Introduction
Major depression is a highly heritable disease with an estimated heritability of 40% (Burmeister et al. Reference Burmeister, McInnis and Zöllner2008). However, identifying genetic risk variants for psychiatric conditions such as depression has turned out to be difficult, which has been related to the current diagnostic systems lacking sufficient foundation in aetiology or pathophysiology. An approach suggested to overcome these problems is the concept of ‘endophenotypes’ (Gottesman & Gould, Reference Gottesman and Gould2003; Hasler & Northoff, Reference Hasler and Northoff2011). Gottesman & Shields (Reference Gottesman and Shields1973) defined an endophenotype as an internal, intermediate phenotype that fills the gap in the causal pathway from genetic variation to the distal disease. Endophenotypes that are predictive for disease risk in healthy individuals are referred to as vulnerability markers. Endophenotypes or vulnerability markers are assumed to have less complex genetic underpinnings than disease phenotypes and are, therefore, more tractable to genetic analysis (Gottesman & Gould, Reference Gottesman and Gould2003; Hasler & Northoff, Reference Hasler and Northoff2011).
A prominent finding in depression is the frequent co-occurrence with somatization (e.g. Ohayon & Schatzberg, Reference Ohayon and Schatzberg2010; Klengel et al. Reference Klengel, Heck, Pfister, Brückl, Hennings, Menke, Czamara, Müller-Myhsok and Ising2011; for a review, see Bair et al. Reference Bair, Robinson, Katon and Kroenke2003). It is assumed that there is probably an amplification of painful somatic sensations in depression, suggesting a functional connection between both (Bair et al. Reference Bair, Robinson, Katon and Kroenke2003). Several studies have pointed to somatic complaints as a vulnerability marker for affective disorders observable already before disease onset (Clayton et al. Reference Clayton, Ernst and Angst1994; Ising et al. Reference Ising, Lauer, Holsboer and Modell2004b). At the physiological level, somatization can be investigated by long-latency somatosensory evoked electroencephalogram (EEG) potentials (SSEPs) reflecting brain activity related to the processing of somatosensory stimuli (Colon & de Weerd, Reference Colon and de Weerd1986). Their late components appear to indicate psychological processes of perception and appraisal (Miltner et al. Reference Miltner, Johnson, Braun and Larbig1989). SSEPs show habituation under recurrent stimulation and it is assumed that this decay of stimulus-evoked brain activity is reflecting the ability to deflect attention from intrusive somatosensory stimuli (Dietl et al. Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001).
Just like somatization, long-latency SSEPs have also been demonstrated to be possible vulnerability markers for affective disorders (Dietl et al. Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001; Ising et al. Reference Ising, Dietl, Dirlich, Vogl, Pollmächer, Nickel, Sonntag, Strian, Lechner, Lauer and Modell2004a). Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001) compared long-latency SSEPs within two time intervals (50–150 and 170–370 ms) after intrusive but not painful electric simulation between patients with major depressive disorder (MDD) and healthy controls. While there was only a tendency for enhanced SSEP responses in the range from 50 to 150 ms after stimulation in MDD patients, differences between the MDD group and the control group turned out to be significant in the 170–370 ms range. Corresponding results were also reported by Ising et al. (Reference Ising, Dietl, Dirlich, Vogl, Pollmächer, Nickel, Sonntag, Strian, Lechner, Lauer and Modell2004a). Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001) argued that enhanced SSEP components as well as their resistance to habituation may indicate a more attentive somatosensory signal processing in MDD patients, possibly contributing to somatic misperceptions and pain syndromes. This perspective is also in line with cognitive abnormalities such as increased attention to threatening and emotionally negative stimuli, which have repeatedly been reported for MDD patients (Leppänen, Reference Leppänen2006).
The assumption that long-latency SSEPs constitute vulnerability markers for affective disorders that could furthermore be suitable for investigation within genetic association studies is also strengthened by the finding that genetic variation contributes significantly to individual differences in evoked potentials (van Beijsterveldt & van Baal, Reference van Beijsterveldt and van Baal2002). Gene regions within the serotonergic, the GABAergic as well as the substance P system can be considered appropriate candidate gene regions for genetic association analyses within the reported context, since extensive evidence points to the importance of these systems for affective disorders, somatosensory processing as well as pain perception (e.g. Ordway et al. Reference Ordway, Klimek, Mann, Davis, Charney, Coyle and Nemeroff2002; Rupniak & Kramer, Reference Rupniak, Kramer, Davis, Charney, Coyle and Nemeroff2002; Bair et al. Reference Bair, Robinson, Katon and Kroenke2003; Campbell et al. Reference Campbell, Clauw and Keefe2003; Holmes et al. Reference Holmes, Heilig, Rupniak, Steckler and Griebel2003; Elhwuegi, Reference Elhwuegi2004; Cryan & Kaupmann, Reference Cryan and Kaupmann2005; Ebner & Singewald, Reference Ebner and Singewald2006; Enna & McCarson, Reference Enna and McCarson2006; Kalueff & Nutt, Reference Kalueff and Nutt2007).
