INTRODUCTION
The advent of rapid DNA sequencing technology has facilitated and enabled analyses of gene expression in parasites via the isolation of partial cDNA sequences, termed expressed sequence tags (ESTs). For instance, Blaxter et al. (1996) used this approach to survey genes expressed in third-stage larvae (L3s) of Brugia malayi (a filarial parasite of humans), and Hoekstra et al. (2000) investigated changes in gene expression in Haemonchus contortus (the barber's pole worm of small ruminants) upon its transition from the pre-parasitic to the parasitic stage. EST sequencing is perfectly suited for automation and is commonly employed for the identification of genes from nematodes of socio-economic significance, particularly for drug discovery and vaccine development (reviewed by Gasser and Newton, 2000; Newton, Boag and Gasser, 2002; Knox, 2004; McCarter, 2004; Nisbet, Cottee and Gasser, 2004; Foster et al. 2005). Despite the usefulness of EST sequencing and the relative ease with which large data sets can be generated, this approach has the disadvantage that many of the sequences obtained represent common ‘house-keeping’ genes (present in all developmental stages) and that many abundantly expressed genes are highly represented, thus generating relatively large amounts of redundant sequence information. Also, genes expressed at low levels may not be detected.
Differential display (originally described by Liang and Pardee, 1992) is a direct PCR-based technique employed for isolating differentially expressed genes, for example, stage- or sex-specific genes. This approach has been used for the isolation of stage-specific cDNAs from both Angiostrongylus cantonensis (a rat lungworm) (see Joshua and Hsieh, 1995) and H. contortus (see Hartman et al. 2001). In the latter study, the advantage of this approach was demonstrated through the identification of adult-specific genes not previously identified by EST sequencing of conventional cDNA libraries prepared from adult H. contortus (see Hoekstra et al. 2000). More recently, a modified differential display approach (i.e. using ‘random primers’) was employed for the isolation and subsequent characterization of a small subset of sex-specific genes from Oesophagostomum dentatum (a porcine nodule worm) (Boag et al. 2000). However, in spite of its usefulness, the differential display approach has had limitations in that it is laborious and time-consuming to carry out, and that frequently ‘false positive’ display products are isolated and subsequently cloned and sequenced (e.g. Liao and Freedman, 2002; Stein and Liang, 2002). The technique of suppressive-subtractive hybridization (SSH) (Diatchenko et al. 1996) overcomes the limitations of differential display. This PCR-based approach allows the effective removal of common (e.g. ‘house-keeping’) genes from the RNA population of interest prior to library construction and also has the advantage that rare transcripts are amplified efficiently (i.e. are enriched), which is not the case for the conventional EST sequencing approach. Despite its utility and effectiveness for isolating differentially expressed genes, the SSH approach has not yet been applied widely to parasitic nematodes (Nisbet and Gasser, 2004; Nisbet et al. 2004). In the present study, we constructed by SSH archives of female- and male-enriched ESTs representing adult stages of O. dentatum, and conducted bioinformatic and microarray analyses of a subset of ESTs from these archives as a foundation for future work on the functional aspects of genes differentially expressed between the sexes of this nematode.
MATERIALS AND METHODS
Parasite material
Third-stage larvae (L3) of O. dentatum were produced from eggs by incubating the faeces from monospecifically infected pigs for 14 days at 20–22 °C (cfTalvik et al. 1997), harvested using the Baermann funnel technique (Ash and Orihel, 1987) and stored at 10 °C for a minimum of 21 days. Helminth-free pigs (Danish Landrace×Yorkshire×Duroc; both sexes; 20 kg live weight) were inoculated by intra-gastric intubation with 6×103 L3. Adults of O. dentatum were recovered from the large intestinal content 22–25 days after inoculation, harvested into physiological saline using an agar gel method (Slotved et al. 1996), the two sexes separated using a dissection microscope, and subsequently snap frozen in liquid nitrogen. The specific identity of adult nematodes was determined using an existing description (Haupt, 1966).
