INTRODUCTION
Much work has been done on the development of a vaccine against H. contortus and several candidate vaccine antigens have been identified, such as H11 (Munn et al. 1993), Haemonchus galactose-containing glycoprotein complex (H-gal-GP) (Smith et al. 1999) and a Thiol-Sepharose-binding fraction (TSBP, Knox et al. 1999). These antigens are found on the microvillar surface of the parasite gut and are not normally seen by the host's immune system, hence they are termed ‘hidden antigens’ (Newton and Munn, 1999). The TSBP fraction is enriched for cysteine proteinases which play a key role in the biology of parasitic nematodes (Knox et al. 1999). Cysteine proteases are involved in immuno-evasion, enzyme activation, virulence, tissue and cellular invasion as well as excystment, hatching and moulting (Sajid and McKerrow, 2002). In particular, cysteine proteinases are thought to play a key role in the digestion of the bloodmeal ingested by haematophagic parasites (Sajid et al. 2003; Dalton et al. 2003). Legumains or asparaginyl proteinases are a novel class of cysteine proteinase, and were originally identified in plants (Chen et al. 1998) and were first identified in animals in the human blood fluke, Schistosoma mansoni (Dalton et al. 1995) which is a voracious blood feeder like H. contortus. They have since been found in mammals (Chen et al. 1997), the parasitic protozoan Trichomonas vaginalis (Leon-Felix et al. 2004), and in the parasitic nematodes Toxocara canis and Brugia malayi (Maizels et al. 2000). Studies in S. mansoni have suggested that legumain may play an indirect role in the digestion of haemoglobin, by activating other proteinases, predominantly cysteine proteinases, which digest the haemoglobin directly (Dalton and Brindley, 1996; Tort et al. 1999). Specific legumain activity has been demonstrated in S. mansoni extracts (Dalton et al. 1995) and activation of native S. mansoni cysteine proteinases by recombinant legumain has also been shown in this parasite (Sajid et al. 2003). This study describes the identification of a legumain homologue within the H. contortus expressed sequence tag (EST) dataset. The full-length coding sequence was compared to known legumain sequences and developmental expression analysed by RT-PCR. The legumain was expressed in an inactive form in Escherichia coli and antibody to the resultant recombinant protein used to define the distribution of the enzyme in Haemonchus extracts and its localization in sections of adult parasites. Native enzyme activity and inhibitor sensitivity were monitored in parasite extracts.
MATERIALS AND METHODS
Identification of the EST clone
An annotation search of the GenBank database identified a contiguous Haemonchus EST sequence (Accession number BF422994) which showed 38% amino acid sequence identity to S. mansoni legumain. This EST clone was kindly provided by the Edinburgh University/Sanger Centre Nematode EST project. Bluescript phagemids were excised from the λ ZAP-XR vector according to the manufacturer's instructions (Stratagene). Plasmid DNA was subsequently isolated using a Wizard SV Mini prep kit (Promega) and was sequenced on an ABI377 automated sequencer.
Cloning and expression in pGEX-6P-3
The full-length coding sequence was identified and cloned into the vector pGEX-6-P3 (Pharmacia Biotech) in frame with the Glutathione-S-Transferase (GST) fusion partner and transformed into competent Escherichia coli BL21 cells (Codon Plus, Stratagene). A colony containing the insert was grown as an overnight culture and induced to express by addition of β-D-thiogalactopyranoside (IPTG, 1 mM final concentration) by standard methods. Bacterial pellets were lysed, treated with DNase and RNase, then centrifuged to produce soluble and insoluble protein fractions, as previously described by Redmond et al. (1997). Fractions were analysed by SDS-PAGE. Aliquots of the fractions were mixed with reducing sample buffer containing β-mercaptoethanol and separated on 4–15% ready gels (BioRad) at 200 V for 45 min and proteins visualized by Coomassie Blue staining. Duplicate gels were also run and transferred to Immobilon P membrane (Millipore, UK) by wet blotting at 80 V for 60 min. Blots were then probed with anti-GST antibody (Pharmacia) to detect the legumain/GST fusion protein.
