Introduction
The cell cycle in eukaryotic organisms is a very well-regulated process, divided in four phases: G1 (cell enlargement), S (DNA replication), G2 and M (cytokinesis) (Suryadinata et al., Reference Suryadinata, Sadowski and Sarcevic2010). In general, cells from differentiated animal tissues, such as hepatocytes, are arrested in the G1–S transition (Nelsen et al., Reference Nelsen, Rickheims, Tucker, Hansen and Albrech2003). The mechanism of arrest involves the retinoblastoma proteins (RBp) which inhibit the activation of the E2F-DP transcription complex. This pathway is conserved in mammals and plants (Lammens et al., Reference Lammens, Li, Leone and De Veylder2009). E2F proteins are a family of transcription factors which regulate expression of genes that participate in the G1–S transition. Cyclin D and E (Nelsen et al., Reference Nelsen, Rickheims, Tucker, Hansen and Albrech2003), cyclin-dependent kinases (CDKs), DNA polymerases, the proliferating cell nuclear antigen (PCNA, part of the replisome complex) (Strzalka and Ziemienowicz, 2011) and many other proteins related to DNA replication are among the proteins that participate in this transition and are regulated by E2F (Lydeard et al., Reference Lydeard, Lipkin-Moore, Sheu, Stillman, Burgers and Haber2010).
Cyclin phosphorylation by mitogen-activated protein kinases (MAPKs) is induced in response to mitogens. This leads to the formation of CDK–cyclin complexes activating a CDK which, in turn, sequentially phosphorylates RBp, dissociating it from the E2F transcription factors. As a consequence, the transcription of genes regulated by E2F is stimulated, and transition to the S phase with DNA replication occurs (Lammens et al., Reference Lammens, Li, Leone and De Veylder2009).
In recent years, it has been reported that insulin and insulin-like growth factors (IGFs) show mitogenic activity in animals. In studies using primary cell cultures and hepatic cell lines, it was observed that insulin not only induced the transcription of genes related to lipid synthesis, transport and carbohydrate metabolism (Mounier and Posner, Reference Mounier and Posner2006; Chakravarty and Hanson, Reference Chakravarty and Hanson2007), but also regulated transcription and translation of genes participating in the cell cycle, in particular cyclin D1 (Nelsen et al., Reference Nelsen, Rickheims, Tucker, Hansen and Albrech2003; Frederick et al., Reference Frederick, Min, Altieri, Mitchell and Wood2007), and had a mitogenic role during corticogenesis in rats (Mairet-Coello et al., Reference Mairet-Coello, Tury and DiCicco-Bloom2009). In addition, there are reports of hepatocyte cell cultures showing that IGFs play a role in homeostasis control, cell growth and development in a similar way as insulin (Sun et al., Reference Sun, Tu and Baserga2007). One of the most studied effects of insulin/IGFs is the regulation of protein synthesis by activation of translational apparatus components through the PI3K-TOR pathway (Patursky-Polischuk et al., Reference Patursky-Polischuk, Stolovich-Rain, Hausner-Hanochi, Kasir, Cybulski, Avruch, Rüegg, Hall and Meyuhas2009), including the translation of specific mRNAs such as those coding for ribosomal proteins, such as S6rp, and translation factors, such as eEF1α (Meyuhas and Dreazen, Reference Meyuhas and Dreazen2009). Furthermore, it has been reported that insulin-like growth factor I (IGF-I) increases ribosomal DNA transcription and ribosome biogenesis (Wu et al., Reference Wu, Tu, Prisco and Baserga2005).
Several studies in maize, Arabidopsis thaliana and other species suggest that the general mechanism for cell-cycle regulation is very similar to that described for animals (Vázquez-Ramos and Sánchez, Reference Vázquez-Ramos and Sánchez2003; Barroco et al., Reference Barroco, Van Poucke, Bergervoet, De Veylder, Groot, Inzé and Engler2005). However, recent studies in Arabidopsis have pointed at some differences, such as the absence of cyclin E in plants (Harashima and Schnittger, Reference Harashima and Schnittger2010) and the control of mitosis entry in plants (Dissmeyer et al., Reference Dissmeyer, Weimer, Veylder, Novak and Schnittger2010).
In the particular case of mature seeds, cells are dehydrated and acquire a quiescent state with very low metabolic activity. The imbibition process reactivates the metabolism and induces seed germination. The cell cycle is among the metabolic events that are reactivated during seed germination (Barroco et al., Reference Barroco, Van Poucke, Bergervoet, De Veylder, Groot, Inzé and Engler2005). In maize embryonic axes, cells are mainly arrested in the G1–S transition (Baíza and Sánchez de Jiménez, Reference Baíza and Sánchez de Jiménez1989). Among the elements participating in this transition, four kinds of cyclin D (Vázquez-Ramos and Sánchez, Reference Vázquez-Ramos and Sánchez2003), RBR protein homologues to the animal RBp (Sabelli et al., Reference Sabelli, Dante, Leiva-Neto, Jung, Gordon-Kamm and Larkins2005), as well as PCNA and several CDKs (Sánchez et al., Reference Sánchez, Gurusinghe, Bradford and Vázquez-Ramos2005) have been characterized. Additionally, the presence of E2F family protein homologue sequences has been identified through expressed sequence tag (EST) analysis (Alexandrov et al., Reference Alexandrov, Brover, Freidin, Troukhan, Tatarinova, Zhang, Swaller, Lu, Bouck, Flavell and Feldmann2009).
