INTRODUCTION
Sponges (phylum Porifera) are the oldest and simplest metazoans still inhabiting aquatic environments (Brain et al., Reference Brain, Prave, Hoffmann, Fallick, Botha, Herd, Sturrock, Young, Condon and Allison2012). Most of them are marine specimens. They are unselective suspension feeders, collecting bacteria, viruses, and any other fine suspended planktons (protists and microalgae) as well as any detrital organic particles fewer than 50 µm in diameter (Hadas et al., Reference Hadas, Marie, Shpigel and Ilan2006). However, bacteria represent the major food source for sponges. Besides this nutritional aspect, a large number of bacteria populate transiently or permanently the mesohyle of almost if it is not all sponges defining these communities of organisms as holobionts (Zilber-Rosenberg & Rosenberg, Reference Zilber-Rosenberg and Rosenberg2011). Sponge symbiotic bacteria sensu de Bary (Reference Bary1879) can represent up to 40–50% of the consortium biomass (Fieseler et al., Reference Fieseler, Horn, Wagner and Hentschel2004; Schmitt et al., Reference Schmitt, Hentschel and Taylor2012) consisting of hundreds to thousands of taxa (Webster et al., Reference Webster, Taylor, Behnam, Lücker, Rattei, Whalan, Horn and Wagner2010; Bruck et al., Reference Bruck, Reed and McCarthy2012; Jackson et al., Reference Jackson, Kennedy, Morrissey, O'Gara and Dobson2012; Schmitt et al., Reference Schmitt, Hentschel and Taylor2012). About 47 phyla of bacteria inhabit the sponges (Reveillaud et al., Reference Reveillaud, Maignien, Eren, Huber, Apprill, Sogin and Vanreusel2014). The vast majority of these bacteria remain uncultivable (Taylor et al., Reference Taylor, Radax, Steger and Wagner2007). Generally, symbionts give to their sponge hosts potentialities to exploit diverse metabolic repertoires such as photosynthetic carbon fixation, nitrification and minerals regulation. For example, ammonia is an end product of the host metabolism. It is likely used by the bacteria as a nitrogen source, while carbohydrates and amino acids should be available as the result of host extensive phagocytosis of bacteria and suspended organic material (Hentschel et al., Reference Hentschel, Usher and Taylor2006; Thacker & Freeman, Reference Thacker and Freeman2012). Another example cited by Müller et al. (Reference Müller, Grebenjuk, Thakur, Thakur, Batel, Krasko, Müller and Breter2004) highlights the effect of the oxygen availability on a symbiotic bacterium, SB2 (Alphaproteobacterium MBIC3368) present in the demosponge Suberites domuncula. The authors established a relationship between the oxygen supply, the enzyme's gene expression, and the bacterium growth. Symbionts also provide secondary metabolites allowing sponges to live in nutrient poor and harsh environments (Wang et al., Reference Wang, Brandt, Thakur, Wiens, Batel, Schroeder and Muller2013). However, in a wider view, the mechanisms which allow relationships to establish, to be maintained and to be coordinated between bacteria-bacteria, bacteria-sponges within this holobiont still remain poorly documented (Gardères et al., Reference Gardères, Taupin, Bin Saidin, Dufour and Le Pennec2012, Reference Gardères, Henry, Bernay, Ritter, Zatylny-Gaudin, Wiens, Müller and Le Pennec2014, Reference Gardères, Bedoux, Koutsouvelli, Créquer, Desriac and Le Pennec2015). On one hand, the complexity of the host metabolism and immune system and, on the other hand, the multitude of bacteria and other microorganisms co-existing in these consortia are probably complicating factors in the understanding of the strengths which govern this established concept (Thacker & Freeman, Reference Thacker and Freeman2012). Nevertheless, by comparing with well-studied bacteria-host interactions such as bacteria-plants and bacteria-animals, whether interactions are symbiotic or pathogenic, N-Acyl Homoserine lactones (AHLs) and other small molecules are among the ones reported as participating in those interactions (Brelles-Marino & Bedmar, Reference Brelles-Marino and Bedmar2001; Lyon & Novick, Reference Lyon and Novick2004; Cui & Harling, Reference Cui and Harling2005; Ryan & Dow, Reference Ryan and Dow2008; Parker & Sperandio, Reference Parker and Sperandio2009; Dobson et al., Reference Dobson, Cotter, Ross and Hill2012; Duncan, Reference Duncan2012; Gardères et al., Reference Gardères, Taupin, Bin Saidin, Dufour and Le Pennec2012, Reference Gardères, Henry, Bernay, Ritter, Zatylny-Gaudin, Wiens, Müller and Le Pennec2014; Hartman & Schikora, Reference Hartman and Schikora2012). Concerning marine invertebrates, the symbiotic association between Vibrio fischeri and the Hawaiian squid Eupryma scolopes provides a model to study the communications between the prokaryote and the eukaryote governed at least partially by AHLs (Fuqua et al., Reference Fuqua, Winans and Greenberg1994). Interactions occurring during the establishment of the symbiosis between these two organisms influence the development of both partners (Geszvain & Visick, Reference Geszvain and Visick2005). Scarce data have been published on other marine bacteria except two studies which demonstrated that bacteria isolated from Porifera were able in vitro to produce uncharacterized AHLs (Taylor et al., Reference Taylor, Schupp, Baillie, Charlton, de Nys, Kjelleberg and Steinberg2004; Mohamed et al., Reference Mohamed, Cicirelli, Kan, Chen, Fuqua and Hill2008), and one proving the existence of three AHLs released within the sponge Suberites domuncula (Gardères et al., Reference Gardères, Taupin, Bin Saidin, Dufour and Le Pennec2012). The objectives of this study were to isolate and identify cultivable bacteria from the demosponge S. domuncula and to demonstrate their ability to produce in vitro AHLs using the reporter strain Escherichia coli pSB406. One hundred and two bacteria were studied. Their kinetic of ALHs productions were compared with the one of the reference strain Pseudomonas aeruginosa PAO1 (pDA224).
MATERIALS AND METHODS
Samples collection
Specimens of S. domuncula (Olivi, 1792) were collected at 0–10 m depth by scuba diving. Three collections were made between December 2008 and February 2009, along the Brittany coasts, nearby Roscoff (48°42′48.17″N 3°54′04.28″S), France.
Bacterial isolation media
Five agar media (15 g L−1) were used to isolate sponge-associated bacteria: Marine Agar 2216 (MA) prepared following the recommendation of the manufacturer (Difco™, France); MA (0.01%) prepared in distilled water supplemented with sea salts (40 g L−1) (Sigma-Aldrich, France); MA (0.01%) complemented with sponge extract (SE) (10 µg mL−1); HEME medium (1 g L−1 glucose, 1 g L−1 NaH2PO4, 40 g L−1 marine salt); HEME containing SE (10 µg mL−1) (Figure 1).