Construction of suppressive-subtractive hybridization (SSH) libraries
Total RNA (2 μg) was extracted from ‘pooled’ adult males or females of O. dentatum using Tripure™ (Roche Molecular Biochemicals) following the manufacturer's protocol. Messenger RNA isolated using the Poly (A) pure™ kit (Ambion) was used to synthesize cDNA (using the PCR-Select™ cDNA subtraction kit; Clontech) for SSH. In brief, cDNA was synthesized from male or female mRNA, digested with Rsa-I and ligated to adapters to produce ‘tester’ cDNA for each gender. The female-tester cDNA was hybridized with an excess of male cDNA (“driver”) to remove common cDNA transcripts from the adapter-ligated female cDNA, generating the ‘female-library’. The reciprocal procedure was performed using male-tester cDNA and female-driver cDNA, thus constructing the ‘male-library’. As controls, both male- and female-testers were hybridized with water instead of driver cDNA to provide ‘unsubtracted’ cDNA representing both genders.
Verification of sex specificity
Southern and Northern blot analyses were carried out to determine the efficiency of cDNA subtraction. For Southern blots, equal amounts (1·8 μg) of the PCR products from each SSH were resolved electrophoretically in an agarose gel (1%), transferred to a positively-charged nylon membrane (Roche Molecular Biochemicals), denatured and cross-linked under ultraviolet light. The membrane was pre-hybridized (at 68 °C for 2 h) in 0·25 M NaHPO4, 1 mM ethylene diamine tetraacetic acid (EDTA), 7% (w/v) sodium dodecyl-sulphate (SDS) and 100 μg ml−1 of denatured herring sperm DNA, and then probed (at 68 °C for 20 h) with 150 ng of Rsa I-digested male- or female-driver cDNA labelled with 32P-αdCTP by random priming (Prime-a-gene, Promega); the blot was then washed at 60 °C in 6× SSC, 0·1% SDS (3 times for 15 min), followed by a wash at 0·1× SSC, 0·5% SDS prior to autoradiography (3 h at −70 °C). For Northern blots, 10 μg of total RNA isolated from male or female O. dentatum and resolved in a 0·8% (w/v) denaturing (2·2 M formaldehyde) agarose gel in 1× MOPS running buffer (40 mM MOPS, 5 mM sodium acetate, 1 mM EDTA pH 7). RNA was transferred for 3 h to a positively charged nylon membrane, using 0·01 M NaOH, 3 M NaCl as the buffer, and hybridized overnight with a 32P-α-dCTP-labelled probe (representing an individual EST). The membrane was washed 3 times for 20 min in 2× SSC, 0·1% SDS at 60 °C prior to autoradiographic exposure usually for 6–12 h.
Sequencing and bioinformatic analyses
Clones from the gender-enriched EST libraries were amplified by PCR, and amplicons column-purified (Wizard™ PCR-Prep, Promega) and ligated into the vector pGEM®-T Easy (Promega). Recombinant plasmids were transformed into competent Escherichia coli (strain JM109; Promega) by heat shock using standard protocols. Individual recombinant clones were picked (based on blue/white selection) and grown overnight in Luria Bertani medium (LB) containing 7% (v/v) glycerol. Ten μl of each culture were used to inoculate 1·3 ml of LB containing 10 mg/ml of ampicillin and grown for 18 h at 37 °C. Cultures were centrifuged (6000 g for 10 min, at 18 °C) and the cell pellets stored at −80 °C until plasmid extraction and automated sequencing (96-well format) using the T7 primer (Promega).
After the removal of flanking vector and adapter sequences, all ESTs from each SSH library were aligned using the Contig Assembly Program (CAP; available at http://bio.ifom-firc.it/ASSEMBLY.html), employing a minimum sequence overlap length ‘cut-off’ of 30 bases and an identity threshold of 95%. ESTs were compared with sequences available in public databases using BLAST algorithms. BLASTn and BLASTx were used to search the non-redundant National Centre for Biotechnology Information databases (available at http://www.ncbi.nlm.nih.gov/blast/), the Parasite genome WU-BLAST2 Nematoda database (from the European Bioinformatics Institute, http://www.ebi.ac.uk/blast2/parasites.html) and WormBase (http://www.wormbase.org/). An EST of O. dentatum was designated a ‘statistically significant gene homologue’ if the P or E values of the sequence alignment (at the amino acid level) were [les ]0·001.