The existence of insulin-like factors has been reported in spinach (Collier et al., Reference Collier, Watkinson, Cleland and Roth1987) and bean (Oliveira et al., Reference Oliveira, Ribeiro, da Cunha, Gomes, Fernández and Xavier-Filho2004). In maize, a 5.7 kDa peptide with a structure similar to that reported for insulin and IGF-I has been purified, characterized and named insulin-like growth factor of maize (ZmIGF) (Rodríguez-López et al., Reference Rodríguez-López, Rodríguez-Romero, Aguilar and Sánchez de Jiménez2011). Even though insulin has not been found in plants, it has been reported that this hormone and IGFs induce similar responses in plants; both effectors induce germination and growth of maize seeds (Sánchez de Jiménez et al., Reference Sánchez de Jiménez, Beltrán-Peña and Ortíz-López1999; García-Flores et al., Reference García-Flores, Aguilar, Reyes de la Cruz, Albores and Sánchez de Jiménez2001) and accelerate growth of sunflower (Goodman and Davis, Reference Goodman and Davis1993) and Canavalia ensiformis seedlings (Oliveira et al., Reference Oliveira, Ribeiro, da Cunha, Gomes, Fernández and Xavier-Filho2004); and in maize both ZmIGF and insulin enhance, by TOR pathway activation, the specific mobilization to polysomes and selective translation of mRNA related to protein synthesis, like S6rp, the translation initiation factor eIFiso4E (Dinkova et al., Reference Dinkova, Reyes de la Cruz, García-Flores, Aguilar, Jiménez-Garcıa and Sánchez de Jiménez2007) and ribosomal proteins (as has been demonstrated recently by microarray analysis; Jiménez-López et al., Reference Jiménez-López, Mancera-Martínez, Donayre-Torres, Rangel, Uribe, March, Jiménez-Sánchez and Sánchez de Jiménez2011). Also, it has been reported recently that ZmIGF/insulin induce mitogenic activation by increasing DNA synthesis in maize tissue cultures, as well as the mobilization into polysomes of mRNA coding for the proteins associated with the cell cycle, like PCNA and cyclin D1 (Sotelo et al., Reference Sotelo, Garrocho-Villegas, Aguilar, Calderón and Sánchez de Jiménez2010) which participate in the G1–S transition, as mentioned earlier.
The co-ordinating mechanisms for growth and cell division during maize embryonic axis germination are not known with precision, but several reports support the presence of a conserved signal transduction pathway, TOR-S6K, that co-ordinates growth and cell proliferation in eukaryotes (Garrocho-Villegas and Sánchez de Jiménez, Reference Garrocho-Villegas and Sánchez de Jiménez2012). Also, it is unknown if all the tissues of the embryonic axis respond in the same way to growth regulators. In this work, regulation of cell-cycle restart in germinating maize embryonic axes in the presence of insulin was studied. As previously mentioned, most of the cells in the embryonic axes of maize seeds are arrested in G1–S; therefore, it was considered relevant to study the effect of insulin on the expression of some of the genes involved in the G1–S transition, such as PCNA and E2F.
Materials and methods
Plant material and treatments
Maize (Zea mays L.) var. Chalqueño seeds were used in all experiments. All reagents were from Sigma-Aldrich (St Louis, Missouri, USA) unless otherwise stated in the text. Seeds were disinfected, imbibed and treated with a pulse of 200 μU ml− 1 of bovine insulin for 2 h, as described by Buentello-Volante et al. (Reference Buentello-Volante, Díaz de León-Sánchez, Rivera-Cabrera, Aguilar Caballero, Ponce-Valadez, Sánchez de Jiménez and Pérez-Flores2010). This insulin concentration and time of treatment has been demonstrated previously to be effective in inducing maize seedling growth (Reyes de la Cruz et al., Reference Reyes de la Cruz, Aguilar and Sánchez de Jiménez2004; Dinkova et al., Reference Dinkova, Reyes de la Cruz, García-Flores, Aguilar, Jiménez-Garcıa and Sánchez de Jiménez2007; Buentello-Volante et al., Reference Buentello-Volante, Díaz de León-Sánchez, Rivera-Cabrera, Aguilar Caballero, Ponce-Valadez, Sánchez de Jiménez and Pérez-Flores2010). Due to its ready availability, insulin was used instead of ZmIGF to stimulate the maize embryonic axes. Germination times are specified for each experiment. Samples used to determine the insulin effect by flow cytometry were processed immediately. Samples to analyse the insulin effect in the regulation of gene expression were frozen with liquid N2 and stored at − 70°C until their analysis. All experiments were done in triplicate.
Effect of insulin on radicle and coleoptile growth of growing maize embryonic axes
To determine the effect of insulin on growth of the embryonic axis, sets of 50 seeds were imbibed for 22 h in moisturized cotton. The embryonic axes were dissected and incubated with or without insulin for 2 h. At the end of the incubation period the axes were rinsed with distilled water and the length of the radicle and coleoptile was measured. Subsequently, embryonic axes were placed in 100-mm Petri dishes with filter paper moisturized with 5 ml of Murashige and Skoog medium (MS) and were incubated at 25 ± 2°C for 72 h, to reach a total incubation period of 96 h. Radicle and coleptile lengths were measured every 24 h and the embryonic axes were transferred to a Petri dish with fresh MS medium.
Determination of S6K-Thr 389
Embryonic axes imbibed for 22 h and treated as previously described with or without insulin were used for this experiment. At the end of the incubation time, radicles and coleoptiles were separated. Total proteins from each part were extracted according to Reyes de la Cruz et al. (Reference Reyes de la Cruz, Aguilar and Sánchez de Jiménez2004). The phosphorylation of ribosomal protein S6 kinase (S6K) was determined by Western blotting. Proteins (25 μg) were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 12% w/v acrylamide gel and transferred to a Nylon membrane, Hybond-P (Amersham, California, USA), using a semi-dry transfer system at 8 mA cm− 2. The membranes were incubated with agitation overnight at 4°C using a primary antibody against human p70S6K phosphorylated at threonine 389 (Sc-11 759, Santacruz Biotechnology Inc., California, USA) at a 1:20 dilution in 1 × phophate-buffered saline (PBS), 0.5% v/v Tween and 8% w/v powdered milk. Subsequently, the membrane was washed twice with 1 × PBS, 0.5% v/v Tween and 8% w/v powdered milk at room temperature for 20 min each time and incubated for 1 h with peroxidase-conjugated anti-rabbit secondary antibody (Sc-2769, Santacruz Biotechnology Inc.) at 1:5000 dilution in 1 × PBS, 0.5% v/v Tween and 0.2% w/v bovine albumin fraction V.
