studies pointed to a higher genetic diversity within Trypanosoma than considered by the current classification, questioning the whole taxonomy of this genus. Thus, how this genus should be organized in subgenera, species and subspecies is still controversial. It seems probable that continued studies on species within the genus may require the subdivision of the trypanosomes into additional subgenera and epidemiologically relevant groups as well as the creation of several new species.
This study focused on understanding the evolutionary relationships and taxonomic position of T. rangeli by inferring phylogenetic relationships among 34 isolates of T. rangeli, T. rangeli-like and allied species (T. saimiri, T. preguici, T. leeuwenhoeki) and isolates previously classified as Megatrypanum that have been revealed to be T. rangeli (T. minasense and T. legeri) (Stevens et al. 1999a; Maia da Silva et al. 2004). Aiming to infer relationships within Herpetosoma, we also analysed 2 T. lewisi isolates and 4 allied species. The inferred trees confirmed that subgenus Herpetosoma is a strongly supported polyphyletic group divided in 2 distant monophyletic clades, allied to either T. rangeli or to T. lewisi. Several trypanosome isolates can be readily ascribed to either one of these clades according to traditional taxonomic criteria, as well as by robust molecular methods, enough to propose a reliable taxonomic revision of Herpetosoma as previously proposed (Stevens & Gibson, 1999; Stevens et al. 1999a, 2001). Molecular data are in agreement with the large differences existing among T. rangeli, T. cruzi, T. lewisi and T. brucei regarding several aspects such as their life-cycles, vectors, transmission routes, and relationships in both vertebrate and invertebrate hosts. All these species could be clearly distinguished from each other by a number of morphological, biological, and biochemical peculiarities (autapomorphies).
T. lewisi and allied species tightly clustered into a well-supported monophyletic clade, thus justifying the continued recognition of the taxon Herpetosoma and the maintenance of T. lewisi as the type species of this subgenus, as stated when Hoare (1964, 1972) created this subgenus. Besides the historical priority, this species presents all major features of Herpetosoma as originally described (Hoare, 1964, 1972; Molyneux, 1976; Añez, 1982).
T. rangeli and allied species were positioned with stercorarian trypanosomes into a different clade (Clade T. rangeli) distinct from the T. lewisi clade and close to taxa classically recognized as Schizotrypanum (see also Stevens et al. 1999b, 2001). T. rangeli and associated taxa share the following major features: (a) wide mammalian host range; (b) geographical distribution from Central to South America; (c) lack of pathogenicity and low parasitaemia in the vertebrate hosts; (d) pathogenicity to triatomine vectors. However, the only exclusive taxonomic parameter for recognizing trypanosomes as a member of this subgenus is the demonstration of metacyclic trypomastigotes in the salivary glands of triatomine bugs. However, the successful establishment of the extra-intestinal cycle of development of T. rangeli depends upon several factors such as vector species, age of isolates maintained in cultures and the method used for bug infection (intracoelomic inoculation or xenodiagnosis) (D'Alessandro & Saraiva, 1992; Maia da Silva et al. 2004).
Our phylogenetic analysis also confirmed that T. rangeli is more closely related to clade T. cruzi, which includes the Schizotrypanum species (T. cruzi, T. cruzi marinkellei and T. dionisii) than to all other trypanosome species. Based on phylogenetic analysis and paleographic evidence, Stevens et al. (1999, 2001) hypothesized that T. rangeli, T. cruzi and a trypanosome from kangaroo diverged from a common ancestor, probably a trypanosome in marsupials on a southern super-continent of South America, Antarctica and Australia. However, divergence on SSU rRNA sequences between T. rangeli and T. cruzi suggested distinct evolutionary pathways of these species within the Americas. In addition, although sharing geographical distribution, mammalian host range and transmission by triatomines, T. rangeli and T. cruzi differ significantly in most morphological and biological aspects.
All data corroborated the fact that transmission by bite is not a homologous character, but a feature acquired independently by distinct lineages of trypanosomes. Apparently, other species can also be transmitted by bite, such as T. (Megatrypanum) theileri, whose development in ticks suggests that it can also be transmitted through both saliva and faeces (Burgdorfer, Schimidt & Hoogstraal, 1973).
All 34 isolates of T. rangeli and allied species investigated in this work were shown to be very closely related and were grouped tighter together in a robust cluster (Clade T. rangeli) by analysis of SSU rRNA sequences. Nevertheless, consistent levels of genetic diversity on both SSU and ITS sequences clearly separated these organisms into 4 major groups. Group A corresponds to that previously described for isolates from northwest South American countries by analysis of zymodemes, RAPD markers (Steindel et al. 1994; Maia da Silva et al. 2004) and mini-exon sequences (Grisard et al. 1999; Vallejo et al. 2003). Group B is formed exclusively by Brazilian isolates from Amazonia and was described before using spliced-leader gene markers (Silva et al. 1999) and RAPD analysis (Maia da Silva et al. 2004). Human isolates from Panama and El Salvador and a dog isolate from Colombia (RGB) were grouped with T. leeuwenhoeki (Group C). Likewise in all other previous studies based on zymodemes, RAPD patterns and mini-exon gene sequences (Steindel et al. 1994; Grisard et al. 1999; Maia da Silva et al. 2004), the isolate SC58 from Southern Brazil was clearly separated from all other taxa by both SSU and ITS gene polymorphism and ascribed to Group D.
There is not a significant degree of genetic polymorphism in either SSU or ITS sequences of the trypanosomes from wild mammals to justify their status as T. rangeli-like or as separate species allied to T. rangeli. However, there are enough molecular, epidemiological and biological data to assert the separation of isolates in high consistent and homogeneous phylogenetic groups, the taxonomic status of which needs further evaluation. For instance, T. leeuwenhoeki (from a sloth from Panama) was named as a different species of Herpetosoma allied to T. rangeli because it does not develop in R. prolixus, whereas it could be transmitted by the bite of R. pallescens (Shaw, 1985). Likewise, some human isolates of T. rangeli from Panama and Colombia can be distinguished by their inability to develop in vector species other than R. pallescens, their sympatric triatomine species (Sousa & Dawson, 1976; Vallejo et al. 2003). T. leeuwenhoeki clustered tightly together with human T. rangeli isolates from Panama, El Salvador and Colombia in previous studies based on ribosomal and spliced leader sequences (Stevens et al. 1999) and on RAPD patterns (Maia da Silva et al. 2004).
Ribosomal (SSU and ITS) polymorphisms reported in this study and in a previous one based on RAPD markers (Maia da Silva et al. 2004) and on SL sequences (Silva et al. 1999; Maia da Silva et al. in preparation) disclosed 4 groups of T. rangeli. Thus, Clade T. rangeli is a complex taxon, constituted by different populations which appear to have evolved with non-random association of independent molecular markers. All groups contain members from different host species (human, domestic or sylvatic mammals), without any vertebrate host-species association. T. rangeli from sylvatic mammals can be very closely related to human isolates, suggesting that the same parasite circulates among humans, sylvatic and domestic animals and triatomines. Our data demonstrated that despite some geographical distance determination of segregation patterns, other factor(s) than geographical isolation are also determining these groups.
Sympatric triatomine species are excellent vectors for T. rangeli isolates, whereas non-sympatric species present different infection rates of salivary glands and can be even completely unsuitable as vectors (Sousa & Dawson, 1976; D'Alessandro & Saraiva, 1992; Machado et al. 2001). Vector and isolate parameters modulate the haemocoelic T. rangeli invasion and its survival in different vector species (Gomes et al. 1999). These data indicate a complex parasite–vector relationship and suggest co-evolution of T. rangeli populations with their local vector species. Recent analysis of Colombian T. rangeli isolates based on kDNA markers suggested either clonal evolution or speciation of T. rangeli populations in their triatomine vectors (Vallejo et al. 2003).
Taken together, biological and molecular features have now been gathered to demonstrate that T. rangeli, T. rangeli-like and allied species from distinct host and geographical origin can be tightly clustered together in the T. rangeli clade. In contrast, data are still insufficient to make definitive interpretations about evolutionary relationships among genetic groups within this clade. This would require comparison of more isolates from different vertebrate and triatomine species, from widespread geographical origins. New molecular markers and biological differences must be investigated before we can make definitive statements concerning both inter- and intra-specific variations and the determining factors of segregation patterns within T. rangeli and related taxa.
This work was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Nacional de Saúde (FUNASA). We are indebted to many persons for supplying us with isolates and for inestimable help in the fieldwork, and to Instituto Leonidas e Maria Deane (Manaus) and Fundação Nacional de Saúde (FUNASA-Barcelos) for facilities. We are grateful to M. C. Brigido and Cibele Bonvicino for blood samples and identification of monkeys. Work in Rondonia was done at the laboratory of the ICB IV, USP. F. Maia da Silva and A. C. V. Junqueira are FAPESP and CNPq graduate student fellows, respectively.