Fig. 1. Flowchart presenting the methodology to perform the bacteria isolation from the sponge Suberites domuncula and the detection of the production of AHLs using the Escherichia coli pSB406 reporter strain. MA: marine agar; MASS: marine agar with sea salts; MASE: marine agar with sponge extract; HEME: Heme medium; HEMESE: Heme medium with sponge extract.
Sponge extract
The sponge extract (SE) was obtained by crushing sponge pieces (~ 0.5 cm3) in 5 mL of cold (+4°C) sterile marine phosphate buffer saline (mPBS) (NaCl 333.7 mm, KCl 2.7 mm, Na2HPO4 8.25 mm, KH2PO4 1.76 mm, pH 7.4) per gram of sponge tissue. Resulting suspension was centrifuged at 10,000 rpm, for 10 min, at 4°C. The supernatant was lyophilized, and finally suspended in ultrapure water prior to filtration (0.22 µm). The protein concentration was measured using the Quick Start Bradford Dye Reagent (Bio-Rad Laboratories, France) and the bovine serum albumin as a standard. Aliquots were frozen at −80°C prior to use. Sponge extract was supplied at 10 µg mL−1 of culture medium.
Isolation of cultivable sponge-associated bacteria
Three protocols were used to isolate bacteria associated with S. domuncula: a direct isolation from sponge biopsies; isolation after a treatment of sponge fragments with 50 µm of L-Dithiothreitol (DTT); and, isolation from primmorphs (Figure 1). Three sponges were used for each experiment. All experiments were carried out in an aseptic environment.
For the direct isolation method a sampling of the sponge surface (~ 3 cm2 × 0.3 cm depth) including the pinacoderm and a part of the mesohyle was made with a sterile scalpel. Each biopsy was individually reduced into smaller pieces (about 2 mm3) using a sterile scalpel. One of the 2 mm3 fragments was transferred into a sterile Eppendorf tube. Three consecutive washes of this piece with 1 mL of 0.22 µm filtered sterile natural seawater were made with gentle shaking for 1 min, at room temperature (RT°C, namely 20 ± 1°C), to remove any loosely attached bacteria. Natural seawater was collected at seashore at Ploemeur (Brittany, France) (N 47.71121 – W 63.47092 S), filtered (0.22 µm) and stored aseptically at 4°C, in the dark, prior to use. Each washing water was separately collected. After the fourth addition of sterile seawater, the sponge piece was crushed using a sterile pipette tip. The resulting suspension was incubated for 1 min, at RT°C, under gentle shaking. Finally, 200 µL of each rinsing water was collected at the end of each stage and 200 µL of the final suspension were seeded individually onto the previously five mentioned agar media.
The DTT treatment occurred during the protocol of sponge cells isolation, a necessary procedure to perform primmorphs in vitro culture. Primmorphs were prepared as described by Le Pennec et al. (Reference Le Pennec, Perovic, Ammar, Grebenjuk, Steffen, Brummer and Müller2003). Note that the addition of antibiotics in solutions of cell dissociation was omitted throughout the experiment to preserve bacteria. Briefly, sponge pinacoderm was cut aseptically into fine pieces in a sterile calcium and magnesium free seawater (CMFSW) containing 2.5 mm of EDTA in order to release cells from sponge tissue. Resulting pieces were then separated into two batches. One was used for the preparation of primmorphs as described in Le Pennec et al. (Reference Le Pennec, Perovic, Ammar, Grebenjuk, Steffen, Brummer and Müller2003); the other batch was incubated on a roller mixer SRT6D (Stuart, Bibby Scientific Ltd, UK), at 20 rpm, in CMFSW with EDTA and supplemented with 50 µm of DTT, for 30 min, at RT°C. After incubation, the cell suspension was centrifuged at 1900 rpm for 5 min, at RT°C, to pellet the cells and to remove any traces of EDTA. The supernatant and the pellet were both used for bacteria isolation on the different agar media described above.
Primmorphs were separated from the media by centrifugation at 1900 rpm, for 5 min, at RT°C, after 4 days of cultivation. The supernatant was directly seeded on the media described above. The resulting primmorph pellets were aseptically crushed using a sterile pipette tip into 1 mL of sterile natural seawater. The resulting supernatant obtained was also spread on bacterial culture media.
Petri dishes were incubated at 20°C and observed daily during 2 weeks. New bacteria colonies were picked up daily, annotated and streaked onto fresh appropriate media. Bacteria were selected according to their colony colour, morphology and consistency, growth time and pigment secreted in the medium. After two subcultures on their respective primary isolation media, all isolates were then successfully maintained on MA. Bacteria were then grown in liquid marine broth (MB, DIFCO™) for 24–48 h, culture time depending on each bacterium: cultures lasted 24 h for fast-growing bacteria, and 48 h for the slow-growing ones. Pure cultures were then stocked into glycerol and MB (v: v), at −80°C, prior to further experiments.
Detection of AHLs
The Escherichia coli pSB406 reporter strain containing luminescent luxCDABE-based plasmid sensor under the control of rhIR promoter was used to detect the production of AHLs (Winson et al., Reference Winson, Swift, Fish, Throup, Jorgensen, Chhabra, Bycroft, William and Steward1998). The strain contains a reporter sensitive to the C4-, C6-, C8-, C12- and C14-HSL (Kumari et al., Reference Kumari, Pasini, Deo, Flomenhoft, Shashidhar and Daubert2006) and does not produce any AHL itself (Lindsay & Ahmer, Reference Lindsay and Ahmer2005). The assay was adapted from Winson et al. (Reference Winson, Swift, Fish, Throup, Jorgensen, Chhabra, Bycroft, William and Steward1998). Briefly, the sensor strain was grown into LB medium at 37°C with an antibiotic selection consisting in 100 µg mL−1 of ampicillin. After 24 h of incubation, the E. coli working density solution was adjusted to 0.2 (OD at 600 nm) in the same LB medium. One hundred microlitres per well were dispersed into a 96-well microplate. Beforehand, sponge-associated bacteria were individually cultured in 10 mL of liquid MB, at 20°C, on a rocking platform (190 rpm). One millilitre of the cultures was harvested for each sponge-associated bacterium every 24 h during 96 h, centrifuged at 6000 rpm for 5 min, at 20°C. Then, 100 µL of the bacterial culture supernatants were added into the wells of the microplate containing 100 µL of the reporter strain. One hundred microlitres of the reporter strain solution with 100 µL of sterile MB were used as a negative control. Duplicates were prepared per bacterium as well as for the control. Plates were incubated on a rocking platform (120 rpm), at 37°C, for 24 h. The luminescence, induced by the presence of already mentioned AHLs at 24, 48, 72 and 96 h, was monitored using the Infinite M200 Pro multiplate reader (TECAN, France) (Figure 1). Mean and standard deviation for each duplicate and each condition were calculated. Results were expressed as the percentage of luminescence against the control.