Based on this evidence, the aim of our study was to investigate genetic associations with long-latency SSEPs in families susceptible for affective disorders in order to reveal vulnerability genes within the serotonergic, the GABAergic as well as the substance P system underlying altered somatosensory processing. To test whether potential findings will be specific for the investigated SSEP markers, the results will be compared with the outcome of a classical genetic approach considering only presence or absence of a history of affective disorders.
Method
Subjects and psychiatric assessment
The study protocol was approved by the local ethical committee of the medical faculty of the Ludwig Maximilians University Munich, and participants gave written informed consent to their participation.
The total study sample was composed of 83 subjects out of twelve families who were participating in the Munich Vulnerability Study, a prospective high-risk study aiming to reveal vulnerability markers for affective disorders. Inclusion criteria for families were the participation of at least two family members with a negative history of affective disorders and of at least two further first-degree relatives suffering from unipolar or bipolar affective disorder suggesting genetic susceptibility for such disorders within these families.
Subjects were diagnosed using the German version of the Structured Clinical Interview for DSM-IV Disorders (SCID-I and SCID-II; Wittchen et al. Reference Wittchen, Zaudig and Fydrich1997). Moreover, self-rating symptom scales were applied for the closer assessment of participants' mental and somatic complaints at time of investigation. Symptoms of depressiveness were evaluated using the German version of the Beck Depression Inventory (BDI; Beck et al. Reference Beck, Rush, Shaw and Emery1986). The tendency to experience somatic and general complaints was assessed using the Complaint List (Beschwerdeliste; von Zerssen, Reference von Zerssen1976). Furthermore, somatic symptoms were also evaluated with the subscale ‘Somatization’ of the revised German version of the Symptom Checklist (SCL-90-R; Derogatis, Reference Derogatis1986).
All participants were Caucasians originating from Germany.
SSEP acquisition and signal analysis
Out of the 83 subjects recruited from families selected within the Munich Vulnerability Study, 69 subjects agreed to participate in EEG recordings in addition to genotyping and psychiatric investigation and therefore underwent the SSEP acquisition procedure. SSEP recordings and parameter calculations were in the main conducted following the paradigm described within the study of Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001) .
During the experiment participants were seated in an electrically- and sound-shielded room and were watching a movie, which was carefully selected for keeping the subjects' attention constant without inducing emotions. SSEPs were evoked through the application of intrusive, but not painful, electric stimuli to the index finger of the non-dominant hand. The sensory threshold was determined beforehand for each subject and the strength of the electric stimulation was set to 2.5 times the individual threshold. This led to a mean strength of electric stimulation of 561.7 mA (s.d.=167.3, range 263–1000 mA). As analyses showed, strength of electric stimulation was not significantly influenced by psychiatric health status (affected, remitted or without life-time diagnosis) when considering age and gender as covariates (p=0.34).
Stimuli were organized as a series of eight blocks each containing eight stimulus cycles. Every cycle consisted of 27 stimuli each lasting 0.2 ms and reoccurring with a frequency of 0.9 Hz. After every cycle there was a stimulus-free interval of 15 s. EEG data were collected using five Ag/AgCl electrodes (Fz, Cz, Pz, C3, C4) which were set up according to the international 10–20 system (Jasper, Reference Jasper1958) and referenced to the averaged mastoid electrodes. Eye movements were recorded using two electrodes set up diagonally at the outer-upper and outer-lower edge of the left and right eye, respectively. Signals were recorded with a DC recording system (Schwind Medizintechnik, Germany) at a sampling rate of 500 Hz and an amplitude resolution of 0.3 μV and filtered offline (0.5–40.0 Hz). EEG impedance at single electrodes was kept as small as possible (<5 kΩ). Time periods with eyeblinks or other eye movement artefacts were detected and corrected computationally using a multiple regression approach after data had been visually screened and motor artefacts had been removed manually.
Data were averaged and SSEP amplitude values were calculated in reference to an artefact-free 50 ms baseline interval prior to stimulus onset. In accordance with the study of Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001), SSEP parameters were calculated for two time intervals, namely 50–150 ms and 170–370 ms after stimulation, both positioned in the long latency range of SSEPs. As suggested by Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001), the standard deviation of the squared SSEP amplitude values within the respective analysis interval was used as a measure of response strength (RS) reflecting SSEP intensity in terms of the average variability of the signal assuming that stronger reactions cause larger deflections.
Besides the average response strength at 50–150 ms (RS1) and 170–370 ms (RS2) after stimulation, we also calculated SSEP habituation within the respective time intervals (50–150 ms: RS1Hab; 170–370 ms: RS2Hab) by calculating the linear component of an orthogonal polynomial across the eight consecutive stimulation blocks.