Microarray analysis
For each gender-enriched library, one EST representing each contig assembly and all singleton ESTs were PCR-amplified using vector-specific primers. The amplicons were purified using 96-well filter plates (Multiscreen PCR, Millipore), desiccated, resuspended in 50% dimethyl sulphoxide (DMSO) and then arrayed on CMT-GAPS aminosilane-coated glass slides (Corning) as 5 separate, replicate grids. The slides were then baked, cross-linked (Stratagene) and probed with cDNA. To generate these probes, total RNA (2 μg) purified from adult male or female O. dentatum was reverse transcribed and the cDNA labelled with Cy3 or Cy5 using the Genisphere 350RP labelling kit (Geneworks). The two probes were combined and used for hybridization. Microarray slides were hybridized (in quadruplicate, using 60 μl of probe) at 65 °C overnight and then washed in 2× SSC, 0·1% SDS at 65 °C for 20 min, 1× SSC at 22–24 °C for 20 min and 0·1× SSC at 22–24 °C for 20 min. ‘Dye swapping’ (cf Smyth, Yang and Speed, 2003) was carried out to control for any bias in hybridization signal between the Cy-labelled probes (produced for 2 distinct mRNA populations). After washing, the slides were dried by centrifugation and scanned using an arrayWorx e scanner (Applied Biosystems). The hybridization signal for each spot in each microarray was measured using ImaGene 6.0 software (BioDiscovery). The signal intensity of each spot was corrected in relation to background, and the data analysis (using the mean signal intensity for each EST tested in quadruplicate) performed using the GeneSight 4.1.6 program. The data were globally normalized by dividing the mean of each hybridization signal value by the mean of all values and log2-transformed. The data were analysed for differential hybridization (of at least 2-fold) between male and female O. dentatum using the confidence analyzer tool of the GeneSight 4.1.6 program (BioDiscovery) using a statistical confidence interval of [ges ]99·9%.
Verification of differential gene expression by reverse transcription-coupled PCR (RT-PCR)
Complementary DNA was synthesized from total RNA from each gender of adult O. dentatum and purified as described by Cottee et al. (2004). Primers designed to amplify each candidate EST were designed using Primer3 (available at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Differential expression was verified by RT-PCR analysis. Each reaction (50 μl) contained 5 mM Tris- HCl, pH 8, 10 mM NaCl, 0·01 M EDTA, 0·1 mM dithiothreitol (DTT), 5% glycerol, 0·1% Triton®X 100; Promega), 50 μM of each dNTP, 3 mM MgCl2, 1 U Taq polymerase (Promega), 50 μM of each gene-specific primer and 2 μl (~1200 ng/μl) of cDNA. Negative (no-DNA) and positive (EST) controls were also included. Cycling parameters were: 1 cycle of 94 °C for 3 min, 25–35 cycles of 94 °C for 30 sec, 50 °C for 30 sec, 72 °C for 30 sec, followed by 1 cycle of each 70 °C for 7 min. The RT-PCR products were detected in ethidium bromide-stained agarose gels (2%) and photographed.
RESULTS
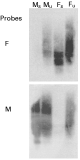
The Southern blot analysis of subtracted and unsubtracted cDNA from male and female O. dentatum probed with female-driver cDNA displayed a strong hybridization signal to unsubtracted-male and both subtracted- and unsubtracted-female cDNAs, whereas only a very faint signal to a minor component in the male-subtracted cDNA was detectable (Fig. 1). When the blot was ‘stripped’ and subsequently probed with male-specific driver cDNA, the reciprocal result was achieved (Fig. 1). Also, the Northern blot analysis of total male- or female-RNA using amplicons derived from selected ESTs as probes showed gender-specific hybridization (Fig. 2). These findings demonstrated the effectiveness of the cDNA subtraction process for library construction.