Kinetic production of AHLs by Pseudomonas aeruginosa PAO1 (pDA224)
Pseudomonas aeruginosa PAO1 (pDA224) was used as a model for the production of AHLs which was compared with the production of AHLs of sponge-associated bacteria. The P. aeruginosa PAO1 (pDA224) luminescent reporter strain contains the luxCDABE-based plasmid sensor which responds to C4-HSL and 3-Oxo-C12-HSL and produces its own AHLs. The method to detect the production of AHLs was the same as described above i.e. 100 µL of P. aeruginosa suspension at OD600nm of 0.2 plus 100 µL of LB medium. The luminescence was recorded each hour during 96 h. The luminescence values were subtracted with the luminescence background (P. aeruginosa PAO1 (pAB133)) as described by Bazire et al. (Reference Bazire, Dheilly, Diab, Morin, Jebbar, Haras and Dufour2005). Experiments were done in triplicate; means and standard deviations were calculated.
DNA isolation
Ten microlitres of each sponge-associated bacterial culture were individually dispensed into a sterile Eppendorf tube and centrifuged at 12,000 rpm for 2 min, at 20°C (Figure 1). Supernatants were discarded and bacteria cell pellets were microwaved three times for 10 s, at 900 W, to disrupt cell membranes. The resulting material was suspended into 50 µL of PCR grade water. Total DNA was kept at −20°C prior to 16S rDNA amplification.
Polymerase Chain Reaction (PCR)
The universal bacterial primers used to amplified the 16S rDNA gene were the 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and the 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Teng et al., Reference Teng, Guan and Zhu2008). Reactions of amplification were performed according to the MasterAMP PCR kit (Epicentre Biotechnologies, USA), as follows (final volume 25 µL): 12.5 µL of MasterAMP premix D, 1.0 µL of each primer (25 µm), 0.1 µL of MasterAMP Taq polymerase, 1.0 µL of DNA template and 10.4 µL of PCR grade water. PCR reactions were completed in a thermal cycler MJ Mini 48-well Personal Thermal Cycler (Bio-Rad, France) under the following conditions: initial denaturation step at 95°C for 10 min, 45 cycles of 95°C for 30 s, 55°C for 45 s and 72°C for 90 s. A final extension step was programmed at 72°C for 4 min. Amplicons were subjected to a 1% agarose gel electrophoresis in Tris/Acetate/EDTA buffer prior to sequencing to control the presence of a single product at the expected size of sim 1100 kb.
Phylogenetic analysis
16S rDNA sequences were manually edited and compared with sequence databases using the Basic Local Alignment Search (BLAST), using the MEGABLAST algorithm option at www.ncbi.nlm.nih.gov/Blast.cgi (Figure 1). Chimeras were checked using Black Box Chimera Check (B2C2) software version 5.1.1 (Gontcharova et al., Reference Gontcharova, Youn, Wolcott, Hollister, Gentry and Dowd2010) with the following parameters: 0–2 were chimeric, 3–4 possible chimeric and 5–7 non-chimeric; other options were set as default. Resulting 16S rDNA amplified sequences were submitted to GenBank for identification.
RESULTS
Identification of AHLs producing bacteria
Among the 102 cultivable isolates, 67 induced the luminescence of the reporter strain E. coli pSB406; 13 did not produce any luminescence. The remaining 22 bacteria were contaminated and unfit for this experiment.
Kinetic models of AHLs production by sponge-associated bacteria
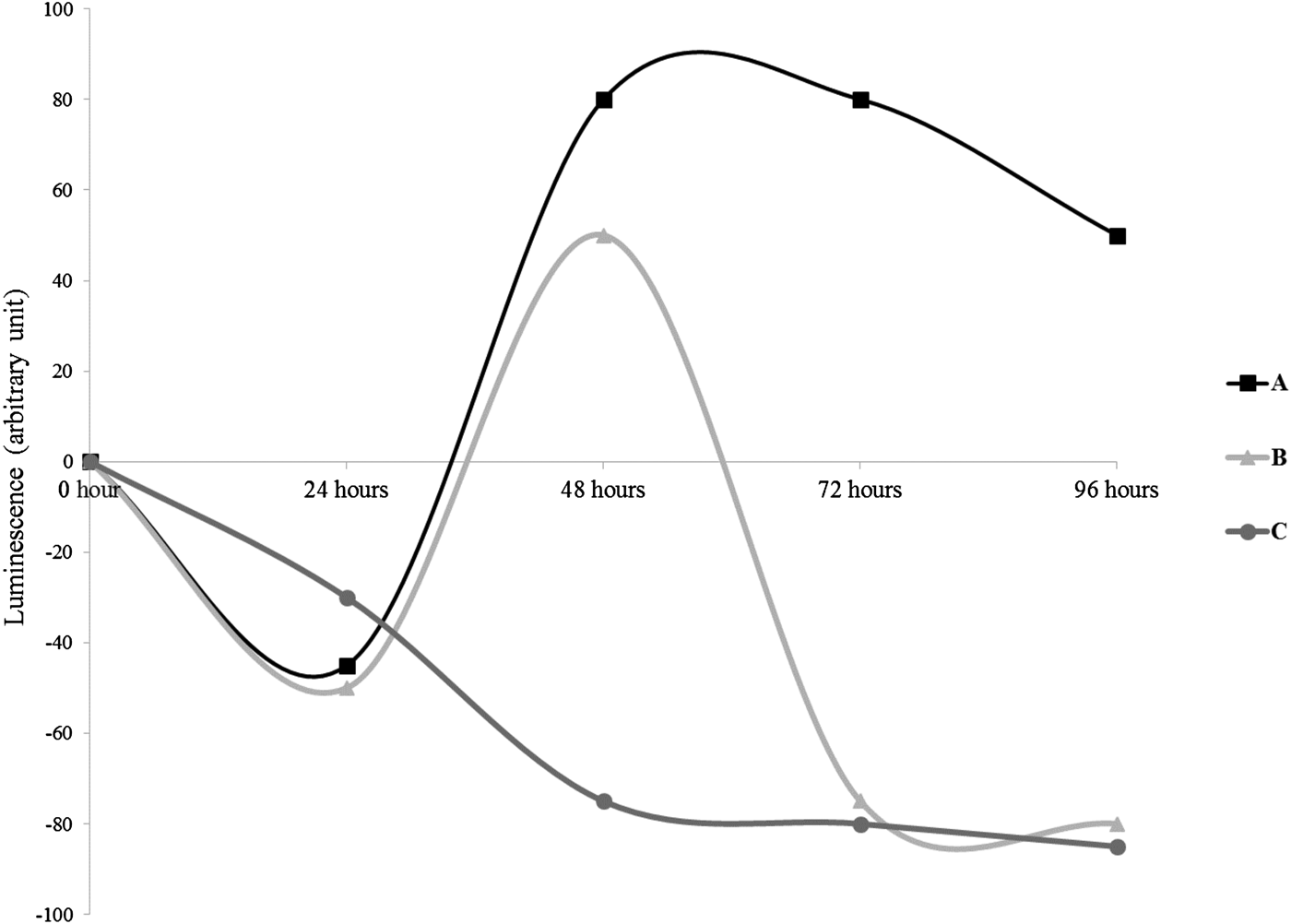
The models of production of AHLs by the cultivable sponge-associated bacteria are presented in Figure 2. The production of AHLs followed three curve profiles. In the first type (curve A), the luminescence decreased during the first 24 h, highly increased from 24 to 48 h and decreased slowly between 48 and 96 h. The curve B profile was as the curve A one except a drastic decrease of the luminescence registered between 48 and 72 h and a stabilization between 72 and 96 h. In both curves A and B, at the time of the culture, between 24 and 48 h, the AHLs production was undetectable. The luminescence decreased slowly in the curve C type, from the beginning of the experiment until 48 h prior to stabilization until 96 h.