SSEP parameters (RS1, RS2, RS1Hab and RS2Hab) were calculated from the average activity of the midline electrodes Fz, Cz and Pz showing the most pronounced SSEP signal. In the case of a significant genetic association with one of the four SSEP parameters, we additionally analysed single electrodes to disentangle the topographic localization of the effect.
Candidate genes and genotyping
In choosing candidate genes we followed an extended candidate gene approach (Bickeböller & Fischer, Reference Bickeböller and Fischer2007) and selected a broad set of genes with attributable function, coding for substructures of the serotonergic, the GABAergic as well as the substance P system and being discussed in connection with affective disorders, somatosensory processing as well as pain perception (for reviews, see the Introduction section). Namely, we chose serotonin receptor genes (HTR1A, HTR1B, HTR2A, HTR2C), genes involved in the synthesis of serotonin (TPH1, TPH2) as well as in the degradation of monoamines (MAOA, COMT), genes involved in the reuptake of serotonin (SLC6A4) and γ-aminobutyric acid (GABA) (SLC6A1), GABA receptor genes (GABRA1, GABRA2, GABRA3, GABRA4, GABRA5, GABRA6, GABRB1, GABRB2, GABRB3, GABRG1, GABRG2, GABRG3, GABBR1, GABBR2), genes involved in the synthesis (GAD1, GAD2) as well as the degradation (ABAT) of GABA, and the gene coding for substance P (TAC1) as well as its most important receptor (TACR1). After the selection process, we had to exclude four genes due to technical reasons, either because of lacking coverage by the gene chip used (HTR1A) or an X-chromosomal gene position (MAOA, HTR2C, GABRA3), which is incompatible with the statistical analysis software. Finally, we ended up with 25 candidate genes.
DNA was extracted from participants' blood samples and 176 single nucleotide polymorphisms (SNPs) from within the candidate gene regions including coding regions and flanking sequences (±10 kb) were genotyped using Illumina BeadChip technology (Human-1, 100k; Illumina, Inc., USA).
As quality criteria for genetic data we chose a minor allele frequency (MAF) of 5% and a call rate of at least 95%. Moreover, we performed exact tests for Hardy–Weinberg equilibrium (HWE) for all SNPs (Wigginton et al. Reference Wigginton, Cutler and Abecasis2005) and checked genetic data for Mendelian inheritance errors using PLINK 1.05 software (Purcell et al. Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, de Bakker, Daly and Sham2007; http://pngu.mgh.harvard.edu/purcell/plink/).
Statistical analysis
Genetic associations with SSEP parameters (RS1, RS2, RS1Hab, RS2Hab) were calculated using qfam (family-based association tests for quantitative traits) which is implemented in the PLINK 1.05 software package (Purcell et al. Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, de Bakker, Daly and Sham2007). qfam is based on the between/within model reported by Fulker et al. (Reference Fulker, Cherny, Sham and Hewitt1999) and Abecasis et al. (Reference Abecasis, Cardon and Cookson2000) . Genetic associations are thereby assessed calculating a linear regression of phenotype on genotype and using a special permutation procedure to correct for non-independence of individuals within families. We set the number of permutations to 100 000 and ran qfam as a total association test considering between-family as well as within-family components of genetic associations (Purcell et al. Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, de Bakker, Daly and Sham2007). Since possible effects of age, gender, strength of electric stimulation and EEG electrode impedance cannot be excluded, we calculated multiple regression residuals of the SSEP parameters corrected for the effects of the covariates.
To test whether potential genetic findings are speci-fic for the investigated SSEP markers, we additionally conducted a classical genetic analysis using disease history information only. For this analysis we applied the dfam module (family-based association for disease traits) of the PLINK software package after defining family members diagnosed as affected or remitted for an affective disorder as disease trait carriers. dfam is based on the transmission disequilibrium test, which is generalized to family structures allowing the inclusion also of discordant sibship data as well as unrelated subjects (Purcell et al. Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, de Bakker, Daly and Sham2007).
In order to correct for multiple testing we calculated false discovery rates (FDRs) across all tests within our two analyses (Benjamini & Hochberg, Reference Benjamini and Hochberg1995) and considered FDR values of less than 0.05 as statistically significant.
For gene regions that turned out to be of special interest in the course of genetic association analysis, we inspected linkage disequilibrium pattern and block structure using Haploview 4.2 (Barrett et al. Reference Barrett, Fry, Maller and Daly2005; http://www.broadinstitute.org/haploview/haploview). Haplotype block definition was performed based on the four-gamete rule (Wang et al. Reference Wang, Akey, Zhang, Chakraborty and Jin2002).