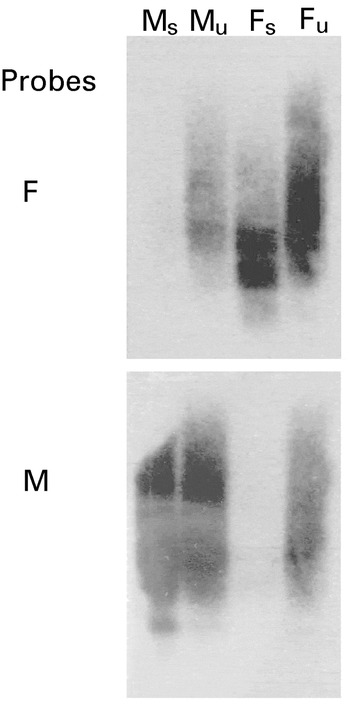
Fig. 1. Verification of suppressive-subtractive hybridization by Southern blot analysis. Equal amounts (1·8 μg) of male-subtracted (Ms); male-unsubtracted (Mu), female-subtracted (Fs); female-unsubtracted (Fu) cDNAs from Oesophagostomum dentatum were resolved by agarose gel electrophoresis, transferred to a positively-charged nylon membrane, pre-hybridized and then hybridized with radio-isotope labelled cDNA from adult female (F) or male (M) O. dentatum.

Fig. 2. Verification of sex-specificity of transcripts by Northern blot analysis. Equal amounts (10 μg) of total RNA from adult males (M) or females (F) of Oesophagostomum dentatum were resolved by electrophoresis, transferred to a nylon membrane and then hybridized with radio-isotope labelled amplicons produced from ESTs Odm001 (Trichostrongylus vitrinus serine proteinase inhibitor homologue, TvSERP1), Odm002 (Caenorhabditis elegans hypothetical protein NM_072012 homologue), Odm003 (with no known homologue) and Odf001 (C. elegans phosphoenolpyruvate carboxykinase homologue, R11A5.4).
In total, 873 ESTs (440 male and 433 female) were obtained from 960 clones selected from the O. dentatum gender-enriched libraries (480 from each), achieving a sequencing success of 91%. Individual sequences were edited (removing both vector and adaptor sequences) and subjected to contig assembly. Subsequently, groups (clusters) of similar sequences were aligned and compared to establish the degree of redundancy within each group. This analysis of the 440 ESTs from the male-library allowed the classification of 73 different groups of non-overlapping contigs and 123 singletons, resulting in 196 unique ESTs (cfTable 1). The analysis of the 433 ESTs from the female-library yielded 55 different groups of non-overlapping contigs and 265 singletons, resulting in 320 unique ESTs (cfTable 1).
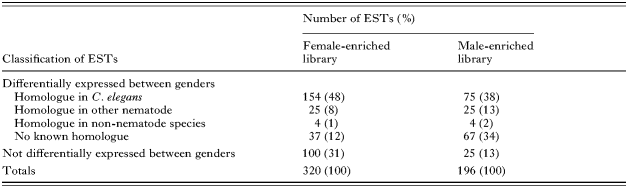
Microarray analysis of the 516 (i.e. 196+320) ESTs representing both gender-enriched libraries revealed differential hybridization for 391 of them (75·8%) (cfFig. 3). Of these 391 ESTs, 220 (56·3%) had significantly greater signal intensity for the female and 171 (43·7%) for the male (cfFig. 3). The other 125 ESTs (24·2%) did not have a differential hybridization signal (cfTable 1). Of the 220 ESTs with a statistically higher signal intensity in the female, 154 (70%) were predicted to possess C. elegans homologues (Table 2), These homologues include molecules involved in nucleic acid synthesis and function (14·3%), gametogenesis (3·9%) and metabolic pathways (11%), molecular binding or transport (8·4%), vitellogenins (9·1%), heat-shock proteins (HSP) (3·9%), protein kinases or signal transduction molecules (5·2%), mitochondrial associated proteins (1·9%) and various proteins/enzymes and putative proteins (e.g. translationally controlled tumor protein homologue, GMP reductase, phosphatidic acid phosphatase) (38·9%) (Table 2). The log2-transformed signal intensities for these 154 ESTs in the microarray analysis ranged from 1·1 to 34·9. In C. elegans, ‘non-wildtype’ RNAi phenotypes (such as Adl, Bmd, Dpy, Egl, Emb, Nmo, Let, Lva, Mvu, Pvl, Rup, Sck, Slu, Ste, Stp and/or Unc; see WormBase: http://www.wormbase.org/) have been recorded for 67 (43·5%) of the 154 homologues (Table 2). Of the 171 ESTs with statistically higher signal intensity in male O. dentatum (Table 1), 75 (43·8%) had C. elegans homologues (Table 3) representing serine proteases or protease inhibitors (35%), major sperm proteins (MSPs) or MSP-like molecules (5%), keratin-like molecules (5%) and various molecules involved in metabolism (~10%), PDZ domain-containing proteins (3%), sugar binding proteins (4%), protein kinases (4%), molecules involved in proteolysis (3%) and various other proteins, including enzymes (e.g. sorbitol dehydrogenase and AAA ATPase), and putative proteins (31%) (Table 3). The log2-transformed signal intensities for these 75 ESTs in the microarray analysis ranged from 1·2 to 2·9. In C. elegans, ‘non-wildtype’ RNAi phenotypes (including Emb, Gro, Lva, Lvl, Pvl, Ste, Stp and/or Unc; see WormBase: http://www.wormbase.org/) have been reported for 8 homologues (i.e. Odm02C09, Odm03D10, Odm05A09, Odm02B03, Odm06H04, Odm03B01, Odm03E03 and Odm02H10) (Table 3).