Phylogenetic tree
Amino acid sequence alignments were performed using Clustal W available at (http://ebi.ac.uk) and the phylogenetic tree constructed using MEGA version 3.1 (http://www.megasoftware.net) (Kumar et al. 2004).
Antiserum production
The remaining insoluble fraction was separated under reducing conditions on a large 10% SDS-PAGE at 90 V overnight with ‘Serva Blue G’ dye in the cathode buffer reservoir. The band containing recombinant legumain was readily identified and excised. The recombinant protein was then electroeluted from the gel slice using an AE-3590 Max Yield GP Electroeluter according to the manufacturer's instructions. A rabbit was inoculated subcutaneously with 50 μg of recombinant legumain in Quil A adjuvant, this being repeated 3 and 6 weeks later. Blood was collected and serum was harvested 1 week after the final inoculation.
Immunolocalization
Adult H. contortus paraffin sections were prepared as previously described (Redmond et al. 1997). The sections were de-waxed and rehydrated, then rinsed in 25 mM Tris, 137 mM NaCl, 2·7 mM KCl, pH 7·4, 0·05% (v/v) Tween 20 (TBST) and blocked in 5% (v/v) horse serum in TBST, overnight at+4 °C. Sections were then incubated in rabbit anti-legumain antiserum in TBST at a dilution of 1 in 200 for 3 h. After incubation with primary antiserum the sections were washed (3×10 min) in TBST and incubated in goat anti-rabbit IgG (whole molecule) FITC conjugate (Sigma) diluted 1 in 100 in TBST for 1 h. Sections were rinsed again in TBST 3×10 min and mounted in Citifluor (Citifluor Ltd). Sections were subsequently visualized under a fluorescence microscope (Olympus BX50).
Immunoblot analysis of parasite extracts
Water-soluble (S1), membrane-associated (S2), membrane-bound (S3) extracts, H-gal-GP and TSBP fractions of H. contortus were prepared as previously described (Smith et al. 1994). The respective extracts were separated by SDS-PAGE on 4–15% ready gels (BioRad) under reducing conditions and transferred onto Immobilon P membrane (Millipore). The blot was blocked in 5% (w/v) Marvel in TBST for 1 h, then washed in TBST (3×10 min). Blots were then incubated in the primary antiserum for 3 h at a dilution of 1 in 200 in TBST, then washed (TBST, 5×10 min) prior to incubation in secondary antibody and development with FastDAB (Sigma), as above.
Developmental expression
Reverse transcriptase (RT-) PCR was used to evaluate the developmental expression of H. contortus legumain mRNA. cDNA was prepared from life-cycle stages using the mRNA Isolation Kit (Stratagene) and subsequently the Superscript First Strand Synthesis System for RT-PCR kit (Invitrogen). Fifty μg of each life-cycle stage cDNA was used as template in PCR under the following conditions. Initial denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 1 min, 55 °C for 2 min and 72 °C for 3 min, followed by a final extension of 72 °C for 7 min. A positive control reaction was also included using 50 μg of plasmid DNA prepared from the previously described legumain clones.
Legumain activity and inhibitor sensitivity assays
Legumain activity was assayed by cleavage of the fluorescent substrate benzyloxycarbonyl-alanine-alanine-asparagine-aminomethyl coumarin (Z-Ala-Ala-Asn-AMC; Sigma, 2 μM final concentration). Assays were performed in the presence of dithiothreitol (DTT), (50 mM) in a final volume of 100 μl at room temperature using a LS5OB Luminescence spectrometer (Perkin Elmer Instruments). The amount of fluorescent AMC released over a 30-min period was measured with excitation and emission wavelengths of 340 nm and 450 nm, respectively. Legumain activity was assayed in the S1, S2, S3 and TSBP fractions of H. contortus.
The pH optimum of legumain activity was determined using S3 extracts of H. contortus. Activity was measured as described above, using buffers in the pH range 3–10 as detailed previously (Knox et al. 1993).