We further tested if the psychiatric health status at the time of investigation has an impact on the SSEPs and calculated an analysis of covariance for RS1, RS2, RS1Hab and RS2Hab including psychiatric health status (without life-time diagnosis v. remitted v. affected) as an independent variable and again considering age, gender, strength of electric stimulation as well as EEG impedance as covariates. No effects of psychiatric health status on the SSEP parameters could be detected (p>0.21) and, thus, health status was not considered in the genetic analysis. This finding was expected, considering the fact that we investigated subjects out of high-risk families and that we were using SSEPs as endophenotype within our study, which should be heritable, state-independent (manifest in an individual whether or not illness is currently active) and overrepresented in unaffected relatives of affected individuals compared with members of the general population (Hasler & Northoff, Reference Hasler and Northoff2011).
All non-genetic analyses were performed using SPSS 16.0 (SPSS Inc., USA), and p values <0.05 were considered statistically significant.
Results
Sample characteristics
The mean age of the subjects was 46.8 (s.d.=16.4) years, 45.8% of the total study sample being female. At the time of the investigation, 11.1% of the total sample fulfilled criteria for an affective disorder, 32.1% were diagnosed as remitted and another 56.8% had no lifetime diagnosis for an affective disorder. More precisely, among those subjects diagnosed as affected or remitted, diagnoses of affective disorders encompassed single or recurrent episodes of MDD (74.3%), bipolar affective disorder (20.0%) or dysthymia (5.7%). Of those without a lifetime diagnosis for an affective disorder, 8.7% were currently affected by another axis I disorder encompassing anxiety disorders and substance-related disorders. Furthermore, 11.5% of those with a remitted affective disorder, had a current axis I diagnosis including anxiety disorders, substance-related disorders or somatoform disorder.
At the time of the investigation, the subjects showed on average marginal symptom intensities, with an average BDI depression score of 5.6 (s.d.=6.8) and an average somatization score of 17.1 (s.d.=12.4) (Complaints list) and of 5.1 (s.d.=5.4) (SCL-90-R Somatization).
Genetic association analysis
No Mendelian inheritance errors were detectable in the data. Out of the 176 genotyped SNPs, one SNP had to be excluded due to a call rate lower than 95%. Another 15 SNPs had to be eliminated from further analysis because of a MAF lower than 5%. None of the SNPs had a p value for deviation from HWE lower than 2.8×10−4 (0.05/176 Bonferroni-corrected p value). Thus, no SNP had to be excluded because of deflection from HWE. (For genome positions, allele frequencies, call rates, MAF and p values for deviation from HWE of all genotyped SNPs, see the Supplementary online information.) One subject had to be excluded from genetic association analysis because the respective call rate was lower than 95%, i.e. more than 5% of the SNPs failed genotyping in this subject. Therefore, further reported results for genetic association analysis are based on 160 SNPs and those remaining 68 subjects for whom SSEP data as well as genotypes were available.
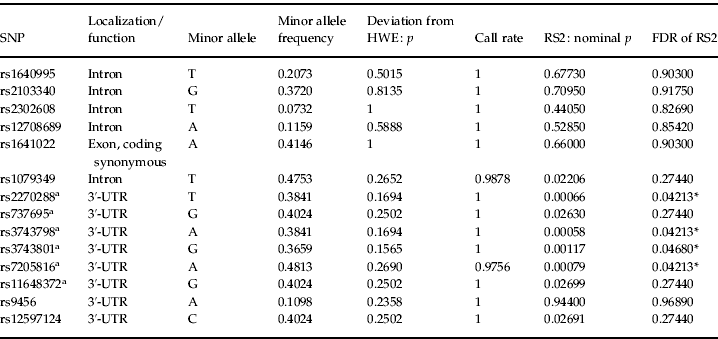
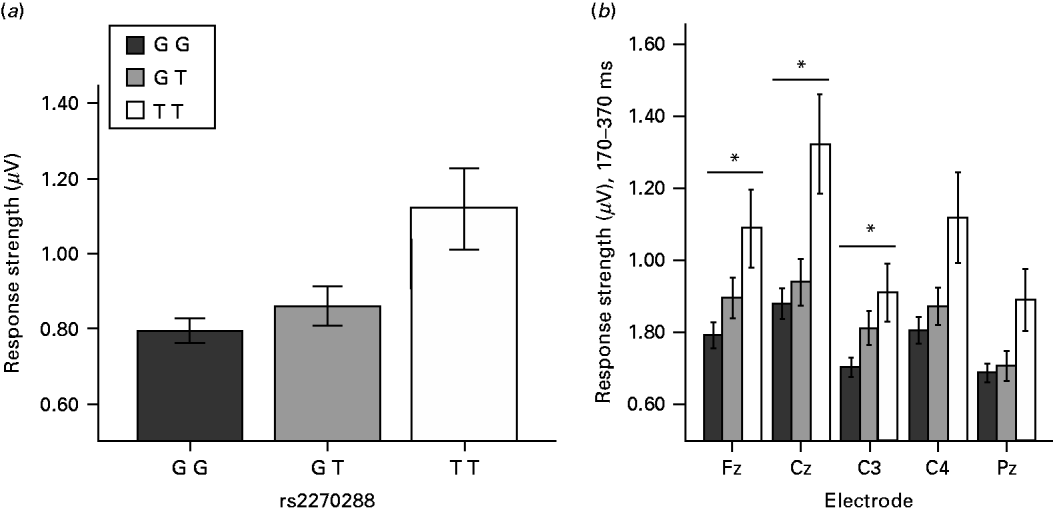
Testing for associations between SSEP parameters (RS1, RS2, RS1Hab and RS2Hab of averaged midline electrodes) and SNPs within the candidate gene regions, we found no significant genetic associations with RS1 (50–150 ms after stimulation) or its habituation (RS1Hab) withstanding correction for multiple testing. However, we found significant associations after correction for multiple testing between RS2 (170–370 ms after stimulation) and four SNPs located in the 3′-untranslated region (3′-UTR) of the GABA transaminase (ABAT) gene. Table 1 gives an overview over all genotyped SNPs within this gene region.