Fig. 3. Display of microarray results for gender-enriched ESTs from Oesophagostomum dentatum. The signal intensity when probed with cDNA from male nematodes was plotted on the abscissa and that of the cDNA from females was plotted on the ordinate. Diagonal lines represent contours along which the hybridization signal is the same (a), 2-fold higher (b) or 4-fold higher (c). Data points shown are mean signal intensities for individual ESTs, derived from replicates on the same microarray, corrected for background, normalised and log2-transformed. These data were reproducible using different mRNA populations and for the ‘dye swap’ (see Materials and Methods section).


Of the 162 ESTs (representing both gender-enriched cDNA libraries) with no known homologue in C. elegans, 50 ESTs (31%) had homologues in other nematodes (Table 4), 8 (5%) ESTs (Odf01D10, Odf02A04, Odf03F04, Odf03H03, Odm02B09, Odm03G08, Odm06C12 and Odm06H05) had homologues in various other organisms, including species of Giardia and Leishmania (protozoa), Neurospora crassa (fungus), Anopheles gambiae (mosquito), Drosophila melanogaster (dipteran), Xenopus laevis (amphibian) and human (Table 5), and 104 ESTs (64%; see Table 1) had no significant homology to any sequence present in the databases interrogated.


In order to independently verify the hybridization results from the microarray analysis, an RT-PCR analysis of selected ESTs using specific primers was conducted (Fig. 4). As predicted from the microarray analysis, male-specific mRNA expression was detected for Odm02A01 (serpin; C. elegans gene code K11D12.6), Odm02B01 (enzyme hatching family member; C. elegans gene code T23H4.3) and Odm04A03 (no known homologue to any organism) (Fig. 4), as was female-specific expression for Odf03G07 (vitellogenin-3; C. elegans gene code F59D8.1), Odf05C03 (heat shock protein 16.48; C. elegans gene code T27E4.3) and Odf01A08 (germline helicase; C. elegans gene code C07H6.5). Also, transcripts were amplified from both sexes using primers to ESTs Odf04F07 and Odf01B11, shown previously by microarray analysis to be present at similar levels in both males and females. Using primers designed to the third intron in the serine/threonine phosphatase gene (Odmpp1) of O. dentatum (see Boag et al. 2003; accession number AF496634), no genomic DNA was detectable by PCR in any of the RNA samples tested.

Fig. 4. Reverse-transcription PCR amplification from cDNA derived from adult male (M) or female (F) Oesophagostomum dentatum using primers designed specifically to selected expressed sequence tags (ESTs) from gender-enriched libraries. Positive genomic DNA (+) and no-DNA (−) controls were included in each set of reactions. Odmpp-1 represents a serine/threonine phosphatase gene of O. dentatum (Accession number AF496634; Boag et al. 2003).
DISCUSSION
The construction of gender-enriched gene libraries for O. dentatum and bioinformatic-microarray analyses have overcome previous limitations of the differential display for isolating and characterizing gender-specific genes (cfBoag et al. 2000). Southern blot and microarray analysis demonstrated the effectiveness of the SSH process in removing common, house-keeping genes and achieved a high degree of enrichment for genes in each sex of adult O. dentatum. After contig assembly, 76% of the 516 ESTs displayed in microarray differential hybridization signals between the sexes, which was higher to the percentage (72·4%) achieved in a study of Trichostrongylus vitrinus (which belongs to the same taxonomic order as O. dentatum) (see Nisbet and Gasser, 2004). The explanation for this difference could relate to a technical difference in the manner of SSH library construction. Given the small size of T. vitrinus compared with the nodule worm, the cDNA was synthesized from total RNA using the SMART™ cDNA synthesis kit (Clontech), which has a long-range PCR amplification step. This step was not included in the present study because, for each sex, substantially more mRNA was isolated from the larger worm (O. dentatum). Also, a comparison of the cDNA subtraction efficiency by Southern blot analysis suggested a greater subtraction efficiency without using the SMART™ kit. Irrespective, both approaches were very effective.