Thereafter the membrane was washed twice with 1 × PBS, 5% v/v Tween and placed in 1 × PBS. The reaction was detected by chemiluminescence with Western Lighting® Plus-ECL (Perkin Elmer Inc., Massachusetts, USA) using a Blue x-Omat Kodak film.
De novo synthesis of DNA
Sets of maize seeds were imbibed for 36 h in moisturized cotton at 25 ± 2°C. Embryonic axes (2.5 g) were dissected and incubated for 12 h in 3 ml of MS media in the presence of 15 μCi ml− 1 of [3H]thymidine, with or without insulin as previously described. At the end of the treatment, axes were washed and the radicles and coleoptiles were separated for the subsequent determinations.
DNA was extracted from coleoptiles and radicles. The tissue was pulverized in liquid N2 and homogenized in 5 ml of hot (65°C) extraction buffer [10% w/v cetyl trimethyl ammonium bromide (CTAB), 100 mM Tris, pH 8, 20 mM EDTA, 0.7 M NaCl]. The homogenate was incubated at 65°C for 1 h, an equal volume of chlorophorm:isoamylalcohol (24:1 v/v) was added and then it was centrifuged at 7500 g (Sorvall RC 5C Plus centrifuge with SS34 rotor type) for 5 min at 4°C. The aqueous phase was obtained and the volume was measured; 10% w/v CTAB in 0.7 M NaCl was added to the supernatant in a 1:10 ratio, mixed and centrifuged in the same conditions. The supernatant was recovered and the DNA was precipitated with 1% w/v CTAB in 50 mM Tris, 10 mM EDTA and incubated overnight at 37°C. The precipitate was centrifuged at 500 g for 5 min at 4°C. The supernatant was decanted and the pellet was resuspended in TE buffer (100 mM Tris, 5 mM EDTA, pH 8). The DNA was re-precipitated with 0.6 volumes of cold isopropanol and incubated overnight at − 20°C. The precipitate was centrifuged at 7500 g for 15 min at 4°C, the pellet was washed with 70% v/v ethanol and centrifuged again in the same conditions, the supernatant was decanted and the remaining ethanol was evaporated. Finally, the pellet was resuspended in 100 μl of TE buffer. DNA was quantified by absorbance at 260 nm and [3H]thymidine incorporation was determined in a Beckman scintillation counter using 50 μl of sample in 5 ml of Bray's solution (dioxane–methanol–ethylene glycol–naphthalene). Results are reported as cpm μg− 1 of DNA.
DNA content by flow cytometry
Sets of 100 maize seeds were imbibed for 13, 16, 22 and 34 h. For 0 h, embryonic axes dissected from the seed without imbibition were used. Entire embryonic axes were dissected and treated with or without insulin as described previously. At the end of the treatment, the radicles were separated. Nuclei were isolated from 0.5 g of 3–4 mm radicle apices which were fixed in acetone for 2 h, pulverized in liquid N2 and homogenized in 5 ml of isolation buffer [10 mM Tris–HCl, pH. 7.2, 1 M sucrose, 5 mM MgCl2, 2 mM β-mercaptoethanol and half a tablet of protease inhibitors COMPLETE® (Roche Diagnostics Deutschland GMBH, Mannheim, Germany) for 100 ml of buffer]. The homogenate was filtered through Miracloth® (Calbiochem, California, USA) with a pore size of 22–25 μm, and subsequently centrifuged at 5000 g for 10 min at 4°C. The pellet was resuspended in 1.5 ml of buffer [10 mM Tris–HCl, pH 7.2, 1 M sucrose, 5 mM MgCl2, 2 mM β-mercaptoethanol, 0.5% v/v Triton X-100 and half a tablet of protease inhibitors COMPLETE® (Roche) for 100 ml of buffer]. Nuclei were purified in a discontinuous Percoll® gradient according to Luthe and Quatrano (Reference Luthe and Quatrano1980). Nuclei were diluted in 1 ml of the isolation buffer and centrifuged at 10,000 g for 20 min at 4°C. The pellet containing the nuclei was carefully resuspended in 360 μl of phosphate saline buffer composed of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, 1 × PBS. Nuclei were incubated with 50 μg ml− 1 ribonuclease A (Promega, Madison, Wisconsin, USA) for 30 min at room temperature; propidium iodide at a final concentration of 50 μg ml− 1 was added in the last 10 min and they were maintained in darkness. Nuclear DNA content was determined in a total volume of 500 μl by flow cytometry using a FACScan cytometer (Beckman, California, USA) equipped with an argon ion laser at 488 nm; fluorescence was detected in a range from 605 to 635 nm. Chicken leucocytes from the kit BD™ DNA QC Particles (BD, Biosciences, San Jose, California, USA) and nuclei from maize leaf obtained as described by Sliwinska et al. (Reference Sliwinska, Zielinska and Jedrzejczyk2005) were used as internal controls. Nuclear samples from radicles were read in the flow cytometer in duplicate with 30,000 events in each determination. The data were analysed with the MODFIT-LT™ software for Mac 3.0. (Verity Software House, Topsham, Maine, USA) The amount of DNA is proportional to the fluorescence signal; 1C value represents the amount of DNA in the haploid chromosome without replication. The frequencies of nuclei in 2C and 4C were calculated with the following formulae: [nuclei 2C/(nuclei 2C+ nuclei 4C)] × 100 and [nuclei 4C/(nuclei 2C+ nuclei 4C)] × 100, respectively, according to Gendreau et al. (Reference Gendreau, Romaniello, Barad, Leymarie, Benech-Arnold and Corbineau2008). The amount of DNA was expressed as the percentage of nuclei in each phase of the cell cycle.
