Introduction
Metabolic adaptations to pregnancy are normal processes aiming at supporting fetal growth. These adaptations include insulin resistance and changes in blood lipid profile.Reference Potter and Nestel1,Reference Desoye, Gauster and Wadsack2 Physiological changes in maternal blood lipids during pregnancy cause a gradual increase in all circulating lipoproteins. So every women, even the ones with normal cholesterol levels before pregnancy, are experiencing temporary hyperlipidemia,Reference Bansal, Cruickshank, McElduff and Durrington3 although normal values defined outside pregnancy might not apply. Interestingly, it was previously suggested that maternal lipid levels during pregnancy were associated with birth outcomes and might predispose the newborns to long-term health consequences.Reference Misra and Trudeau4,Reference Palinski, Yamashita, Freigang and Napoli5 Specifically, low maternal lipid levels may reflect an inadequate physiological response to pregnancy and have been associated with preterm delivery and lower birth weight.Reference Edison, Berg and Remaley6 In contrast, maternal hypercholesterolemia has been linked to poorer birth outcomes and increased cholesterol deposition in the fetal aorta that might therefore influence newborns’ risk of long-term cardiovascular disease (CVD) and morbidity.Reference Leiva, Fuenzalida and Westermeier7–Reference Napoli, D’Armiento and Mancini9
Placenta expresses many lipoprotein receptors, including the ATP-binding cassette transporter A1 (ABCA1), low-density lipoprotein receptor (LDLR), the LDL receptor-related protein 1 (LRP1), and scavenger receptor class B type 1 (SRB1; encoding by SCARB1 gene).Reference Hentschke, Poli-de-Figueiredo, da Costa, Kurlak, Williams and Mistry10, Reference Wadsack, Tabano and Maier11 These receptors are key players for binding of lipoproteins and internalization of cholesterol efflux from maternal to fetal circulation.Reference Wadsack, Tabano and Maier11–Reference Stefulj, Panzenboeck and Becker13
Recent studies have suggested that epigenetic modifications might explain, to some extent, the link between in utero environment and long-term cardiovascular disorders risk for newborns.Reference Lillycrop and Burdge14 Epigenetics refers to DNA modifications regulating gene transcription without changes of the primary DNA sequence. DNA methylation (DNAm) is the most stable and studied epigenetic markReference Henikoff and Matzke15 and is generally associated with gene expression repression.Reference Bird16 Importantly, the newborn epigenetic signature acquired in the womb might be persistent throughout adult life,Reference Heijmans, Tobi and Stein17, Reference Tobi, Lumey and Talens18 although this very likely not affects every gene similarly.
Changes in DNAm have been shown to be associated with in utero environment, including maternal lipid levels and glycemic profile during pregnancy.Reference Finer, Mathews and Lowe19–Reference Ruchat, Houde and Voisin22 Previous studies carried by our group showed that high-density lipoprotein cholesterol (HDL-C) levels during pregnancy were associated with DNAm levels at ABCA1 and lipoprotein lipase (LPL) gene in the placenta.Reference Houde, Guay and Desgagne20, Reference Houde, St-Pierre and Hivert23 Moreover, previous works have shown that maternal lipid levels during pregnancy were associated with changes in LDLR placental protein levels.Reference Ethier-Chiasson, Duchesne and Forest12
Therefore, we hypothesized that changes in maternal blood lipids in response to pregnancy prime placental cells through changes in DNAm to optimize materno-fetal lipid transfer and fetal growth. Accordingly, this study was undertaken to assess the association between maternal lipid level changes in pregnancy, placental DNAm at LDLR, LRP1, and SCARB1 gene loci, and anthropometric characteristics of the newborn at birth.
Materials and methods
Sample and clinical data
Sixty-nine normoglycemic women with a singleton pregnancy were selected from a cohort recruited in a founder population of French–Canadian origin from the Saguenay–Lac-Saint-Jean region of Québec (Saguenay city area, Québec, Canada). The women included in this study were aged between 18 and 40 years old, with no personal history of pregestational type 1 or 2 diabetes, polycystic ovary syndrome, or familial hypercholesterolemia. Women with a positive history of alcohol or drug abuse during the current pregnancy were also excluded. All women included in the current study were normoglycemic as defined as a 2 h-post oral glucose tolerance test (OGTT) glucose concentration < 7.8 mmol/l following a 75-g OGTT performed at 24–28 weeks’ gestation according to the WHO criteria.Reference Ahima24
A written informed consent was obtained for all women before their inclusion in the study, and all clinical data were de-identified. This project received the approval by the Chicoutimi Hospital Ethics Committee, in accordance with the Declaration of Helsinki.