By comparison with C. elegans, similar groups of molecules were classified for O. dentatum as for T. vitrinus (see Nisbet and Gasser, 2004), although a smaller number of ESTs was analysed herein. Nonetheless, there were some distinct differences between the two species, such as the presence/absence of predicted phosphoenolpyruvate carboxykinase(s) and keratin-like proteins in adult O. dentatum females and males, respectively. Also, serine protease inhibitors (serpins) were well represented in male O. dentatum, whereas no serpin homologues were detected previously in T. vitrinus (see Nisbet and Gasser, 2004), although serpins have been isolated and characterized for the latter species (MacLennan, McLean and Knox, 2005). Conversely, only a small number of ESTs predicted to represent protein kinases and phosphatases was detected in O. dentatum, whereas 29 different clusters of male-specific ESTs representing these molecules were identified in T. vitrinus (see Nisbet and Gasser, 2004). Such differences could relate to variation in the abundance between the two species of bursate nematode, but, again, it is also possible that the absence from or inclusion of an amplification step in the SSH library construction (leading to a biased representation of some molecular groups over others) may have a significant effect on the nature and abundance of ESTs in the archives. This aspect should be examined by direct comparison of the two approaches using the same species of nematode.
In this study, O. dentatum ESTs from gender-enriched cDNA libraries were compared with genes of C. elegans (considering relevant information available in WormBase), in order to be able to suggest the biological relevance of some molecular groups. In the female-library, for example, a group of 14 vitellogenin gene homologues (i.e. C. elegans isoforms vit-1 to vit-6) was identified. Vitellogenins, large (~200–700 kDa) lipid-binding proteins synthesized in the maternal intestinal epithelia of C. elegans, represented ~9% of the O. dentatum ESTs from the female-library. The transportation of vitellogenins to the developing embryo in the gonad precedes their uptake into the oocyte via receptor-mediated endocytosis (Grant and Hirsh, 1999). In the developing embryo, the vitellogenins are utilized as a source of amino acids (Chen, Sappington and Raikhel, 1997) and are known to be ‘rate-limiting’ in the development of invertebrate eggs (Dhadialla and Raikhel, 1990). Heat-shock protein genes were another group of female-enriched molecules, including the small heat-shock protein (smHSP) homologues (n=6). Like the larger molecular weight heat-shock proteins, smHSPs are known to have molecular chaperone activities that can be upregulated in response to environmental stimuli (e.g. heat stress) (Arrigo and Landry, 1994; Haslbeck et al. 2005). The smHSPs have been identified in several nematode species, including T. vitrinus (see Nisbet and Gasser, 2004), H. contortus (see Hartman et al. 2003), C. elegans (see Ding and Candido, 2000a,b), B. malayi (see Raghavan et al. 1999), B. pahangi (see Thompson, Martin and Devaney, 1996) and Dirofilaria immitis (see Lillibridge, Rudin and Philipp, 1996). The expression of the smHSP gene family members has been localized in H. contortus and C. elegans to the reproductive tissues (Hartman et al. 2003; Ding and Candido, 2000a,b). In particular, C. elegans HSP16.48 is expressed abundantly in the vulva and spermatheca of hermaphrodites in response to heat stress (Ding and Candido, 2000a,b). While the significance of smHSPs is not yet understood in parasites, such as O. dentatum, it is possible that their increased expression is linked to environmental stimuli (such as body temperature and pH in the host) encountered by parasites when they undergo their transition from the free-living to parasitic life-cycle stages. Another group of female-enriched O. dentatum molecules had C. elegans homologues associated with gametogenesis (n=6). An example is the gene homologue (cgh-1) representing a germ-line helicase. This ste13/ME31B/RCK/p54 DEAD-box RNA helicase is involved in reproductive/germ line processes in C. elegans (Navarro et al. 2001), D. melanogaster, X. laevis and the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae (see de Valoir et al. 1991; Maekawa et al. 1994; Ladomery, Wade and Sommerville, 1997; Moriya and Isono, 1999; Navarro et al. 2001). Silencing of cgh-1 in C. elegans by RNAi affected the gametes of both sexes. Mature, male germ cells had shortened pseudopods (rendering them immobile), whereas the proximal oocytes of the hermaphrodite were deformed and infertile (Navarro et al. 2001). The high level of amino acid sequence identity between O. dentatum and C. elegans and the apparent evolutionary and functional conservation for these DEAD-box RNA helicases in different phyla (Navarro et al. 2001) suggest that the O. dentatum homologue may play the same or a similar functional role. Given the gene silencing phenotype of cgh-1 in C. elegans (see Navarro et al. 2001), it would be informative to perform RNAi studies of fourth-stage larvae of O. dentatum, to transplant them into a naïve host (e.g. via rectal intubation; see Christensen et al. 1996) and to subsequently monitor the reproductive potential of treated worms in vivo. Understanding the function of this (cgh-1) and related genes and gene products, which are involved in fundamental aspects of nematode biology could be important in the discovery of drugs against parasites (cfNisbet et al. 2004; Nisbet and Gasser, 2004).