The inhibitor profile of legumain was determined at pH 7·0 using S3 extracts. The inhibitors tested and the final reaction concentrations were the cysteine protease inhibitors E64 (1 μM), Z-Phe-Ala-CHN2 (10 μM) and leupeptin (1 mM), the alkylating agent, N-ethylmaleimide (2 mM), the serine protease inhibitor, 4-(2-aminoethyl) benzenesulphonyl fluoride AEBSF (5 mM), the aspartyl protease inbibitor, pepstatin A (100 μM) and the metalloprotease inhibitor 1,10 phenanthroline (5 mM). Activity was measured as above.
Legumain activity – antibody inhibition studies
IgG from sheep vaccinated with TSBP, cystatin-binding proteins, recombinant HMCP 1, 4 and 6 or control sera (Redmond and Knox, 2004) were used to determine the effect of antibodies on legumain activity in S3 extracts. IgG was purified using protein G-agarose (Todorova et al. 1995). S3 (10 μl) was incubated at room temperature for 5 min with 10 μl of the purified IgG and then the other assay components, described above, were added. The amount of fluorescence released was measured continuously over 30 min using the time drive function on the LS50B Luminescence Spectrometer.
RESULTS
Sequence analysis
The EST DNA sequence from the databases (HCC00265, NEMBASE@nematodes.org.uk) appeared to be full length by alignment and further sequencing confirmed this. The full coding sequence was 471 amino acids with a predicted molecular weight of 49 kDa. There is a predicted signal peptide cleavage site between residues 19 and 20 and an additional site between residues 29 and 30 (↓ in Fig. 1A). Moreover, comparisons with the S. mansoni sequence indicated that the C-terminus extension is cleaved between 291 and 292 (↓, Fig. 1A). The Haemonchus protein has 3 predicted N-linked glycosylation sites (underlined in Fig. 1A). Alignment (Altschul et al. 1990) of the amino acid sequence (Fig. 1B) showed the similarities of the Haemonchus sequence to other legumains. The H. contortus legumain was found to show 40%, 38% and 34% identity to legumains of rat and human, S. mansoni and jackbean origin respectively. Also the residues associated with catalysis (Chen et al. 1998) were conserved through these 5 highly divergent species including a histidine, glycine and cysteine, essential for catalysis (bold, marked with #, Fig. 1B). In addition, consistent with other proteinases of this class, 2 blocks of 4 predominantly hydrophobic residues (bold, marked with * Fig. 1B), one N-terminal to the active-site HG residues, the other N- terminal to the active-site cysteine, are also conserved.

Fig. 1. (A) The predicted amino acid sequence of Haemonchus contortus legumain showing the predicted cleavage sites (↓) utilized to yield the mature enzyme as well as the active site histidine, glycine and cysteine (in bold, *), the 2 short hydrophobic domains (in bold) and predicted N-linked glycosylation sites (underlined). (B) Amino acid sequence Clustal W alignment of legumain homologues from human (Q99538), rat (Q9R0J8), H. contortus (AM177177), S. mansoni (P09841), and jackbean (P49046).
Phylogenetic tree
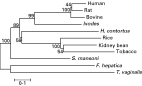
Phylogenetic analysis of the entire coding sequences (Fig. 2) failed to show any clear relationship between the Haemonchus sequence and those from the blood-feeding trematodes S. mansoni and F. hepatica. The sequences of mammalian and plant origin formed discrete groups with good bootstrap confidence and Haemonchus legumain was more closely related to plant legumains although bootstrap confidence was limited. The relationships were unaltered when the analysis was applied to the mature enzyme coding sequences, which include the more conserved active-site domains.