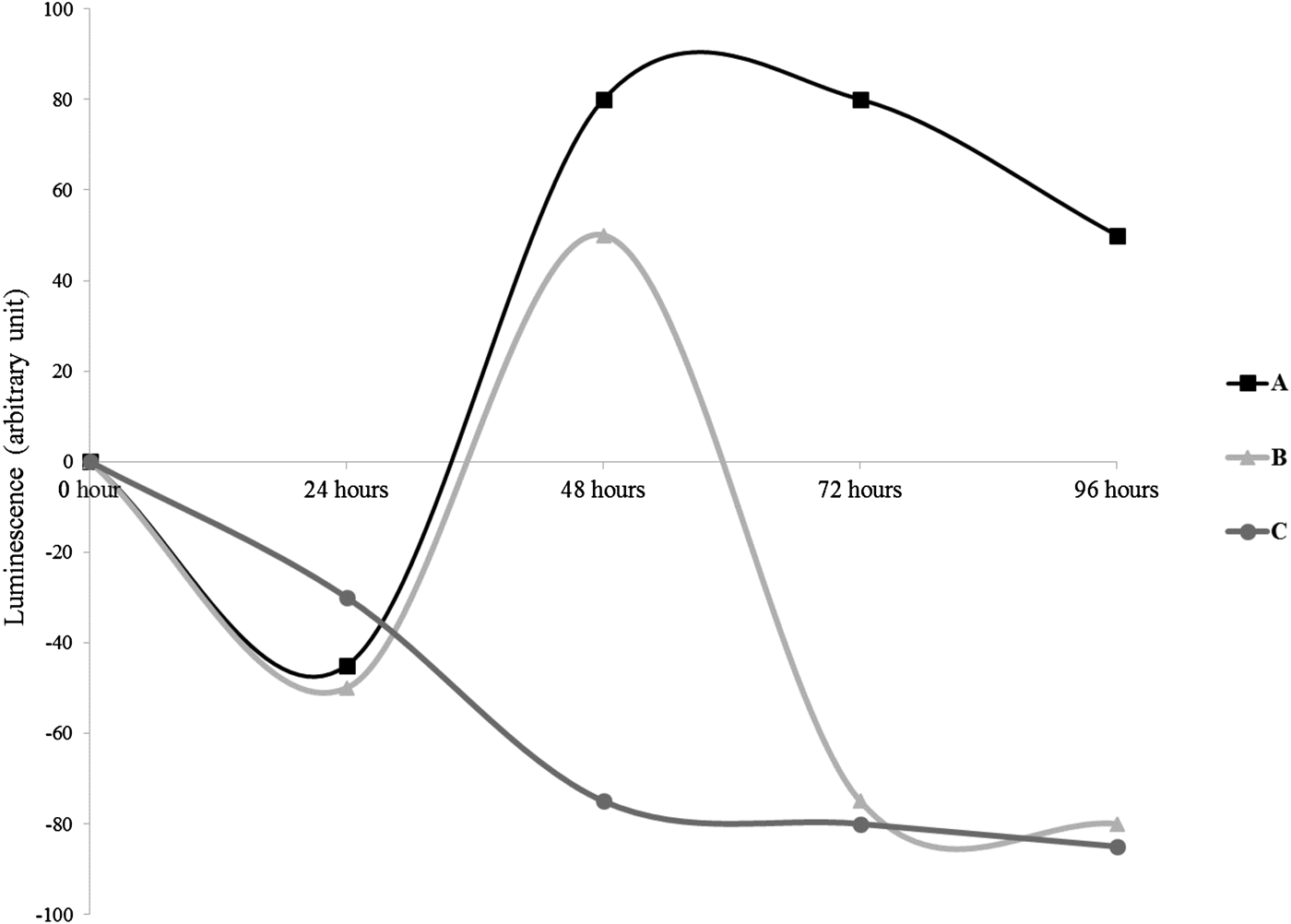
Fig. 2. Profiles of the production of luminescence of the reporter strain E. coli pSB406 in presence of AHLs in Suberites domuncula-associated bacteria supernatants. Three profiles were observed: A: decrease of luminescence during the first 24 h, followed by an induction from 48 to 96 h with a highest values between 48 and 72 h. B: decrease of luminescence during the first 24 h; after 24 h until 48 h an induction of the luminescence was observed prior to a new diminution continuing until 96 h. C: continuous inhibition of the luminescence until 48 h prior to stabilization between 72 and 96 h.
Twenty-one bacteria (31%) belonged to the profile of the curve A, 28 (35%) fitted to the curve B and 14 (18%) to the curve C (Figure 2).