Extraction of total RNA and polysomal fraction RNA from maize radicles and coleoptiles
Maize seeds were imbibed for 13 or 22 h and treated with or without insulin as described previously. After the treatment the radicles and coleoptiles were separated. Total RNA was isolated from 0.1 g of 2–4 mm long radicle or coleoptile apices pulverized in liquid N2, using the Trizol reagent (BRL, Life Technologies, Invitrogen, Carlsbad, California, USA) with the modifications recommended for plant tissues. RNA from the polysomal fraction was obtained from 0.5 g of 2–4 mm long radicle or coleoptile apices. Polysomes and RNA were obtained by differential centrifugation following the technique reported by Dinkova et al. (Reference Dinkova, Reyes de la Cruz, García-Flores, Aguilar, Jiménez-Garcıa and Sánchez de Jiménez2007). Both total RNA and RNA from the polysomal fraction were resuspended in 50 μl of RNAse-free water. RNA purity was determined by the A260nm/A280nm ratio and its concentration was quantified using a Nanodrop 2000 (Thermo Scientific, Wilmington, Delaware, USA). Sample integrity was verified using 2 μg of total RNA or RNA from the polysomal fraction according to the methodology described by Sambrook et al. (Reference Sambrook, Fritsch and Maniatis1989).
Analysis of the regulation of the transcript levels by RT-PCR and qRT-PCR
To verify the effect of insulin on mRNA accumulation in the polysomal fraction, S6rp was amplified by semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) from total RNA and RNA obtained from the polysomal fraction. The primers used for amplifying S6rp were FW: 5′-TGATGCTCTGGGTGAGGAGTTAAA-3′ and RV: 5′-TTCCTTCAGCCTCTGGGCAAGAAG-3′, obtaining an amplification product of 550 bp. Glyceraldehyde-3-phosphate dehydrogenase, G3PDH, was used as reference gene; the primers for its amplification were FW: 5′-CCTTGACCGCAGCCTTGATCCTACATC-3′ and RV: 5′-ACCCATCCTCGTTTCCTCCGTCTAG-3′, obtaining an amplification product of 800 bp. For each pair of primers the cycle number was adjusted for a linear range.
For the quantitative reverse transcription-polymerase chain reaction (qRT-PCR) reactions of E2F and PCNA, 2 μg of total or polysomal RNA were used. The retrotranscription reaction (RT) was performed with 1 U of the M-MLV RT (Moloney Murine Leukemia Virus Reverse Transcriptase; Promega, California, USA) enzyme and 1 μg of the RNA samples treated with 1.5 U of DNAse (Promega) according to the manufacturer's instructions. After this, samples were stored at − 20°C until further use.
Quantitative polymerase chain reaction (qPCR) was performed in a Rotor Gene 3000 (Corbett Research Quiagen, California, USA), using 1 μl of the RT reaction obtained earlier. The amplifications were performed with the One Step qPCR kit® (Invitrogen, Carlsbad, California, USA). The primers used for the amplification were designed based on the following maize sequences: for E2F (Accession Number EU-973980), FW: 5′-TCTCCTCAAATTGACTAACTA-3′, RV: 5′-ATAGGAGTTTAACTGATTGAT-3′, obtaining an amplification product of 535 bp; for PCNA (Accession Number NM_00111991.1), FW: 5′-TGCCGACTTTTTCTCCTCTT-3′, RV: 5′-AAATCCATGGACACACAGCA-3′ and the amplification product was 146 bp long. S13rp (Accession number NM_0011122.38) was used as the reference gene, the primers for amplifying this sequence were FW: 5′-AAAGAAGCTTCCACCCACCT-3′, RV: 5′-GATGAGGTCCGTTGTCCATT-3′ and the amplification product was 151 bp long. Amplification conditions were for E2F, 5 min at 94°C, 30 cycles 94°C for 30 s, 58°C for 30 s and 72°C for 50 s for PCNA, 5 min at 94°C, 30 cycles 94°C for 20 s, 58°C for 20 s and 72°C for 20 s. Before performing the assays for analysing the regulation of gene expression by insulin, it was checked that the amplification reaction efficiency for each of the analysed genes was between 95 and 100% according to Bustin et al. (Reference Bustin, Benes, Garson, Hellemans, Huggett, Kubista, Mueller, Nolan, Pfaffl, Shipley, Vandesompele and Wittwer2009). Also, it was verified that treatment with insulin and germination time did not affect the reference gene expression using the genNorm software version 3.5 (http://medgen.ugent.be/~jvdesomp/genorm/). The reference gene, S13rp, was amplified in the same run using the mentioned conditions for each of the studied genes, without affecting its amplification efficiency. Results were standardized according to Willems et al. (Reference Willems, Leyns and Vandesompele2008).
De novo synthesis of PCNA protein
Sets of maize seeds were imbibed for 13 h on moisturized cotton at 25 ± 2°C. Embryonic axes (2.0 g) were dissected and incubated for 2 h in 3.5 ml of MS media with or without insulin 200 μU ml− 1 in the presence of 450 μCi ml− 1 of [35S]methionine. At the end of the treatment, axes were washed and radicles were separated. PCNA protein was isolated from radicles with 35 μg of maize PCNA antibody (Sc-7907, Santacruz Biotechnology Inc.) bonded to magnetic beads (Dynabeads®, Invitrogen Life Technologies) following the kit instructions. [35S]Methionine incorporation was quantified in a Beckman scintillation counter using 25 μl of sample in 5 ml of ACS® (Amersham Biosciences). The protein concentration was determined by the method of Bradford (Reference Bradford1976) using a 15 μl aliquot of the sample and bovine serum albumin (BSA) as a standard. Results are reported as percentage of incorporation in relation to control tissue.