Fig. 2. Phylogenetic tree of the evolutionary relationship of Haemonchus contortus legumain with others from the animal and plant kingdoms. The protein sequences were obtained from GenBank using the Accession numbers given below and alignments conducted using Clustal W. Phylogenetic analysis was performed using Mega (http://www.megasoftware.net) to produce a neighbour-joining bootstrapped tree. The scale bar shows the number of nucleotide substitutions per site and the numbers on the tree are the bootstrap values. Sequences compared and their Accession numbers were legumains (also known as asparaginyl endopeptidases) from human (Q99538), rat (Q9R0J8), bovine (Q95M12), Ixodes ricinus (Q6PRC7), Rice (Q8GS39), kidney bean (O24325), tobacco (Q707T9), Schistosoma mansoni (Q9NFY9), Fasciola hepatica (Q71182), Trichomonas vaginalis (Q6EHZ6) and Haemonchus contortus (AM177177).
Cloning and expression of recombinant legumain
Recombinant legumain was expressed in pGEX-6P-3 as a fusion protein with GST. The fusion protein appeared as a band of approximately 75 kDa, which is the expected molecular mass for the fusion protein, (GST 26 kDA+49 kDa predicted molecular mass of legumain) and was recognized by anti-GST antibody (Pharmacia Biotech, Fig. 3A).

Fig. 3. (A) Western blot of expression samples probed with anti-GST antibody. Lanes 1-molecular weight markers, 2-uninduced pellet, 3-uninduced supernatant, 4-induced pellet, 5-induced supernatant. Band of expressed fusion protein indicated by arrow. (B) Western blot of S1, S2, S3, H-galGP and TSBP fractions probed with legumain antiserum. Lanes 1 – molecular weight markers, 2 – S1, 3 – S2, 4 – S3, 5 – H-gal-GP, 6 – TSBP. Legumain immunoreactivity indicated by arrow.
Immunoblot and immunolocalization
Antiserum to the recombinantly derived legumain recognized faintly peptides in S3, H-gal-GP and TSBP fractions of adult parasites. A 36 kDa band (predicted Mr of the mature enzyme) was recognized faintly in S3 with slightly larger bands being evident in the H-gal-GP and TSBP fractions (Fig. 3B). The antiserum did not recognize Haemonchus-derived GST (Mr 26 kDa, Sharp et al. 1991).
The antiserum was used to probe transverse sections of the adult parasite and immunofluorescence was observed in the microvillar layer lining the gut lumen with no immunofluorescence observed using control sera (Fig. 4).

Fig. 4. Paraffin wax sections of adult Haemonchus contortus probed with rabbit anti-legumain antiserum (A) and control serum (B). Immunofluorescence (indicated by arrows, A) was restricted to the microvillar surface of the intestine (A) and was absent in controls (B).
Developmental regulation of expression
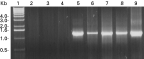
RT-PCR, using cDNA prepared from various life-cycle stages of H. contortus, showed that legumain was transcribed in L4, 11-day, 22-day and 28-day worms but not in eggs or L3 (Fig. 5).

Fig. 5. Developmental expression of Haemonchus contortus legumain. Agarose gel electrophoresis of RT-PCR products amplified from sscDNA of respective life-cycle stages. PCR was carried out from eggs, L3, exsheathed L3, L4 11-, 22- and 28-day H. contortus and positive control and the results shown in lanes 2 to 9, respectively. Lane 1 molecular weight markers.
Legumain activity and inhibition assays
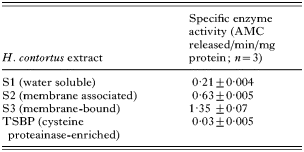
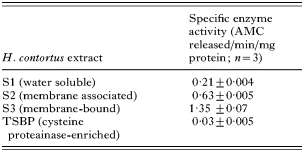
Legumain activity was monitored in different extracts of H. contortus with optimal activity in each extract being detected at pH 7·0 (not shown). In general terms, activity was associated with the membrane-bound S3 fraction being 6-fold higher than that observed in the water-soluble S1 fraction. No significant legumain activity was detectable in the S3 proteins which bound tp Thiol-Sepharose (TSBP, Table 1). The inhibition profile of legumain activity in S3 extract of H. contortus at pH 7·0 was compared to that of S. mansoni (Dalton et al. 1995). In general terms, both enzymes showed similar inhibitor sensitivities with H. contortus activity being only partially inhibited by E64, Z-Phe-Ala-CHN2 and leupeptin (27·1%, 40·8% and 47·3% respectively), but completely inhibited by N-ethylmaleimide. AEBSF inhibited 45·8% of legumain activity, pepstatin A inhibited 8·4% and 1,10 phenanthroline inhibited 27·5% of legumain activity. Activity was relatively unaffected by DTT (not shown).