Production of AHLs by P. aeruginosa
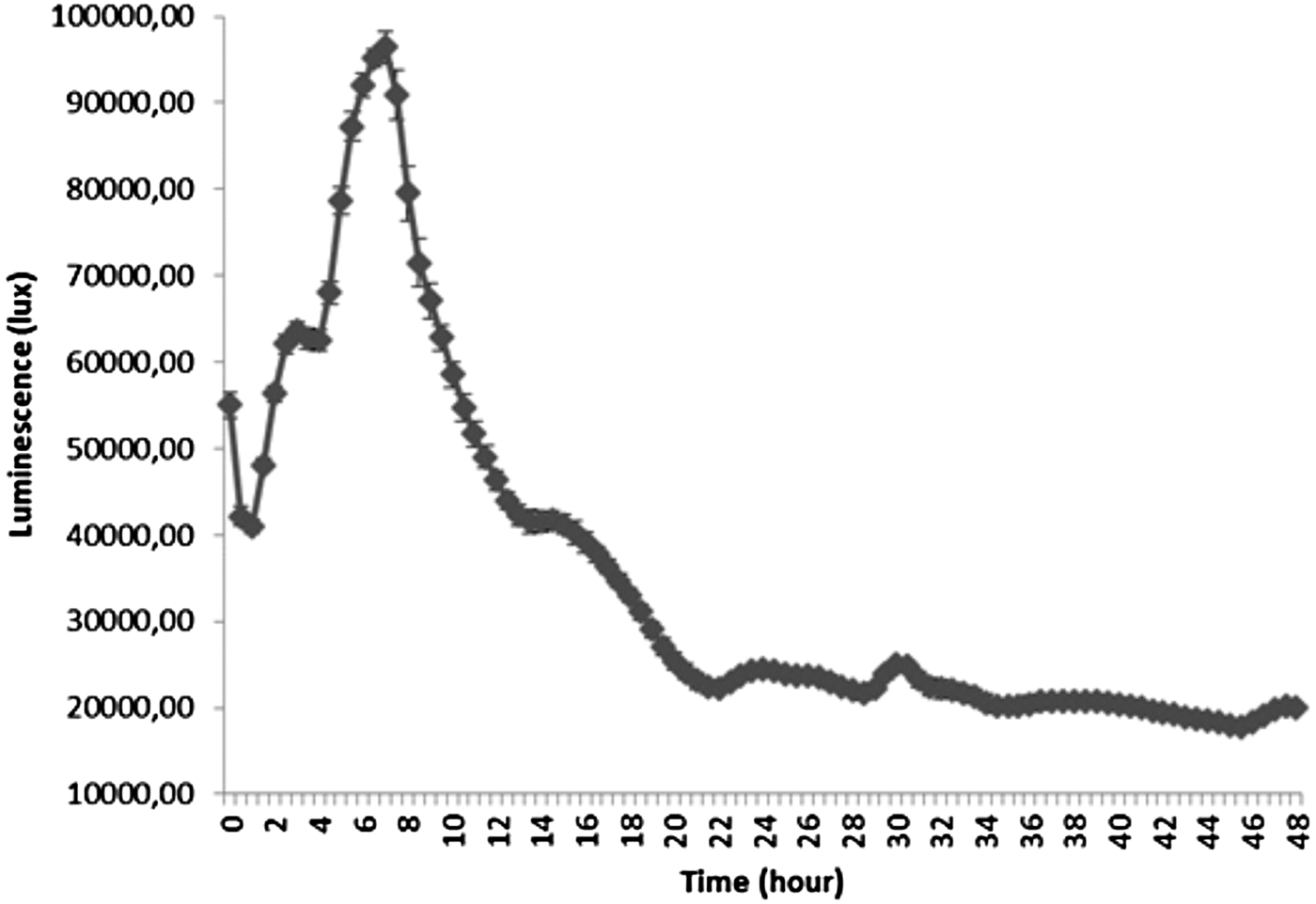
The profile of luminescence related to the production of AHLs by P. aeruginosa PAO1 (pDA224) is presented in Figure 3. The maximum of luminescence was obtained after 7.5 h of culture after which it decreased continuously until 21 h.
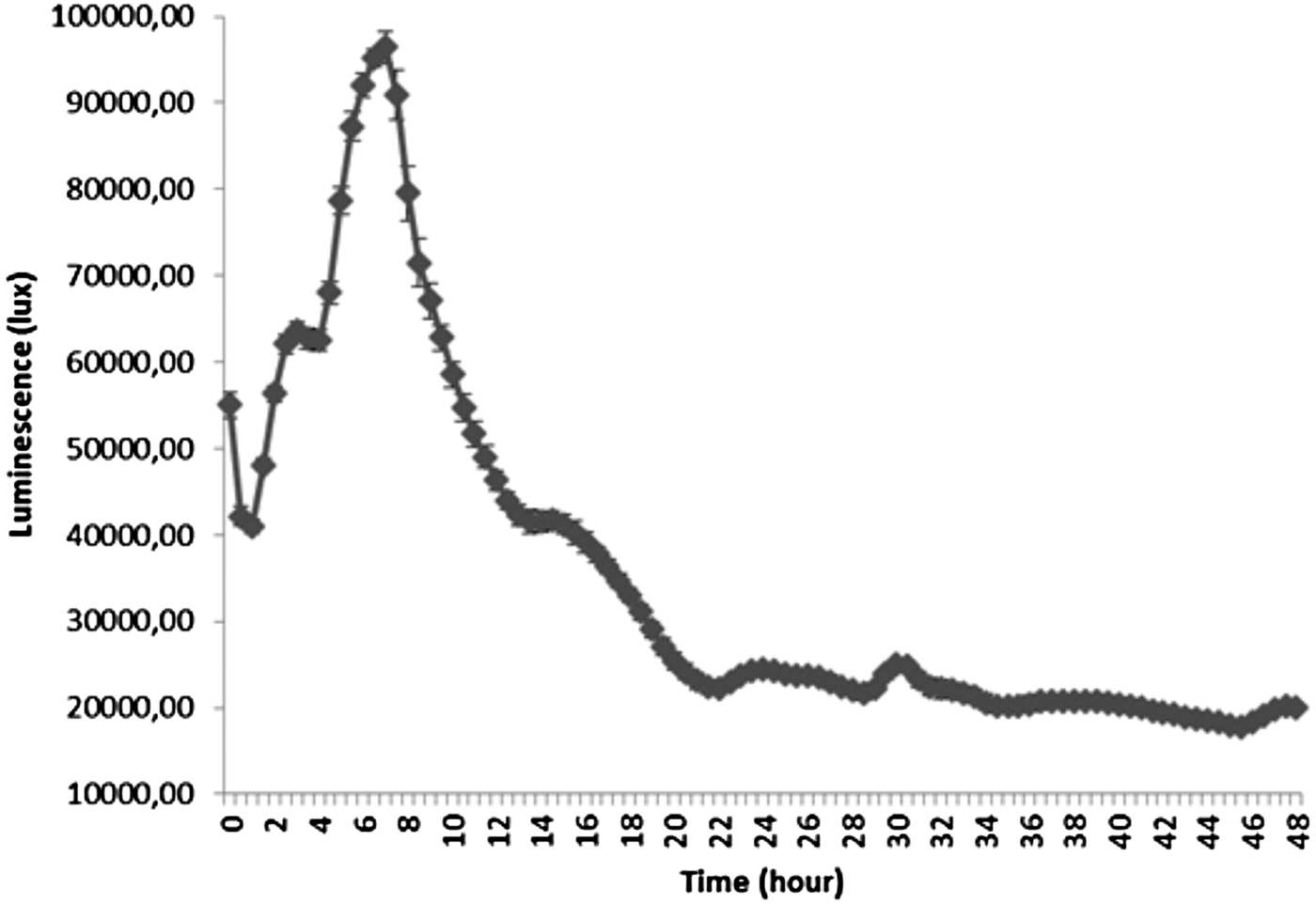
Fig. 3. Luminescence related to the production of AHLs by Pseudomonas aeruginosa PAO1 (pDA224) during 96 h of culture in LB medium.