Women were met at the ECOGENE-21 Clinical Research Center at the first, second, and third trimester of pregnancy. A registered nurse performed a complete anthropometric and metabolic profiling. Anthropometric variables and blood pressure were measured using standardized procedures.Reference Norgan25, Reference Lohman, Roche and Martorell26 Blood samples were obtained after a 12-h overnight fast from the antecubital vein into vacutainer tubes containing EDTA at each gestational trimester and at the time of delivery (cord blood). Plasma cholesterol, TG, HDL-C, and glucose levels were enzymatically measured using Beckman analyzers (models CX7 for lipids and DXC for glucose). LDL-C levels were calculated using the Friedewald formula.Reference Friedewald, Levy and Fredrickson27 Total gestational weight gain was computed by subtracting maternal weight at first trimester of pregnancy to that at the third trimester. Maternal blood lipid changes between trimesters were calculated by subtracting values at third to that at first trimester of pregnancy (ΔT3-T1 blood lipid level). ΔT3-T1 blood lipid levels represent global variation of blood lipid levels throughout pregnancy. Insulin measurements were performed using a radioimmunoassay method (Advia Centaur, Siemens). Insulin resistance levels were assessed using homeostatic model assessment – insulin resistance (HOMA-IR) [fasting glucose (mmol/l) × fasting insulin (mU/l)/22.5] at each trimester of pregnancy.
Clinical information about newborns was collected at birth from medical files and included gestational age, sex of the newborn, and anthropometric measurements. The gestational age was calculated at the first visit from the date of the last menstrual period and was corrected afterward, as recommended, based on the date of the ultrasound scans. Birth weight Z-score was assessed using the 2013 Fenton growth chart.Reference Fenton and Kim28 Newborns’ ponderal index (kg/m3) was calculated as: newborn weight (kg)/newborn length3 (m3). Cord blood lipid levels were assessed as described for the mother. In addition, cord blood leptin levels, measured by ELISA as previously described (B-Bridge International) were used as a marker of adiposity as previously described.Reference Bouchard, Thibault and Guay29,Reference Cote, Gagne-Ouellet and Guay30
Placenta samples and nucleic acid extraction
Placenta samples were taken within minutes of placenta expulsion (on average < 15 min) by clinicians. Biopsies were taken near the insertion point of the umbilical cord and consisted of the chorionic plate with the chorionic villi. Biopsies were washed in PBS 1× to remove cord/maternal blood, dissected to remove conjunctive tissues, and kept in RNAlater (Qiagen, #76106) at −80°c until nucleic acid extraction. Placental DNA and RNA were purified using the All Prep DNA/RNA/Protein Mini Kit (Qiagen, #80004). The ratio of absorbance at 260–280 nm with the Ultrospec 2000 UV/Visible Spectrophotometer (Pharmacia Biotech, USA) was used to confirm the DNA purity.Reference Cote, Gagne-Ouellet and Guay30 Placenta RNA quality was assessed with Agilent 2100 Bioanalyzer RNA Nano Chips (Agilent Technologies, USA, #5067-1511) based on RNA integrity number (RIN).
DNA methylation measurement
We used the gold-standard pyrosequencing technology to determine base-specific cytosine methylation levels.Reference Marsh31 Pyrosequencing assays combine sodium bisulfite DNA conversion chemistry (EpiTech Bisulfite Kits, Qiagen, USA; #59104), polymerase chain reaction (PCR) amplification (PyroMark PCR Kit, Qiagen; #978703), and sequencing by synthesis assay of a target region (PyroMark Gold Q24 Reagents, Qiagen; #978802). Each pyrosequencing runs included a negative PCR control and sequencing primer control as well as a sodium bisulfite conversion control. Additionally, pyrosequencing quality control was assessed for each sample using PyroMark Q24 Analysis Software, Qiagen (v.1.0.10.134). PCR primers were selected using the PyroMark Assay Design software v2.0.1.15. The PCR and pyrosequencing primers for the amplification of the loci are described in Supplementary Table 1.