Thirty-eight percent of the O. dentatum ESTs from the male-library had homology (at the amino acid level) to genes from C. elegans, many of which had been shown previously to display increased mRNA levels in male C. elegans compared with hermaphrodites (Reinke et al. 2000; Jiang et al. 2001). These molecular groups were represented by the major sperm proteins (MSPs), MSP-like molecules and protein kinases/phosphatases, which are also reported to be differentially-expressed in T. vitrinus (see Nisbet and Gasser, 2004). The MSPs are nematode-specific, cytoskeletal proteins involved in the motility of the amoeboid sperm (reviewed by Roberts and Stewart, 2000; Cottee et al. 2004) and can also act as bipartite signalling molecules, activating pathways linked to oocyte production and maturation in some free-living and parasitic nematodes (see Miller et al. 2001, 2003). The intrauterine maturation of immotile spermatids to motile spermatozoa has been reported to commence without concomitant transcription of mRNA or translation into protein (Muhlrad and Ward, 2002), which would explain the absence of msp transcripts from RNA extracted from adult female O. dentatum and the existence of multiple msp transcripts in the male-library constructed herein. Also the MSP-domain containing proteins (MDPs) were well represented within the male-library. Unlike the MSPs, which are entirely nematode-specific, the MDPs have been identified in [ges ]21 species of nematode, including C. elegans, T. vitrinus, B. malayi and Ascaris suum as well as in plants, fungi, protozoa, flatworms and mammals (see Nisbet and Gasser, 2004; Tarr and Scott, 2005). While the biological roles of these molecules are not yet known for nematodes, 2 MDPs in A. suum have been implicated in the regulation of MSP cytoskeleton formation (Buttery et al. 2003). However, the nature of the MSP-MDP interaction in this and other nematodes and its involvement in cytoskeleton dynamics remain to be determined. Another group of male-enriched molecules with significant homology to C. elegans protein phosphatase or protein kinase sequences was considered interesting and relevant. The expression of these 2 classes of molecules in sperm-producing germline tissue has been shown to be substantial in C. elegans, with ~50% of the protein phosphatases being abundant therein (see Reinke et al. 2000). These molecules are proposed to play key roles in regulating sperm maturation by post-translational modification, after the expulsion of the organelles involved in protein synthesis from maturing spermatids (Reinke et al. 2000). The functional analysis of C. elegans homologues of a male-enriched protein phosphatase gene, Od-mpp1, from O. dentatum by RNAi in C. elegans has been shown to result in sterility of hermaphrodites (Boag et al. 2003) through impaired sperm function (cfHanazawa et al. 2001). Such phosphatases and kinases may also be involved in signalling cascades or protein modification within oocytes, following fertilization (cfBoag et al. 2003). Various other molecules from the O. dentatum male-library had significant enzyme homologues (e.g. pyruvate kinases and hexokinases) involved in carbohydrate metabolism. Hexokinases catalyse the ATP-dependent phosphorylation of aldo- and keto-hexose sugars (glucose, fructose, sorbitol and glucosamine) to the hexose-6-phosphate, the first step in a number of metabolic pathways, including glycolysis. For instance, 2 non-overlapping contigs from the male-library had sequence homology to a gene F14B4.2, a predicted hexokinase from C. elegans that is not enriched in germline tissues (Reinke et al. 2000). Hexokinases have been reported previously for parasitic nematodes (e.g. Schmitt-Wrede et al. 1999), and this is the second report of a gender-specific hexokinase from a parasitic nematode (cfNisbet and Gasser, 2004). It has been suggested that most enzymes in the glycolytic pathway have specific equivalents involved in spermatogenesis, although their functional roles are not yet clear (Nakamura et al. 2003). The relative abundance of enzymes involved in carbohydrate metabolism in O. dentatum suggests a significant dependence on carbohydrate-based energy metabolism, which may be associated with high-energy requirements for the production and motility of sperm.