Table 1. Genotyped SNPs within the GABA transaminase (ABAT) gene region and their association with RS2 of averaged midline electrodes

SNP, Single nucleotide polymorphism; GABA, γ-aminobutyric acid; RS2, response strength at 170–370 ms after stimulation; HWE, Hardy–Weinberg equilibrium; FDR, false discovery rate; UTR, untranslated region.
a SNPs located within one block of high linkage disequilibrium (see the ‘Linkage disequilibrium and gene expression within the ABAT gene region’ section and Fig. 2).
* Significant FDRs (p<0.05).
The four SNPs withstanding correction for multiple testing were rs2270288 (FDR=0.04213), rs3743798 (FDR=0.04213), rs3743801 (FDR=0.04680) and rs7205816 (FDR=0.04213). Respective minor alleles of these four SNPs were associated with higher levels of RS2 (see Fig. 1 for rs2270288). No genetic associations withstanding correction for multiple testing could be observed for RS2Hab. For nominal p values and FDRs of associations between each SSEP parameter (RS1, RS1Hab, RS2 and RS2Hab) and every analysed SNP, see the Supplementary online information.

Fig. 1. Response strength (RS) of somatosensory evoked electroencephalogram (EEG) potentials (SSEPs) at the averaged midline electrodes (a) as well as at the single electrodes (b) 170–370 ms after somatosensory stimulation (RS2) for the genotypes of rs2270288 (GG, GT, TT). Values are means, with standard errors represented by vertical bars. * False discovery rate <0.05.
When taking a closer look at the topographical localization of the ABAT effect we observed significant associations withstanding correction for multiple testing between all four SNPs and RS2 at electrodes Fz (rs2270288 FDR=0.01344; rs3743798 FDR=0.01344; rs3743801 FDR=0.01344; rs7205816 FDR=0.01344), Cz (rs2270288 FDR=0.01624; rs3743798 FDR=0.01624; rs3743801 FDR=0.03402; rs7205816 FDR=0.01624) and C3 (rs2270288 FDR=0.02738; rs3743798 FDR=0.02738; rs3743801 FDR=0.04226; rs7205816 FDR=0.02738). Moreover, there was a significant association between RS2 at electrode Pz and SNP rs7205816 (FDR=0.03268). Altogether, these results suggest a potential fronto-central focus for the ABAT association with RS2.
In order to explore the reported effect more closely on a phenotypical level, we calculated correlations between scores in self-rating symptom scales and RS2, both for averaged midline electrodes and for single electrodes showing significant associations with the SNPs reported above. We again included the same covariates as in the genetic association analyses. Analyses were conducted one-tailed following the hypothesis of a positive association between response strength and self-rating symptoms. None of the correlations between scores in symptom scales (BDI, Complaint List, SCL-90-R Somatization) and RS2 for averaged midline electrodes reached significance (p>0.195). However, correlations between self-rated general and somatic complaints and RS2 at electrode Fz only scarcely failed significance (Complaint List: r=0.19, p=0.065; SCL-90-R: r=0.16, p=0.101), thus further pointing to a potential fronto-central topographical specificity of the effect by trend.
To test whether the genetic associations are specific to the investigated SSEP parameters, we additionally conducted a classical genetic analysis using disease history information (i.e. affected and remitted as compared with unaffected by affective disorders) only. Out of 160 investigated SNPs, 17 SNPs were associated with the disease trait at a nominal level of significance (p<0.05), but no effect withstood correction for multiple testing (FDR >0.16). The four SNPs located in the 3′-UTR of the ABAT gene showing associations with RS2 were not associated with the disease trait, with respective nominal p values exceeding 0.89.