16S rDNA identification
Ninety-three amplicons were successfully sequenced. The sequence similarity search using BLAST indicated that 63% (63 isolates) belong to the class of Gammaproteobacteria, 18% (18 isolates), Firmicutes, 12% (12 isolates) Alphaproteobacteria and 7% (7 isolates) Bacteroides (Figure 4). The Gammaproteobacteria belong to four orders: Alteromodales, Oceanospirillales, Pseudomonadales and Vibrionales and eight families (Pseudoalteromonadaceae, Cowelliaceae, Shewanellaceae, Alteromonadaceae, Hahellaceae, Halomonadaceae, Pseudomonadaceae and Vibrionaceae). Only one order was representative of the Alphaproteobacteria, Firmicutes and Bacteroidetes, respectively the order of Rhodobacterales (one family: Rhodobacteraceae), Bacilli (two family: Bacillaceae and Staphylococcaceae) and Flavobacteriales (one family: Flavobacteriaceae).

Fig. 4. Distribution (%) of the bacteria isolated from the sponge Suberites domuncula according to the Class, the family and the order.
Twelve cultivable bacteria were isolated from the primmorphs of S. domuncula after 4 days of culture (Figure 5). Five isolates belong to the genus Pseudovibrio, family of Rhodobacteraceae, four to the genus Tenacibaculum, family of Flavobacteriaceae, two to genus Endozoicomonas, family of Hahellaceae and one to Flavobacterium, family of Flavobacteriaceae. One family was not encountered in sponge biopsies, the Hahellaceae.
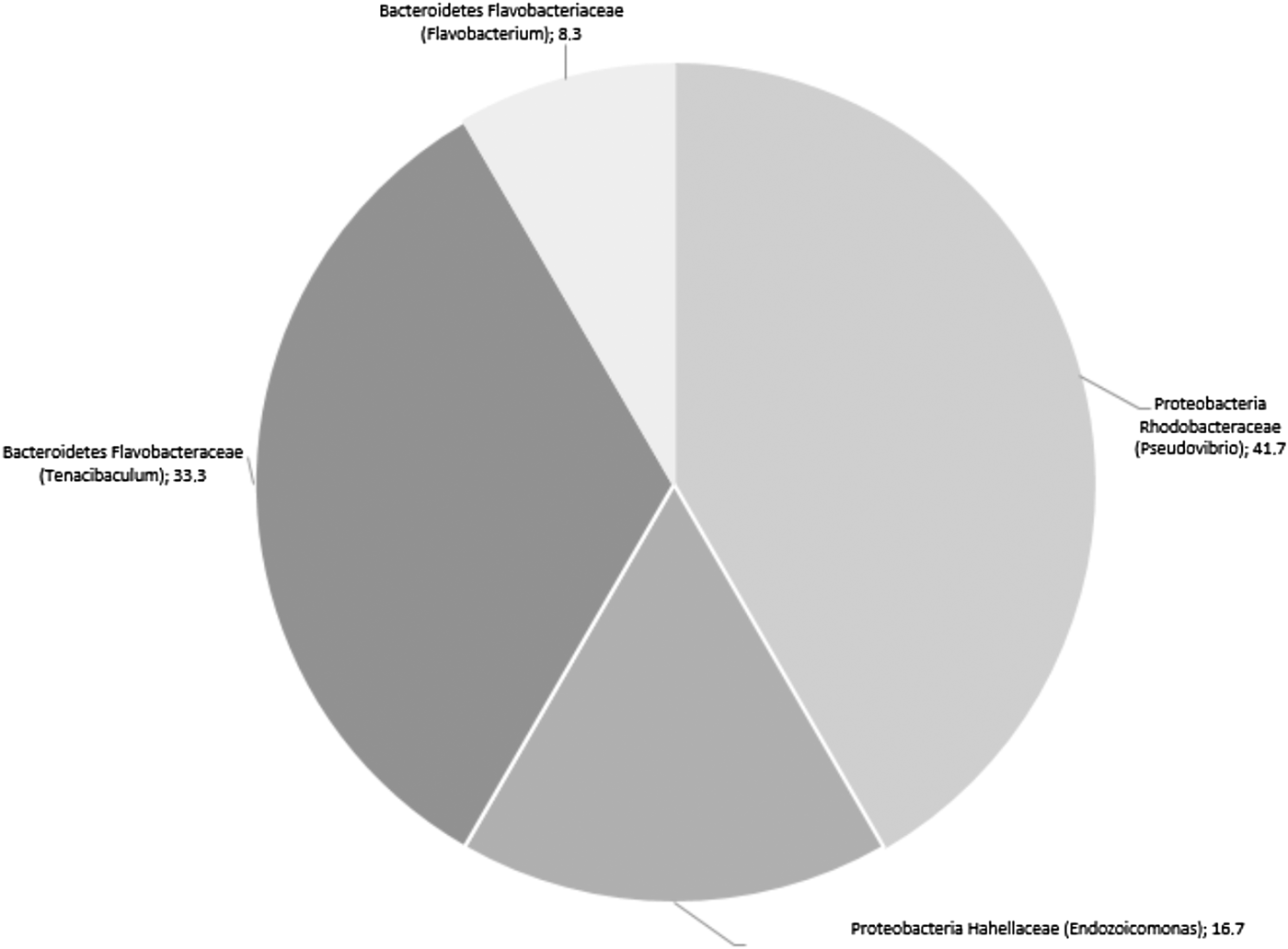
Fig. 5. Distribution (%) of the bacteria isolated from 4-day-old primmorphs prepared from the sponge Suberites domuncula according to the class, the family and the genus.