Statistical analysis
To obtain the parameters of descriptive statistics (mean and standard deviation), as well as to determine significant differences among treatments, the Dunett test of mean comparison against a control, with a significance level of α ≤ 0.05, was used in the NCSS software (NCSS, Utah, USA).
Results
Effect of insulin on the growth of radicles and coleoptiles of germinating maize embryonic axes
It has been reported previously that treatment with bovine insulin or maize insulin-like growth factor (ZmIGF) accelerates maize seed germination and growth (García Flores et al., Reference García-Flores, Aguilar, Reyes de la Cruz, Albores and Sánchez de Jiménez2001). To determine the specific effect of insulin on different parts of the embryonic axis, an insulin pulse was applied. Axes were dissected from seeds imbibibed for 22 h and treated with or without insulin for 2 h. The length of both the entire embryonic axis and the radicles and coleoptiles was measured every 24 h until 96 h. Embryonic axes treated with a pulse of insulin showed a significant increase of approximately 20% in growth after 24 h of treatment compared to the control, which was maintained during the entire period of the study (Fig. 1A). In radicles, the average effect of insulin treatment was about a 25% increase in growth from 24 h onward, while the coleoptile showed a significant effect in response to the treatment after 48 h, with an average of 12% increase compared to the control (Fig. 1B and C). These results suggest that the different parts of the embryonic axis respond in a different way to insulin and, in particular, the radicle showed an earlier and slightly higher response than the coleoptile. Based on these results, in subsequent experiments the insulin effects on both radicle and coleoptiles were analysed.

Figure 1 Effect of insulin on growth of maize embryonic axes during germination (A) length of the entire embryonic axis; (B) length of the radicle and (C) length of the coleoptile. Values are the averages of three independent experiments and are expressed as the length ± SD. * Indicates significant differences among treatments (α = 0.05).
Effect of insulin on S6K phosphorylation and S6rp mRNA expression and accumulation in the polysomal fraction from radicles and coleoptiles
The signal transduction pathway activated by insulin or ZmIGF in maize is very similar to that reported for animals. As part of the activation by insulin, the PI3K-TOR pathway is induced and S6K is phosphorylated at threonine 389 (S6K-Thr 389) (Meyuhas and Dreazen, Reference Meyuhas and Dreazen2009). In the present work it was shown that radicles of embryonic axes respond to insulin treatment faster and in a higher proportion than coleoptiles. Therefore, the insulin effect on S6K phosphorylation was evaluated in radicles and coleoptiles by Western blotting using a specific antibody against the S6K phosphorylated at threonine 389 (Reyes de la Cruz, et al., Reference Reyes de la Cruz, Aguilar and Sánchez de Jiménez2004). Insulin induced a significant increase of 30% in the S6K-Thr 389 phosphorylation in maize radicles, but this effect was not observed in coleoptiles (Fig. 2A). Also, in the maize radicles treated with insulin, accumulation of S6rp mRNA in the polysomal fraction was observed (Fig. 2B). In coleoptiles, no change was detected in the expression of the S6rp mRNA when treated with insulin.

Figure 2 Effect of insulin on S6K phosphorylation, on S6rp mRNA accumulation in the polysomal fraction from radicles and coleoptiles and on de novo synthesis of DNA in radicles and coleoptiles of germinating maize embryonic axes. (A) Western blot of the phosphorylated S6K-Thr 389 form in total proteins (20 μg) isolated from radicles and coleoptiles and separated by SDS-PAGE; (B) S6rp gene expression in radicles and coleoptiles as determined by semi-quantitative RT-PCR. Values are the average of three independent experiments and are expressed in relative units ± SD. (C) De novo synthesis of DNA in radicles and coleoptiles. Values are the average of three independent experiments and are expressed as incorporation in cpm μg− 1 of DNA ± SD. * Indicates significant differences among treatments (α = 0.05).
Effect of insulin on de novo DNA synthesis in radicles and coleoptiles of germinating maize embryonic axes
Tissue growth can be the result of an increase of cell size and/or cell proliferation. However, in both cases proteins constitute the main component of the cell dry mass (Baserga, Reference Baserga2007). De novo DNA synthesis in radicles and coleoptiles of maize embryonic axes germinated for 36 h was measured using the radioactive precursor [3H]thymidine. In control samples, the values of [3H]thymidine incorporation were higher in radicle (1536 cpm μg− 1 DNA) than in coleoptiles (325.5 cpm μg− 1 DNA), showing that de novo DNA synthesis was 4.7-fold higher in the radicles during the studied period. Incubation with insulin increased [3H]thymidine incorporation in the radicles (2446 cpm μg− 1 DNA) by 60% as compared to the control (1536 cpm μg− 1DNA). In coleoptiles no significant differences in the incorporation of the radioactive precursor were observed in the insulin-treated axes compared to the control (Fig. 2C).
These results suggest that insulin has a tissue-specific effect since it preferentially induced de novo DNA synthesis in radicles. Therefore, subsequent experiments were only performed in this part of the embryonic axis.