Linkage disequilibrium and gene expression within the ABAT gene region
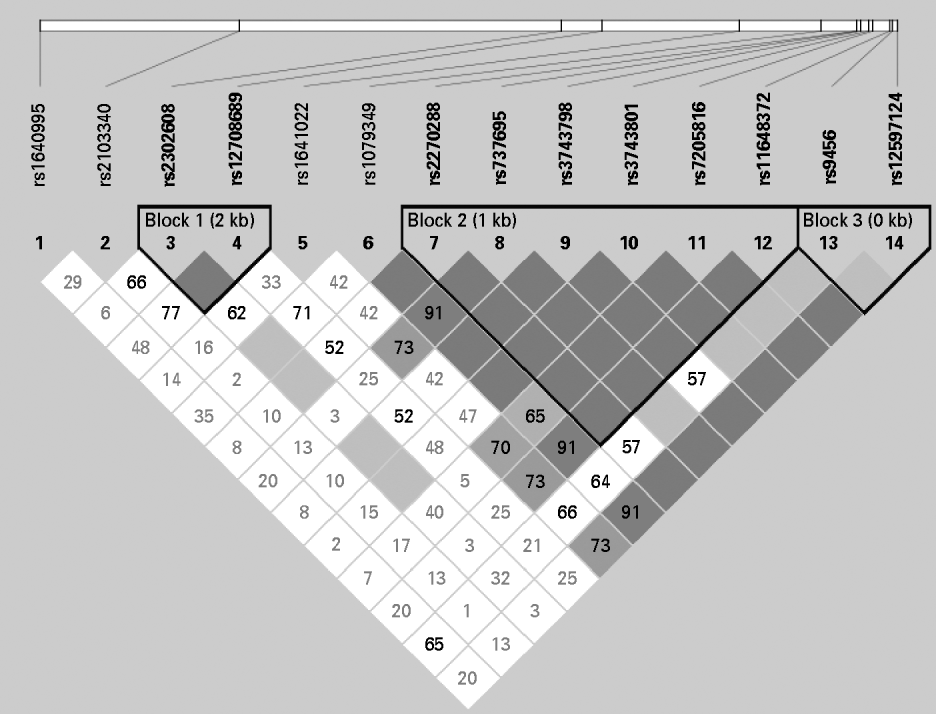
As shown in Fig. 2, the four SNPs showing significant associations with RS2 (rs2270288, rs3743798, rs3743801, rs7205816) belong to a single block of high linkage disequilibrium located in the 3′-UTR of the ABAT gene.

Fig. 2. Linkage disequilibrium (LD) pattern and block structure of the γ-aminobutyric acid transaminase (ABAT) gene region. As a measure of LD, D' is indicated as a percentage within each box. For boxes lacking a number, D'=1.0.
Since the 3′-UTR is not coding for the actual gene product but known to contain transcription and translation regulating sequences (National Center for Biotechnology Information, 1999), we additionally searched for associations between the four SNPs and ABAT gene expression. Therefore, we used data from a publicly accessible database containing gene expression data from Epstein–Barr virus-transformed lymphoblastoid cell lines in the HapMap population (Stranger et al. Reference Stranger, Nica, Forrest, Dimas, Bird, Beazley, Ingle, Dunning, Flicek, Koller, Montgomery, Tavaré, Deloukas and Dermitzakis2007) as well as HapMap genotype data (The International HapMap Consortium, 2003). The gene expression dataset was produced using Illumina whole-genome expression technology (Sentrix Human-6 Expression BeadChip version 1; Illumina, Inc., USA). We downloaded normalized expression data and SNP genotypes from ftp://ftp.sanger.ac.uk/pub/genevar/. In order to ensure a population structure comparable with our German sample, we restricted our analyses to the CEU sample containing data from 60 unrelated individuals living in Utah with ancestry from northern and western Europe collected by the Centre d'Etude du Polymorphisme Humain. However, we found no significant associations between the four SNPs and ABAT messenger ribonucleic acid (mRNA) expression levels in lymphoblastoid cell lines (p>0.05).
Discussion
In this study we investigated the associations between long-latency SSEPs and polymorphisms in candidate genes of the serotonergic, GABAergic as well as the substance P system in families at high risk for affective disorders. We found significant associations withstanding correction for multiple testing between SSEP response strength at 170–370 ms after stimulation (RS2) and four polymorphisms located in the ABAT gene region. These associations could mainly be observed at electrode sites Fz, Cz and C3. Thus, one could speak of a strong effect of the ABAT gene on fronto-central somatosensory processing. Further-more, a role of frontal regions could also be suggested considering our finding of correlations between reported somatization symptoms and RS2 at electrode Fz only scarcely failing significance. Consistent with the assumption of a crucial role of fronto-cortical regions in ABAT-mediated somatosensory processing, there is extensive evidence suggesting prefrontal cortical areas to be essential in neurocognitive aspects of pain perception (e.g. Wiech et al. Reference Wiech, Ploner and Tracey2008) as well as in regulation of aversive emotional states, which in turn is thought to be disrupted in patients with affective disorders (e.g. DeRubeis et al. Reference DeRubeis, Siegle and Hollon2008).