Loci within LDLR, LRP1, and SCARB1 gene regions were selected based on a previous study reporting these regions were showing DNAm variability in the placenta.Reference Ruchat, Houde and Voisin22 Gene promoter regions and Cytosine-phosphate-Guanine (CpG) islands were also selected based on previous studies.Reference Guay, Brisson and Lamarche32, Reference Guay, Brisson, Lamarche, Gaudet and Bouchard33 Overall, 43 CpG dinucleotides within LDLR, LRP1, and SCARB1 gene loci were epigenotyped: 17 at LDLR gene locus (4 different regions: LDLR-CpGA to LDLR-CpGD), 9 at LRP1 gene locus (4 different regions: LRP1-CpGA to LRP1-CpGD), and 17 at SCARB1 gene locus (3 different regions: SCARB1-CpGA to SCARB1-CpGC) (Supplementary Table 1). When DNAm levels were found to be generally well correlated among the CpG dinucleotides from a same region (r > 0.7), a mean DNAm level for these CpGs was computed and used in the subsequent statistical analysis. Otherwise, the CpG dinucleotides were analyzed separately.
Relative placental mRNA level measurement
Forty-six placental RNA samples were available for mRNA quantification. They were of good quality based on a RIN > 7. Complementary DNA (cDNA) was generated from the placental total RNA using a random primer hexamer provided with the High Capacity cDNA Archive kit (ThermoFisher Scientific, USA; #4368814). Equal amounts of cDNA were run in duplicated and amplified in a 20 ul reaction containing 10 ul of 2× Universal PCR Master Mix (ThermoFisher Scientific; Life Technologies, #4366072). Primers and TaqMan probes were designed to cover exon boundaries (Life Technologies, LDLR: Hs01092524_m1, LRP1: Hs00233856_m1, and SCARB1: Hs00969821_m1). Each sample was calibrated to the YWHAZ housekeeping gene (endogenous control; YWHAZ: Hs00237047_m1). Relative quantification estimations were performed using an Applied Biosystem 7500 Real Time PCR System as recommended by the manufacturer. LDLR/YWHAZ Ct ratio (1/x), LRP1/YWHAZ Ct ratio (1/x), and SCARB1/YWHAZ Ct ratio (1/x) were used in the analyses.
Statistical analyses
Normal distribution of all variables was assessed using the Kolmogorov–Smirnov test. The following variables were found normally distributed after they were log10-transformed: TG levels, insulin levels, and HOMA-IR. Maternal blood lipid levels from each trimester of pregnancy were compared using a paired Student’s T-test. Partial Pearson correlations were used to investigate the association between the main variables of interest: DNAm levels, ΔT3-T1 maternal blood lipid levels, newborns’ anthropometric measurements, and cord blood lipid and leptin levels. The statistical models were controlled for the following confounding factors: maternal age, maternal BMI at first trimester of pregnancy, total gestational weight gain, maternal blood lipids level at second and third trimester of pregnancy, gestational age, and newborn gender (see Figure’s and Table’s footnotes for details). Impacts of other confounding factors (smoking, gravity, parity, mode of delivery, and time of visits) were tested but not retained in the statistical models as results remained unchanged. Association between DNAm levels and relative mRNA levels was assessed using a partial Pearson’s correlation controlling for gestational age and newborn gender. Results were considered statistically significant when p-values were <0.05 (two-sided).
Stepwise multivariable linear regression was used to identify predictors of the placental LDLR and LRP1 DNAm level variability. The input-independent variables were maternal age, maternal BMI at first trimester of pregnancy, maternal total weight gain, ΔT3-T1 maternal total cholesterol levels, maternal total cholesterol levels at third trimester of pregnancy, gestational age, and newborn gender. Variables with a p-value < 0.10 were retained in the regression model.
We estimated the direct association between maternal characteristics and newborn characteristics and the mediating effect of DNAm using a nonparametric bias-corrected bootstrapping procedure as previously described.Reference Cote, Gagne-Ouellet and Guay30 This allowed us to assess whether DNAm at LDLR and LRP1 gene loci mediated the association between maternal total cholesterol level changes throughout pregnancy and cord blood lipid and leptin levels. Potential confounders were considered in the statistical models: maternal age, maternal BMI at first trimester of pregnancy, maternal total weight gain, maternal total cholesterol levels at third trimester of pregnancy, gestational age, and newborn gender. Results were considered significant if p-values for mediation models were <0.05. Statistical analyses were performed with the IBM SPSS Statistics 24 software, USA (release 24.0.0).