Analysis of the cell cycle in radicles of maize embryonic axes and its stimulation by insulin during germination
The analysis to determine the percentage of cells in each phase of the cell cycle was performed by flow cytometry in the root meristem which, as mentioned earlier, is the first to restart the cell cycle. Apices from radicles were dissected from embryonic axes without imbibition (0 h) and imbibed for 15, 18, 24 and 36 h and treated or non-treated (control) with a 2 h pulse of insulin. In quiescent seeds, at 0 h of germination, 88% of the cells present in the radicular apex were arrested in the G1–S transition (nuclei 2C) and the 12% of cells remaining were found to be in the G2–M transition (nuclei 4C). The 4C/2C ratio was 0.14 (Table 1). In the control samples, an increase in the 4C/2C ratio to 0.38 was observed after 15 h of germination, followed by a decrease after 24 h to 0.17 and increasing again to 0.70 after 36 h of germination. In the insulin-treated samples, the 4C/2C ratio followed the same pattern, increasing with time of germination. However, in general they presented higher values than the control samples. After 18 h, the increase was 55%, after 24 h 170% and 135% after 36 h, as compared to the corresponding control. These results show that the increase in radicle growth in response to insulin can be attributed, at least in part, to the induction of cell proliferation in radicular apices of germinating maize seeds (Table 1).
Table 1 Percentage of 2C, 4C nuclei and 4C/2C ratio. Radicle tips of embryonic axes isolated from dry seeds and germinated for 15, 18, 24 and 36 h of which the last 2 h consisted of incubation with or without 200 μU ml−1 of insulin. Frequencies of 2C and 4C nuclei were calculated with the following formulae: [2C nuclei/(2C nuclei +4C nuclei)]×100 and [4C nuclei/(2C nuclei +4C nuclei)]×100, respectively. Populations of 2C and 4C nuclei were measured in 30,000 nuclei
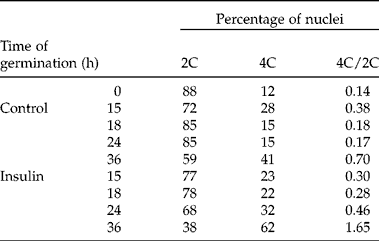
The percentage of nuclei in each phase of the cell cycle was determined with the purpose of identifying how insulin affects the cell cycle (Fig. 3). The percentage of nuclei in G1 decreased during germination, compared to the nuclei in this phase observed in non-germinating seeds. This decrease was significantly higher in the presence of insulin at 24 and 36 h of germination and there was a concomitant and significant increase of the percentage of nuclei in G2. Insulin also induced an increase in the nuclei in the S phase after 18 h of germination (Fig. 3B). These results suggest that insulin stimulates the radicular cells arrested in the G1–S transition to continue the cell cycle.

Figure 3 Flow cytometry. The relative content of DNA in the nuclei from the radicular apices of maize seed without imbibition (0 h) or imbibed for 15, 18, 24 and 36 h, treated with or without 200 μU ml− 1 of insulin during the last 2 h of germination. (A) Representative histogram of the DNA content. The first peak indicates the 2C nuclei (G1) and the second peak the 4C nuclei (G2). The results were obtained with the MODFIT-L software using 30,000 events in each determination. (B) Changes in the percentage of nuclei in each phase of the cell cycle are shown. Values are the average of three independent experiments and are expressed as the percentage of nuclei in each phase ± SD. + Indicates significant differences among germination times and * indicates significant differences among treatments (α = 0.05).
Effect of insulin on the transcript levels of proteins that participate in the G1–S transition of the cell cycle
The results described in the previous sections show that the effect of insulin on growth of the embryonic axis was localized in the radicles. For that reason, it was relevant to study the effect of insulin on the gene expression of markers of the G1–S transition: E2F and PCNA, this last one dependent on E2F. For this purpose, total RNA was obtained from the radicular apices of embryonic axes imbibed for 15 and 24 h and incubated with insulin for the last 2 h. It was confirmed by qRT-PCR that the messages for E2F and PCNA are stored in the radicle of the dry quiescent seed. For PCNA, amplification was detected in the twenty-first cycle and for E2F it was detected in the twenty-second cycle. These results show that both transcripts are found in a similar proportion among the mRNA stored in quiescent seeds (data not shown). The presence of insulin increased the overall level of both mRNAs with respect to the control at 15 and 24 h upon seed germination (Fig. 4A). At 15 h, E2F increased 8.8-fold, while the increase in PCNA was 13-fold, indicating that the effect of insulin was probably at the transcriptional and/or stability levels. At 24 h, the increase in the overall mRNA level in response to insulin was 2.6-fold for E2F and 7.2-fold for PCNA. It is interesting to mention that the effect of insulin was greater for PCNA than for E2F at 15 and 24 h. To corroborate that the stimulation of PCNA mRNA levels by insulin results in an increase of this protein, de novo synthesis of PCNA protein was measured using a maize PCNA antibody. Insulin induced a stimulation of approximately 50% in de novo PCNA synthesis in germinating maize radicles at 15 h (Fig. 4B).

Figure 4 Effect of insulin on E2F and PCNA in maize radicles. (A) E2F and PCNA mRNA relative expression determined by qRT-PCR. Values are the average of three independent experiments and are expressed as the number of times the expression increased in with regard to the control, which was considered as 1. (B) PCNA de novo synthesis in radicles at 15 h of germination. Values are the average of three independent experiments and are expressed as percentage of incorporation in cpm mg− 1 of protein ± SD.
These results show that insulin regulates E2F and PCNA gene expression at the transcriptional level since 15 h of germination resulted in an increase of PCNA protein levels (Fig. 4B), thus accelerating the G1–S transition as observed by flow cytometry after 18 h of germination.