In the mammalian brain, the ABAT gene expresses the primary catabolic enzyme for the elimination of the inhibitory neurotransmitter GABA. Thus, it is not surprising that inhibition of GABA transaminase has been shown to produce increased GABA levels in the brain, which corresponds with a number of behavioural effects including anxiolysis and sedation (Sherif & Ahmed, Reference Sherif and Ahmed1995).
GABAergic transmission plays a key role not only in anxiety, but also in depression (Kalueff & Nutt, Reference Kalueff and Nutt2007; Sanacora & Saricicek, Reference Sanacora and Saricicek2007; Hasler & Northoff, Reference Hasler and Northoff2011; Luscher et al. Reference Luscher, Shen and Sahir2011) as well as in other neuropsychiatric disorders (Sherif & Ahmed, Reference Sherif and Ahmed1995). Low GABA levels in blood plasma, cerebrospinal fluid, and brains of deceased patients with depression have been reported (e.g. Luscher et al. Reference Luscher, Shen and Sahir2011; Sanacora & Saricicek, 2011). Berrettini et al. (Reference Berrettini, Umberkoman-Wiita, Nurnberger, Vogel, Gershon and Post1980) proposed platelet ABAT expression as a vulnerability marker for affective illness and furthermore suggested the importance of genetic factors in its regulation. Moreover, a role of ABAT for affective disorders as well as somatosensory processing is also suggested by the fact that ABAT inhibitors such as valproate or vigabatrin show effectiveness as mood stabilizers (Gajwani et al. Reference Gajwani, Forsthoff, Muzina, Amann, Gao, Elhaj, Calabrese and Grunze2005) as well as antinociceptive (Enna & McCarson, Reference Enna and McCarson2006) and anxiolytic compounds (e.g. Lang & de Angelis, Reference Lang and de Angelis2003), in addition to their antiepileptic properties.
Besides its role in mood disorders, GABAergic neurotransmission is involved in mediation and perception of somatosensory and nociceptive stimuli (Jasmin et al. Reference Jasmin, Rabkin, Granato, Boudah and Ohara2003; Enna & McCarson, Reference Enna and McCarson2006; Xie et al. Reference Xie, Huo and Tang2009). GABA receptor systems are located in peripheral and spinal cord pathways important for the origination and transmission of nociceptive impulses, but also in higher associative brain regions (Enna & McCarson, Reference Enna and McCarson2006). There is profound evidence for an involvement of GABAergic neurotransmission also in descending modulatory pathways interacting with other neurotransmitters such as serotonin and opioids (Xie et al. Reference Xie, Huo and Tang2009). For instance, Jasmin et al. (Reference Jasmin, Rabkin, Granato, Boudah and Ohara2003) reported GABAergic neurotransmission in the rostral agranular insular cortex (RAIC) to make an impact on descending inhibition of spinal nociceptive neurons in rodents. Furthermore, they reported projections from RAIC neurons to the amygdala, which is not only involved in fear but also part of a pain-facilitating circuit. From studies in rodents there is also evidence for reduced GABA neurotransmission to be involved in stress-induced hyperalgesia (e.g. Quintero et al. Reference Quintero, Cardenas and Suarez-Roca2011). Human brain-imaging studies described GABA-mediated resting state levels as predictors of induced cortical activity including the perigenual anterior cingulate cortex (Northoff et al. Reference Northoff, Walter, Schulte, Beck, Dydak, Henning, Boeker, Grimm and Boesiger2007; Muthukumaraswamy et al. Reference Muthukumaraswamy, Edden, Jones, Swettenham and Singh2009). On a subcortical level, Neto et al. (Reference Neto, Ferreira-Gomes and Castro-Lopes2006) described GABA receptors in the thalamus, known as an important target for sensory information and a relay to cortical areas. Psychological processes such as emotion, cognition and the stress response have been shown to be critically influenced by GABAergic transmission (e.g. Hasler & Northoff, Reference Hasler and Northoff2011; Luscher et al. Reference Luscher, Shen and Sahir2011). Consequently, there are multiple routes of GABAergic transmission and its modulators such as GABA transaminase to make an impact on somatosensory processing and respective alterations in SSEPs.
In contrast to response strength at 170–370 ms after stimulation (RS2), no significant associations could be found for the other phenotypes, namely response strength at 50–150 ms after stimulation (RS1) and for both habituation parameters, RS1Hab and RS2Hab. Lacking genetic associations with RS1 and RS1Hab is plausible, considering a few facts. First, candidate gene regions were chosen because of their postulated role for the (late) processing of somatosensory stimuli and pain perception, which is represented rather by RS2 than RS1. Second, RS2 has to be considered a more appropriate vulnerability marker for affective disorders a priori, given that Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001) as well as Ising et al. (Reference Ising, Dietl, Dirlich, Vogl, Pollmächer, Nickel, Sonntag, Strian, Lechner, Lauer and Modell2004a) reported differences in SSEP responses between MDD patients and healthy controls to be more pronounced in the time-frame 170–370 ms in comparison with 50–150 ms after stimulation. Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001) argued that a more attentive somatosensory signal processing might contribute in making depressed patients susceptible to somatic misperception and pain syndromes. This also makes sense in the light of a study of Miltner et al. (Reference Miltner, Johnson, Braun and Larbig1989) reporting P200 and P300, but not N150 of SSEPs to be significantly influenced by the subjects' directed attention towards somatosensory stimuli.