DISCUSSION
Production of AHLs by sponge-associated bacteria
Approximately two-thirds of the bacteria isolated from S. domuncula produced some AHLs. Among the ‘non AHLs-producers’ some may also release AHLs undetectable by the reporter strain E. coli pSB406, for example the 3-oxo-C12HSL the production of which was already shown in a previous study concerning the sponge holobiome (Gardères et al., Reference Gardères, Taupin, Bin Saidin, Dufour and Le Pennec2012). Others may secrete one or more compounds inhibiting the reporter strain which would mask the detection of AHLs. Bacteria belonging to the genus of Pseudomonas (Brelles-Marino & Bedmar, Reference Brelles-Marino and Bedmar2001), Pseudoalteromonas (Wang et al., Reference Wang, Ikawa, Okaue, Taniguchi, Osaka, Yoshimoto, Kishida, Arakawa and Enomoto2008; Guo et al., Reference Guo, Zheng, Zhou, Cui, Han, Tian and Wang2010), Pseudovibrio, Vibrio (Mohamed et al., Reference Mohamed, Cicirelli, Kan, Chen, Fuqua and Hill2008) and Tenacibacullum (Romero et al., Reference Romero, Avendano-Harrera, Magarinos, Camara and Otero2010) secrete AHLs but are also well known for their ability to produce antibacterial compounds (Dietrich et al., Reference Dietrich, Teal, Price-Whelan and Newman2008; Isnansetyo & Kamei, Reference Isnansetyo and Kamei2009; Dheilly et al., Reference Dheilly, Soum-Soutera, Klein, Bazire, Compere, Haras and Dufour2010; Guo et al., Reference Guo, Zheng, Zhou, Cui, Han, Tian and Wang2010; Klein et al., Reference Klein, Soum-Soutera, Guede, Bazire, Compère and Dufour2011). For example, the Firmicutes produce an auto-inducer of type 2, the AI-2 (Ryan & Dow, Reference Ryan and Dow2008). The AI-2 quorum system is shared in both Gram-negative and positive bacteria (Ryan & Dow, Reference Ryan and Dow2008). Therefore, it is possible to consider that the Firmicutes interfere with E. coli. As a result, the assay indicates that the Firmicutes do not produce any AHL. Furthermore, false negative results using AHL biosensor strains can occasionally occur because of the bacteriostatic effect of compounds produced by the bacteria under investigation (Steindler & Venturi, Reference Steindler and Venturi2007). Hence, for several reasons, it is not possible to conclude from the non-appearance of the luminescence that the bacterium tested does not produce any AHL. Further investigations must explore the possibility of using this assay jointly with the reporter strain E. coli pSB406 as a tool to screen AHLs and bacteriostatic or bactericidal production simultaneously. The use of new biosensors covering all the range of AHLs must also be envisaged.
In the present study, the diminution of the luminescence illustrated by the bacteria belonging to the curves A and B profiles during the first 24 h may be indicative of the release of at least one compound interfering with the production of E. coli luminescence or to a partial mortality of the reporter strain. It may be speculated that bacteria belonging to this group might have adopted a strategy to eliminate or to reduce the competitors before monopolizing the medium. The bacteria following the curve B profile displayed a new decrease of the luminescence at 72 h, possibly due to the presence of a newly synthesized antagonist of E. coli or interfering with the induction of luminescence. Bacteria might have sensed their environment and perceived the growth of a competitor strain, i.e. E. coli in this experiment. In answer to this phenomenon, the strains may produce an inhibitor compound. There is also a possibility that, at the same time, AHLs were produced, but the production cannot be observed due to the death of the reporter strain. Between 24 and 48 h, the inhibitor compound(s) production may be lowered or cancelled allowing an outbreak in the inhibition of the production of luminescence. Finally, the curve C profile displayed a continuous decrease of the luminescence until 48 h before stabilizing at its lowest level. Bacteria of this group might have produced at least one E. coli inhibitory compound in a progressive and increasing manner from 24 until 48 h, a production which lasted until 96 h.
By comparing the luminescence of P. aeruginosa PAO1 to the one of sponge-associated bacteria it can be concluded that sponge-associated bacteria relied on different strategies for their production of AHLs. Note must be taken of the possible role of the composition of the culture medium which was different between P. aeruginosa PAO1 and the marine strains and which may influenced the growth of bacteria and their metabolic production.
Bacterial identification of cultivable bacteria
In this study 102 cultivable bacteria were isolated from the demosponge S. domuncula according to three methods and five cultivation media and characterized at the genus level. None of the methods or media was preferable to isolate a specific order, family or genus of sponge-associated bacteria. The same conclusion was drawn with supplemented media with sponge extracts. Furthermore, after a few subcultures, the growth of all the bacteria was successfully made on MA. This seems to indicate that the sponge-associated bacteria isolated in the present study have a low degree of symbiosis with the sponge. The majority might be opportunistic bacteria, a few of them low commensal. The complementation of bacterial isolation media with sponge extract did not seem decisive in isolating commensal bacteria.
Gammaproteobacteria were the most plentiful class identified in this study, followed by the Firmicutes, Alphaproteobacteria and Bacteroidetes respectively. Flemer et al. (Reference Flemer, Kennedy, Margassery, Morrissey, O'Gara and Dobson2011) in a study concerning a close sponge species to S. domuncula, Suberites carnosus, showed that Gammaproteobacteria prevailed as also demonstrated in the present study. However, the authors indicated a lower representation, 36% instead of 63%. The other phyla present in S. carnosus were Alphaproteobacteria (34%), Actinobacteria (11%), Firmicutes (10%) and Bacteroidetes (7%). In the present study, cultivable Actinobacteria were not represented. The alphaproteobacteria were underrepresented compared with the study of Flemer et al. (Reference Flemer, Kennedy, Margassery, Morrissey, O'Gara and Dobson2011) while the Firmicutes were nearly twice as present. More generally, it can be concluded that the majority of cultivable bacteria isolated from marine sponges were either Gammaproteobacteria (Flemer et al., Reference Flemer, Kennedy, Margassery, Morrissey, O'Gara and Dobson2011; Jackson et al., Reference Jackson, Kennedy, Morrissey, O'Gara and Dobson2012; Steinert et al., Reference Steinert, Whitfield, Taylor, Thoms and Schupp2014) as evidence in this study or Alphaproteobacteria (Bruck et al., Reference Bruck, Reed and McCarthy2012). Actinobacteria, Firmicutes and Bacteroidetes are the following classes in order of abundance. This is in agreement with the data described in the literature concerning the presence of a common bacterioflora over time and localization in sponges (Schmitt et al., Reference Schmitt, Tsai, Bell, Fromont, Ilan, Lindquist, Perez, Rodrigo, Schupp, Vacelet, Webster, Hentschel and Taylor2013). The bacteria composition from this study is almost identical with bacteria that were found in seawater dominated by Gammaproteobacteria, followed by Alphaproteobacteria, Bacteroides and Firmicutes (Ji et al., Reference Ji, Zhao, Yin, Zhao, Liu, Xiao and Zhang2012).