Discussion
Dehydrated maize seeds are in a quiescent state and their cells are arrested in the G1–S transition (Vázquez-Ramos and Sánchez, Reference Vázquez-Ramos and Sánchez2003). Seed imbibition reactivates metabolism and initiates germination, which ends in radicle protrusion. Cell-cycle reactivation is among the most relevant events that occur during germination (Barroco et al., Reference Barroco, Van Poucke, Bergervoet, De Veylder, Groot, Inzé and Engler2005). This process is required for plant growth and development. Likewise, cell growth and division are stimulated by phytoregulators such as auxins and benzyladenine (Herrera et al., Reference Herrera, Sánchez-de la Paz, Molinab, Plasencia and Vázquez-Ramos2000). Recently, an insulin-like growth factor named ZmIGF was reported in maize seeds. This peptide exhibits a well-defined α-helical structure by circular dichroism analysis, similar to that reported for insulin or for insulin-like growth factor I (IGF-I) (Rodríguez-López et al., Reference Rodríguez-López, Rodríguez-Romero, Aguilar and Sánchez de Jiménez2011). Previous studies reported that insulin, at the concentration used in the present study, had similar effects as the endogenous factor ZmIGF, by accelerating germination and stimulating growth of maize embryonic axes (García-Flores et al., Reference García-Flores, Aguilar, Reyes de la Cruz, Albores and Sánchez de Jiménez2001; Buentello-Volante et al., Reference Buentello-Volante, Díaz de León-Sánchez, Rivera-Cabrera, Aguilar Caballero, Ponce-Valadez, Sánchez de Jiménez and Pérez-Flores2010; Sotelo et al., Reference Sotelo, Garrocho-Villegas, Aguilar, Calderón and Sánchez de Jiménez2010; Rodríguez-López et al., Reference Rodríguez-López, Rodríguez-Romero, Aguilar and Sánchez de Jiménez2011).
The treatment with a pulse of insulin used in this work was sufficient to induce growth of maize embryonic axes during germination, in a similar way to that previously reported for continuous treatments with this effector (García-Flores et al., Reference García-Flores, Aguilar, Reyes de la Cruz, Albores and Sánchez de Jiménez2001; Dinkova et al., Reference Dinkova, Reyes de la Cruz, García-Flores, Aguilar, Jiménez-Garcıa and Sánchez de Jiménez2007). Our results show that the radicle presents an earlier and higher response than the coleoptile (Fig. 1). In previous dose–response studies with the endogenous factor ZmIGF, radicles also showed a higher growth than coleoptiles at all the tested concentrations (Dinkova et al., Reference Dinkova, Reyes de la Cruz, García-Flores, Aguilar, Jiménez-Garcıa and Sánchez de Jiménez2007). It has been demonstrated that insulin and ZmIGF activate the TOR-S6K signalling pathway in maize embryonic axes (Dinkova et al., Reference Dinkova, Reyes de la Cruz, García-Flores, Aguilar, Jiménez-Garcıa and Sánchez de Jiménez2007; Garrocho-Villegas and Sánchez de Jiménez, Reference Garrocho-Villegas and Sánchez de Jiménez2012). In the present work, the activation of this pathway was determined by phosphorylation of S6K-Thr 389 and the preferential accumulation of the S6rp transcript in the polysomal fraction, observed only in radicles but not in coleoptiles. In this work, it is shown for the first time that the maize seed response to insulin is differential, depending on the region of the embryonic axis, with the radicle as the target tissue with a higher capacity of response to the effector (Figs 1 and ). Another possible explanation for these results is that insulin uptake and/or metabolism (degradation) are different for each region analysed. Future work is needed to clarify these possibilities. These results correlate with previous reports in which a large body of physiological experiments has pointed at the root tip as the site for perception and integration of many of the signals, including detection of gravity, water and nutrients, during seed germination (Santisree et al., Reference Santisree, Nongmaithem, Vasuki, Sreelakshmi, Ivanchenko and Sharma2011).
Plant growth is the result of an increase of cell size and/or cell proliferation. Cell size plays an indirect role in cell proliferation, since cells duplicate their size before dividing. The amount of proteins, which determines to a large extent the cell biomass, is influenced by the number of ribosomes (Baserga, Reference Baserga2007) and, as a consequence, ribosomal biogenesis is tightly coupled to growth (Urban et al., Reference Urban, Soulard, Huber, Lippman, Mukhopadhyay, Deloche, Wanke, Anrather, Ammerer, Riezman, Broach, De Virgilio, Hall and Loewith2007). Our results show that insulin induced DNA synthesis in a preferential manner (Fig. 2C), supporting previous observations that IGFs stimulated cell proliferation by regulating synthesis of DNA and regulatory proteins of the cell cycle, such as cyclin D1 and G1–S transition (Mairet-Coello et al., Reference Mairet-Coello, Tury and DiCicco-Bloom2009). Sotelo et al. (Reference Sotelo, Garrocho-Villegas, Aguilar, Calderón and Sánchez de Jiménez2010) also reported that insulin or ZmIGF induces cell division in maize callus. In this regard, the results of the present work corroborate previous reports about the role of insulin in the co-ordination of cell growth and division by the TOR-S6K pathway (Dinkova et al., Reference Dinkova, Reyes de la Cruz, García-Flores, Aguilar, Jiménez-Garcıa and Sánchez de Jiménez2007; Garrocho-Villegas and Sánchez de Jiménez, Reference Garrocho-Villegas and Sánchez de Jiménez2012) and, besides, they demonstrate that cells respond to the inducer effect in a selective manner, as shown when the effects were compared in radicles and coleoptiles (Figs 1 and 2).