Legumain activity – antibody inhibition study
Post-vaccination IgG from TSBP-vaccinated sheep had more inhibitory effect on native legumain activity than post-vaccination IgG from sheep vaccinated with an unrelated group of proteins prior to challenge infection (data not shown). This was not, however, statistically significant according to analysis with a general linear model (F3,1=0·27, P=0·612).
DISCUSSION
Cysteine proteases are thought to be involved in various key roles of parasite biology including digestion of the bloodmeal (Williamson et al. 2003, 2004). Recent work has shown that a cathepsin B from S. mansoni with haemoglobinase activity is activated by cleavage of the pre-pro-region by an asparaginyl proteinase (legumain) to yield the mature protein (Sajid et al. 2003). Given the presumed importance of cathepsin B-like proteinases for bloodmeal digestion in Haemonchus (Baig et al. 2002), this study was initiated to establish whether or not H. contortus possessed a legumain-like enzyme and if so, to characterize this enzyme more fully. Indeed, the cathepsin B-like cysteine proteinases of H. contortus are themselves leading vaccine candidates, with a cocktail of 3 recombinant cysteine proteases (HMCP 1, 4 and 6) inducing a 38% reduction in worm burdens in a recent vaccination trial (Redmond et al. 2004). If an enzyme that was potentially capable of processing these molecules did exist, then it could be a viable vaccine candidate or drug target in its own right.
Asparaginyl endopeptidases are a novel family of cysteine proteinases that were initially characterized in leguminous plants, hence the name legumain (Dalton et al. 1995). The first legumain was identified and characterized from Canavali ensiformis, the jack bean, and since then they have been identified in many plants and also some mammals and the blood-fluke S. mansoni (Sajid et al. 2002). They are cysteine-class enodopeptidases assigned to the CD clan (Caffrey et al. 2000) but are not susceptible to E64 inhibition. They are present in mammals, helminth and protozoan parasites and in plants (see http://merops.sanger.ac.uk/pepcards/c13p004.htm and Brindley and Dalton, 2004). We readily identified an EST sequence from the Haemonchus dataset (NEMBASE, Parkinson et al. 2004) which showed homology to existing legumain sequences. Comparisons of the predicted full-length peptide sequence with other legumain sequences showed that H. contortus legumain was 40% identical to rat and human, 38% identical to the blood-fluke S. mansoni and 34% identical jackbean legumains. Also key histidine, glycine and cysteine residues required for catalysis, were found to be conserved through these 5 highly divergent species. In addition, this analysis indicated that, in common with the S. mansoni enzyme (Caffrey et al. 2000) and other legumains of animal and plant origin, the Haemonchus sequence encodes a putative N-terminal pro-domain and a C-terminal extension. The latter may inhibit the active-site until activity is required (Caffrey et al. 2000). The predicted molecular weights of the full-length coding sequence and mature enzyme for Haemonchus legumain are 49 kDa and 30 kDa respectively. The putative mature protein was detected in parasite extracts at ~36 kDa. The discepancy in molecular weights could be explained by glycosylation at one or more of the 3 predicted N-linked glycosylation sites. In addition, it is notable that protein in the S3 extract appears as a doublet, possibly indicative of differing glycoforms. Moreover, slight differences in the molecular weights of the immunoreactive peptides in S3, H-gal-GP and TSBP were noted, possibly indicating differing processed forms of the enzyme in these fractions. These observations are generally consistent with the enzyme being processed in a similar manner to the S. mansoni legumain.