Of the molecules isolated that did not have C. elegans homologues, 13% of the male-enriched and 8% of the female-enriched O. dentatum ESTs possessed significant homology (at the amino acid level) to known sequences from other parasitic nematodes. This finding is likely to reflect the differences in the developmental and reproductive biology as well as ecology between parasitic and free-living nematodes. Also, 12–34% of the ESTs had no known homologue in any organisms represented in current sequence databases, in accordance with previous studies of parasitic nematodes (e.g. Hoekstra et al. 2000; Nisbet and Gasser, 2004). Obviously, these groups of molecules pose the greatest challenge in terms of defining the functions of gene and/or gene products, but are likely to be the most interesting biologically and also in relation to searching for new intervention targets.
The bioinformatic-microarray analysis focused predominantly on comparisons with homologues from C. elegans, because members of the order Strongylida (Clade V) are considered to be relatively closely related to C. elegans (see Blaxter et al. 1998), because the entire genome sequence of C. elegans has been determined (C. elegans Sequencing Consortium, 1998) and because there is a wealth of information on the localization of molecules (cfhttp://www.bcgsc.ca/gc/celegans/) as well as the function of genes in this nematode by gene silencing (i.e. RNAi) (Fire et al. 1998) and gene knock-out (cfhttp://www.wormbase.org/; http://elegans.bcgsc.bc.ca/knockout.shtml). The functions of most genes in C. elegans have been assessed through RNAi (e.g. Maeda et al. 2001; Kamath et al. 2003; Mello and Conte, 2004; Sugimoto, 2004) in the hermaphroditic stage, whereas functional information for parasitic nematodes is scant because of the limitations with their maintenance and propagation (exclusively) in vitro. Nonetheless, given the recent progress and successes in the application of RNAi to parasitic nematodes (Aboobaker and Blaxter, 2004), particularly the development of an effective RNAi approach for T. colubriformis (utilizing an electroporation step to transfer dsRNA into the larval stage of the nematode) (Issa et al. 2005), there is considerable prospect for an RNAi assay for Oesophostomum dentatum, provided that this nematode has the appropriate ‘RNAi machinery’ (van Roessel and Brand, 2004). The attributes of O. dentatum, including a short life-cycle (usually ~21–25 days; Talvik et al. 1997), the production of large numbers of progeny, the availability of a non-surgical transplantation procedure (Christensen et al. 1996) to manipulate, for example, the ratio of male and female parasites or the number of worms per host individual (intensity of infection) and the ability to maintain the parasite in culture in vitro for 28 days (Daugschies and Watzel, 1999), indicate that this system will be particularly useful for testing (both in vitro and in vivo) the functional roles of gender-specific genes represented in the EST archives constructed herein. Future investigations will focus on isolating and characterizing selected genes.
The authors sincerely thank Niels-Peter Hansen of the Danish Centre of Experimental Parasitology for producing O. dentatum and for support. Thanks also to Dr Ben Ong for discussions and help. P.A.C. is a recipient of a scholarship from The University of Melbourne. This work was supported by grants from the Australian Research Council (Linkage Project IDs LP0346983 and LP0667795), Genetic Technologies Limited and Meat and Livestock Australia (project AHW.022) (to R.B.G.).