Dietl et al. (Reference Dietl, Dirlich, Vogl, Nickel, Sonntag, Strian and Lechner2001) stated that they were not able to disentangle whether enhanced long-latency SSEP responses were rather due to differing initial potential amplitudes or differences in the habituation rate. The fact that we could not observe any significant associations between SNPs within our candidate gene regions and RS2 habituation suggests that mean SSEP response strength over time possibly represents a better vulnerability marker for affective disorders and co-morbid somatic complaints than SSEP habituation. However, this issue remains to be further investigated.
On the molecular level, the four SNPs associated with RS2 are located within a single block of high linkage disequilibrium in the 3′-UTR of the ABAT gene region. A 3′-UTR constitutes a region not coding for the gene product but known to contain gene expression regulation sequences (National Center for Biotechnology Information, 1999). Therefore, it is possible that the reported associations between the four polymorphisms and RS2 are based on the effect of elements within the 3′-UTR on ABAT expression, which could possibly further make an impact on somatosensory processing. However, we did not find any significant associations between the four SNPs and ABAT mRNA expression levels. Yet, it has to be emphasized that this result refers to gene expression in lymphoblastoid cell lines and that it is unsure whether it would hold as well for expression in the brain which we were unfortunately not able to investigate. Moreover, it is known that sequences within 3′-UTRs do not only influence gene transcription and therefore mRNA expression levels, but that they also make an impact on gene expression in various other ways, including the regulation of translation efficiency, amongst others (e.g. Conne et al. Reference Conne, Stutz and Vassalli2000).
Therefore, at the moment it remains unclear via which exact mechanisms the four SNPs or other genetic variants being in linkage disequilibrium with these SNPs make an impact on GABA catabolism and further on the processing of somatosensory stimuli. This should be the issue of further molecular biological investigations.
No genetic association was found with the classical disease trait approach. This is in line with the failure to observe associations between SSEP measures and the psychiatric health status (affected, remitted, without life-time diagnosis). Considering that subjects have been recruited from families being at high risk for affective disorders, the majority of participants might be expected to carry the respective genetic risk. This common genetic background could explain why we did not find any relationships between psychiatric health status and SSEP measures and why correlations between self-rated depressive as well as somatic complaints and SSEP measures did not reach significance. Furthermore, one has to consider that we were using SSEP measures as an endophenotype (vulnerability marker) which should be heritable, state-independent (manifest in an individual whether or not illness is active) as well as overrepresented in unaffected relatives of affected individuals compared with members of the general population (Hasler & Northoff, Reference Hasler and Northoff2011).
As a limitation of our study we have to mention the moderate sample size. We tried to address this limitation by using qfam, a family-based association test, which outperforms classical linkage analysis in terms of statistical power (Purcell et al. Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, de Bakker, Daly and Sham2007). Furthermore, all participants underwent extensive phenotyping including structured clinical interviews to assure highly reliable phenotype data. One should further notice that, although we used strict methods to control for errors due to multiple testing, our results still harbour the possibility of a false-positive finding. Therefore, our results should be regarded as tentative knowledge until their replication, as Sullivan (Reference Sullivan2007) suggested for results of single genetic association studies.
In summary, our results point to an involvement of an altered GABA catabolism in the central processing of somatosensory stimuli in individuals at high risk for affective disorders, which, in turn, is associated with elevated somatization symptoms. This finding complements previous results from our group suggesting the suitability of long-latency SSEP response strength in the time-frame of 170–370 ms after stimulation as a vulnerability marker for affective disorders. We, furthermore, showed that the genetic analysis based on the investigated SSEP markers, following the endophenotype concept, outperforms the classical disease trait approach in detecting associations in families at high risk for affective disorders within the selected candidate genes. Further studies are warranted to investigate the exact role of the ABAT gene and of GABA catabolism for somatosensory processing characteristics within this context.
Note
Supplementary material accompanies this paper on the Journal's website (http://journals.cambridge.org/psm).
Acknowledgements
We are grateful to Elisabeth Binder, M.D., Ph.D., Max Planck Institute of Psychiatry, for her assistance in recruiting and screening of the subjects and to Professor Martin Schalling, Karolinska Institute, Stockholm, for his help in designing the study and conducting the genotyping. We further want to thank Johannes Huber for continuous excellent technical support. The study was funded by the Max Planck Institute of Psychiatry. There was no further financial support or any external funding.
Declaration of Interest
None.