Despite this similarity, phylogenetic analysis revealed that the legumains from the blood-feeding helminths were divergent from each other, possibly reflecting differing functions, substrate specificities or, perhaps, immunological pressures.
Antiserum to the recombinant fusion protein strongly recognized a band at ~75 kDa and further immunoblot analysis showed that the protein had a native molecular weight of ~36 kDa and is not water soluble, being undetectable in the S1 fraction. Notably, this analysis indicated that the protein was present in TSBP despite only trace levels of enzyme activity being detectable. Legumain activity assays confirmed that native asparaginyl protease activity is mostly present in the S3 fraction of H. contortus although this analysis did indicate that some activity was also present in the water-soluble fraction.
The antiserum used was raised to the legumain fused to S. japonicum GST raising the possibility that immunoreactivity could also be ascribed to cross-reaction between S. japonicum and H. contortus GSTs. However, the immunoblot analysis clearly showed that there is no recognition below 30 kDa, Haemonchus GST having a known molecular weight of 26 kDa (Sharp et al. 1991). These authors also commented on the apparent lack of immunological cross-reactivity between GSTs from different sources despite sequence similarities.
Immunohistochemical analysis showed that the legumain was expressed on the microvillar surface of the intestinal cells in adult worms. This coincides with the location of a family of cysteine proteinases thought to be involved in blood-feeding (Skuce et al. 1999). It is possible that the legumain is involved in the activation of these proteinases in a similar way to the activation of S. mansoni cathepsin B (Sajid et al. 2003). At least 3 Haemonchus cathepsin B sequences aligned from the databases contain an asparagine residue at the predicted junction of the pro-region with the mature enzyme (D. P. Knox, unpublished observations) and this could be a cleavage site for the Haemonchus legumain.
The developmental expression studies showed that H. contortus legumain is expressed in L4, immature and mature adult stages, all of which imbibe host blood. Expression is coincident with that of gut-derived cysteine proteinases (Skuce et al. 1999) further suggesting that the legumain could indeed activate these enzymes. Immunolocalization studies in S. mansoni have shown that legumain and several of the cysteine proteinases (cathepsins L, B and D) are also expressed in the gut (Dalton et al. 1996) and, as noted above, further studies have shown the activation of cysteine proteases by legumain in S. mansoni (Sajid et al. 2003).
The H. contortus legumain shares similar biochemical characteristics with that from S. mansoni. The pH optimum of the former is pH 7 and the latter is pH 6·8 (Dalton et al. 1995). The inhibitor profile of the two enzymes is also similar and follows the inhibition profile typical of the asparaginyl endopeptidase family (Dalton et al. 1995). Both are partially inhibited by E64, Z-Phe-Ala-CHN2 and leupeptin and almost completely inhibited by N-ethylmaleimide. AEBSF reduces activity by about half and pepstatin A inhibits approximately 10% of activity for both enzymes. 1,10 phenanthroline partially inhibits the H. contortus legumain.
Similarities in the pH optima and inhibitor profiles between the H. contortus and S. mansoni legumains further suggest that H. contortus legumain may play a similar role in the activation of cysteine proteases. If this is the case, then legumain would be an ideal candidate for a vaccine or be a viable drug target. By inactivating legumain, activation of the cysteine proteases might be blocked and digestion of the bloodmeal would be impaired with likely detrimental effects on the parasite. Indeed, antibodies from sheep that had been partially protected following vaccination with TSBP (Redmond et al. 2004) did partially inhibit native H. contortus legumain activity, although this was not statistically significant (data not shown).
In conclusion, this study has identified one of the first legumains to be found in a parasitic nematode. However, much work still remains to characterize H. contortus legumain more fully and to evaluate it as a candidate vaccine antigen or drug target. The first step would be to test whether recombinant legumain has any enzymatic activity and can indeed process H. contortus cysteine proteases as has been reported in S. mansoni (Caffrey et al. 2000).
We thank Claire Whitton of the Nematode EST project for kindly supplying the EST. The financial support of the Scottish Executive Environment and Rural Affairs Department (SEERAD) is gratefully acknowledged.