Because of the previous observations, the effect of insulin on the cell-cycle phases in root meristems was analysed between 0 and 36 h of germination by flow cytometry. The results revealed that most of the cells were arrested in the G1–S transition before imbibition, confirming previous reports in maize (Sánchez et al., Reference Sánchez, Gurusinghe, Bradford and Vázquez-Ramos2005), Arabidopsis (Barroco et al., Reference Barroco, Van Poucke, Bergervoet, De Veylder, Groot, Inzé and Engler2005) and barley seed (Gendreau et al., Reference Gendreau, Romaniello, Barad, Leymarie, Benech-Arnold and Corbineau2008). In animals, it has been reported that this transition is the only part of the cell cycle that depends on growth factors (Blagosklonny and Pardee, Reference Blagosklonny and Pardee2002). The results of the present work showed that after 24 h of germination, insulin induced a decrease in the percentage of nuclei in G1 from 68.7 to 47.8% (approximately 30%) and an increase in the percentage of nuclei in G2 from 11.8 to 22.1% (approximately 87%) in the root meristems of germinating maize seeds, as compared to the control (Table 1, Fig. 3B). These results are consistent with previous studies showing that cell-cycle reactivation during germination of maize embryonic axes is associated with a decrease in the percentage of cells in G1 and an increase in the percentage of nuclei in G2 (Sánchez et al., Reference Sánchez, Gurusinghe, Bradford and Vázquez-Ramos2005). The induction of the G1-S transition by insulin observed in the present work was higher than the one reported for maize in response to the mitogenic synthetic hormone benzyladenine where a decrease in the percentage of nuclei in G1 from 68.4 to 58.3% (approximately 14.7%) and an increase in the percentage of the nuclei in G2 from 19.9% to 29.7% (approximately 67%) was observed compared to the control (Sánchez et al., Reference Sánchez, Gurusinghe, Bradford and Vázquez-Ramos2005). These results show that insulin acts as a mitogen in the meristem of maize embryonic axis radicles during germination. This is consistent with previous observations in maize cells in suspension cultures, in which both insulin and ZmIGF considerably increased the cell mitotic index (Sotelo et al., Reference Sotelo, Garrocho-Villegas, Aguilar, Calderón and Sánchez de Jiménez2010), and in rat cells in which IGF-I promotes G1/S cell-cycle progression (Mairet-Coello et al., Reference Mairet-Coello, Tury and DiCicco-Bloom2009).
The induction by insulin of the expression of genes that regulate the G1–S transition, such as E2F and PCNA, was confirmed by qRT-PCR in maize radicles. Insulin increased E2F and PCNA transcription both after 15 and 24 h of germination (Fig. 4A). As a result, after 15 h of germination insulin had an effect on the de novo protein synthesis of PCNA (Fig. 4B). Over the past years, the cell cycle in plants has been intensively investigated (Vázquez-Ramos and Sánchez, Reference Vázquez-Ramos and Sánchez2003; Barroco et al., Reference Barroco, Van Poucke, Bergervoet, De Veylder, Groot, Inzé and Engler2005). Genes that participate in restarting and progression of the cell cycle have been studied. E2F transcription factors are part of the RBR-E2F pathway which participates in apoptosis, cell differentiation and development, and plays a crucial role in the regulation of the expression of genes involved in the G1–S transition and in DNA replication (Lammens et al., Reference Lammens, Li, Leone and De Veylder2009), shortening replication and cell division times (Magyar et al., Reference Magyar, De Veylder, Atanassova, Bakó, Inzé and Bogre2005). E2F proteins promote the transcription of several genes, including PCNA (Sozzani et al., Reference Sozzani, Maggio, Giordo, Umana, Ascencio-Ibañez, Hanley-Bowdoin, Bergouioux, Cella and Albani2010). The PCNA gene has been correlated with DNA replication, and in recent years with DNA repair and chromatin remodelling (Strzalka and Ziemienowicz, 2011). The higher expression of PCNA compared with E2F (Fig. 4A), observed in this work, might be associated with the different functions in the cell suggested for this protein. In this regard, previous studies reported that in maize seeds imbibibed for 15 h, thymidine incorporation was associated with repair events (Baíza and Sánchez de Jiménez, Reference Baíza and Sánchez de Jiménez1989); it was also found that the mitogenic hormone benzyadenine induced PCNA protein accumulation (Herrera et al., Reference Herrera, Sánchez-de la Paz, Molinab, Plasencia and Vázquez-Ramos2000). In the present study, PCNA and E2F mRNAs were detected by qRT-PCR among the messengers stored in the quiescent seeds (data not shown), although in previous studies (Sánchez et al., Reference Sánchez, Gurusinghe, Bradford and Vázquez-Ramos2005) PCNA mRNA was not found among the transcripts stored in the seed, which might be due to the sensitivity of the technique used.
In conclusion, the results of the present work, using insulin as an effector, generate knowledge that can contribute to description of the possible mechanism by which endogenous ZmIGF induces growth in germinating maize embryonic axes. The insulin effect is tissue specific, with the radicle particularly sensitive to growth induction, DNA synthesis and cell proliferation. The results show that growth was due, at least in part, to reactivation of the cell cycle. The insulin effect on cell division is due to the acceleration of the G1–S transition, as in eukaryotic animal cells (Thomas, Reference Thomas2002; Mairet-Coello et al., Reference Mairet-Coello, Tury and DiCicco-Bloom2009). In the present study we demonstrated that in maize this occurs, at least partially, through the induction of the E2F transcription factor and the E2F-dependent PCNA gene expression, as well as de novo protein synthesis.
Acknowledgements
This work forms part of the PhD dissertation by A.X.A.A. The research was supported by Universidad Autónoma Metropolitana (UAM), Consejo Nacional de Ciencia y Tecnología (CONACyT; 61424), Programa de Mejoramiento del Profesorado (PROMEP; 34775) and partially by Dirección General Asuntos del Personal Académico PNP Programa Nacional de Posgrados (DGAPA-212910). We also thank CONACyT (18554) for their financial support to A.X.A.-A. for her PhD studies (185590, Programa Nacional de Posgrados (PNP) Experimental Biology Program). We thank Dr V. Garrocho-Villegas for her help in the establishment of methodologies; MSc R. Aguilar and B.E.A. Hernández-López for their technical assistance, and Dr Mónica Ponce for critical reading of this manuscript.