The 4-day cultivation of primmorphs in the absence of antibiotics revealed the presence of bacteria in contact with eukaryote cells. This confirms the absolute necessity of the addition of antibiotics in primmorphs culture medium to avoid bacterial development (Le Pennec et al., Reference Le Pennec, Perovic, Ammar, Grebenjuk, Steffen, Brummer and Müller2003). The family Hahellaceae was only isolated from primmorphs while the others were common to the sponge and to the primmorphs. The process of cell isolation and primmorphs cultivation might have favoured the highlighting or the enrichment. The genus Endozoicomonas which belongs to this family is rarely found in marine invertebrates except for one sponge species, Arenosclera brasiliensis, to which the genus is plentiful (Rua et al., Reference Rua, Trindade-Silva, Appolinario, Venas, Garcia, Carvalho, Lima, Kruger, Pereira, Berlinck, Valle, Thompson and Thompson2014).
Production of AHLs
The profile of production of AHLs allowed division of the sponge-associated bacteria according to the three models of curves. However, no definitive pattern of distribution was observed according to the order, family or genus of bacteria. Furthermore, the comparison of the production of AHLs between sponge-cultivable bacteria and P. aeruginosa PAO1 (pDA224) showed that marine bacteria mainly reached the maximum of AHLs concentration at 48 or 72 h while P. aeruginosa PAO1 (pDA224) achieved it at 7.5 h. This was probably due to two factors: sponge-associated bacteria may first adapt their growth strategy to the culture medium i.e. in absence of their native sponge environment; the growth rates of those bacteria were slower compared with P. aeruginosa PAO1 (pDA224), then AHLs, produced at minute quantity, accumulated proportionally to population densities (Ravn et al., Reference Ravn, Christensen, Molin, Givskov and Gram2001; Zan et al., Reference Zan, Fuqua and Hill2011), that was slow for marine bacteria. It is also interesting to note that some bacteria divided according to the profiles of curves A and B did not release any investigated AHL in the culture medium during the first 24 h but arose later. Therefore, in further studies, in order to not underestimate the numbers of AHLs producers via the method described here, the bacterial culture supernatants aged more than 24 h should also be systematically tested.
Thirty-four per cent of the sponge-cultivable bacteria inhibited the production of luminescence of E. coli pSB406 throughout the culture time, probably because of a bacteriostatic or a bactericide effect. It may be relevant to examine culture supernatants of those specific bacteria in search of compounds producing these effects to value them in a protocol of antibacterial fight. Indeed, Muscholl-Siberhorn et al. (Reference Muscholl-Siberhorn, Thiel and Imhoff2007) described that among 112 bacteria isolated from S. domuncula collected from the Adriatic Sea 46% of them showed an antimicrobial activity.
The results from the present study showed that the majority of Gamma- and Alphaproteobacteria produced some AHLs. Nevertheless, the results validate or invalidate an ability and a real-time production of AHLs by these bacteria within their host, S. domuncula. In their native environment, there is a multitude of factors that may influence the AHLs production, such as bacteria-bacteria interactions (competition, cooperation …) and/or host-bacteria interactions (predation, immune system response …). As an example, eight out of 11 bacteria isolated from the primmorphs, which may be considered as the most related to S. domuncula in this study, produced some AHLs in vitro, but no AHL was directly detected in S. domuncula primmorph (Gardères et al., Reference Gardères, Taupin, Bin Saidin, Dufour and Le Pennec2012) even if the preparation of primmorphs and culture conditions used for both experiment were same.
Among the bacteria isolated from 4-day-old primmorphs, three belonging to the genus Pseudovibrio (JX436408, JX436413 and JX436419) and one Endozoicomonas (JX442224) did not produce any AHL but produced an E. coli inhibition compound. Thakur et al. (Reference Thakur, Thakur, Indap, Pandit, Datar and Müller2005) also isolated a bacterium from a S. domuncula primmorph which represented an unidentified novel species of Pseudomonas. Butanol extracts of this bacterium exhibited antiangiogenic, antimicrobial, haemolytic and cytotoxic properties. The bacteria extracts were also strongly active against multidrug-resistant clinical strains of Staphylococcus aureus and Staphylococcus epidermidis, isolated from hospital patients. Mitova et al. (Reference Mitova, Tommonaro, Hentschel, Müller and De Rosa2004) reported also on a bacterium isolated from S. domuncula primmorphs and which shared 98% of similarity with Ruegeria atlantica. Cyclopeptides isolated from R. atlantica were active against Bacillus subtilis. This latest bacterium has not been encountered in the present study. Its presence, since the sponge species was the same, may be attributed to the location from which they come.
Endozoicomonas is an interesting genus. Among eight bacteria isolated from S. domuncula and related to the genus Endozoicomonas, seven of them (JX436366, JX436367, JX442223, JX442221, JN624846, JX442219.1 and JX442229) produced some AHLs at least once during the culture time. The last one inhibited the production of luminescence of E. coli reporter strain throughout its growth. This study is the first to report on AHLs production by an Endozoicomonas-related genus. It is also the second study mentioning more than one Endozoicomonas species present in an invertebrate (Rua et al., Reference Rua, Trindade-Silva, Appolinario, Venas, Garcia, Carvalho, Lima, Kruger, Pereira, Berlinck, Valle, Thompson and Thompson2014).
To conclude, the present study has demonstrated the possibility to cultivate bacteria isolated from the marine sponge S. domuncula. This cultivable bacterioflora was genetically characterized at the genus level. Two-thirds of those bacteria were able to enhance the production of the luminescence of the reporter strain E. coli pSB406 at least at one time-point of the culture revealing the production of C4-HSL, C6-HSL, C8-HSL, C12-HSL or C14-HSL. Those AHLs were thus produced in vitro, in the absence of the host. The question of the necessity for sponge-associated bacteria of producing some AHLs must be addressed: are those molecules intended for them to use, for example for their own density regulation within host niches, and/or to communicate with it? Gardères et al. (Reference Gardères, Taupin, Bin Saidin, Dufour and Le Pennec2012, Reference Gardères, Henry, Bernay, Ritter, Zatylny-Gaudin, Wiens, Müller and Le Pennec2014) gave indications on the role of such bacterial molecules on sponge physiology. Furthermore, the present study allowed the detection of bactericidal and/or bacteriostatic compounds in bacterial culture supernatants.
Further efforts must be undertaken to describe the production and regulation of AHLs by sponge-associated bacteria within the holobiome, their role(s) in bacterial communities, within a dialogue with the host and in the maintenance of the equilibrium of such a complex association.
FINANCIAL SUPPORT
JBS was the recipient of doctoral fellowship of Ministry of High Education, Malaysia. This work was funded by the Axis 1 ‘Genomics and blue chemistry’ of the GIS Europôle Mer, and European FEDER. The sequencing analysis was funded by Institute of Marine Biotechnology, Universiti Malaysia Terengganu, Malaysia.