Table 1. Trypanosoma rangeli isolates and allied species and trypanosome species of Trypanosoma lewisi group used in this study showing host species and geographical origins

Fig. 1. Geographical origin of Trypanosoma rangeli isolates and allied species employed in this study. Groups of T. rangeli defined in this study: A ([squf ]), B ([bull ]), C ([lozf ]) and D (*).

Fig. 2. Schematic diagram of the ribosomal gene showing the annealing sites for oligonucleotides (in box) used for PCR amplifications and DNA sequencing.

Fig. 3. (A) Phylogenetic tree based on bootstrapped Maximum Likelihood analysis of SSU (V7-V8) rRNA sequences from 52 trypanosomes. (B) Dendrogram of 34 Trypanosoma rangeli isolates and allied species constructed by Maximum Likelihood analysis of V7-V8 SSU rRNA sequences. The numbers in parenthesis refer to the bootstrap values of the clusters in 100 replicates. T. rangeli groups: A ([squf ]), B ([bull ]), C ([lozf ]) and D (*). Source of trypanosomes: h, human; w, wild mammals; t, triatomine bugs; d, dog.

Table 2. Grouping of Trypanosoma rangeli isolates and allied species based on RAPD, SSU rRNA and ITS analysis

Fig. 4. PCR-RFLP analysis as a tool for quick genotyping of Trypanosoma rangeli. Agarose gel (2%) of the restriction DNA fragments generated by digestion of amplified V7-V8 regions of SSU rRNA with Rsa I enzyme and stained with ethidium bromide.

Fig. 5. Agarose gel (2%) showing amplified fragments of whole ITS (ITS1+5.8S+ITS2) from Trypanosoma rangeli isolates and allied species stained with ethidium bromide (A, D). Southern blotting of the gel in (A) hybridized with probes constituted by amplified ITS sequences of T. rangeli isolates: ITS-TrA from SA (B) or ITS-TrB from AM80 (C).

Fig. 6. (A) Agarose gel (2%) showing amplified fragments of ITS1 from Trypanosoma rangeli isolates and allied species stained with ethidium bromide. (B) Dendrogram representing the genetic relatedness among 7 T. rangeli isolates and T. saimiri constructed by maximum likelihood analysis of ITS1 rRNA sequences. The numbers in parenthesis refers to the bootstrap values of the clusters in 100 replicates. T. rangeli groups: A ([squf ]), B ([bull ]), C ([lozf ]) and D (*).