Maternal lipid levels at third trimester of pregnancy were included as confounding variables in all of our statistical models in order to evaluate the independent contribution of the ΔT3-T1 maternal lipid levels to DNAm level variability and newborn characteristics. In addition, maternal blood lipid levels at third trimester are more likely representative of the lipid environment to which the placenta and newborn were exposed at birth, when the samples were collected.
Results
The characteristics of the mothers and their newborns are shown in Tables 1 and 2, respectively. At first trimester of pregnancy, women were on average 28.0 ± 3.8 years old, had a mean body mass index (BMI) of 23.4 ± 3.2 kg/m2, fasting glucose of 4.38 ± 0.34 mmol/l, and lipid levels within the normal ranges (based on reference values in nonpregnant women) (Table 1). Maternal blood lipids tended to increase between first, second, and third trimester of pregnancy (Table 1). Babies were 47.8% male (n = 33), had a mean gestational age of 39.4 ± 1.1 weeks, and had a mean ponderal index of 27.6 ± 2.6 kg/m3 (Table 2). Mean cord blood total cholesterol and leptin levels were 1.70 ± 0.43 mmol/l and 10.8 ± 8.5 ng/ml, respectively (Table 2).
Table 1. Maternal characteristics and blood lipid levels throughout pregnancy (n = 69)

BM1, body mass index; LDL, low-density lipoprotein; HDL, high-density lipoprotein; HOMA-IR, homeostatic model assessment – insulin resistance; OGTT, oral glucose tolerance test; SD, standard deviation.
a Geometric mean is presented for this variable.
b Significantly different from first trimester blood lipid levels (p < 0.001).
c Significantly different from second trimester blood lipid levels (p < 0.05).
d Significantly different from second trimester blood lipid levels (p < 0.001).
Table 2. Characteristics of newborns (n = 69)

HDL, high-density lipoprotein; LDL, low-density lipoprotein; SD, standard deviation.
a Geometric mean is presented for this variable.
We first assessed whether changes in maternal blood lipids in pregnancy were associated with newborn’s physical and metabolic characteristics (Table 3). We found that higher maternal total cholesterol changes throughout pregnancy (ΔT3-T1) were correlated with higher cord blood total and low-density lipoprotein cholesterol (LDL-C) as well as lower newborns’ ponderal index and cord blood leptin levels.
Table 3. Association between maternal blood lipid level changes in pregnancy and newborns’ characteristics

HDL, high-density lipoprotein; LDL, low-density lipoprotein.
p-values were obtained after consideration for maternal age, maternal IMC at first trimester of pregnancy, total gestational weight gain, gestational age, newborn’s gender, and blood lipid level at third (ΔT3-T1). Statistically significant results are shown in bold.
To determine whether the placental epigenetic signature is associated with maternal lipid profile and correlated to newborn anthropometric and metabolic profiles, we assessed DNAm levels at 43 CpG sites within LDLR, LRP1, and SCARB1 gene loci. Only the CpG sites with DNAm levels between 10% and 90% and showing interindividual variability [standard deviation > 3%] were considered for further analysis. Among the remaining CpGs, only those correlated with their mRNA levels (r > 0.25 and p < 0.1; mean LDLR-CpGA, LRP1-CpGA2, LRP1-CpGC1, and LRP1-CpGD1; Figs. 1B, 2D, and 3C, Supplementary Figs. 1 and 2, and Supplementary Table 2) were retained for further analyses. Associations were then tested between changes in maternal blood lipids and placental DNAm levels at these four CpGs. We found that increases in total cholesterol and LDL-C levels throughout pregnancy (ΔT3-T1) were associated with lower DNAm at LDLR-CpGA gene locus (Fig. 1A and Supplementary Table 3). At LRP1-CpGA2 gene locus, increases in total cholesterol, LDL-C and HDL-C levels throughout pregnancy (ΔT3-T1) were associated with higher DNAm levels (Fig. 2A and Supplementary Table 3). All the previous associations were independent of maternal blood lipids levels at second and third trimester of pregnancy (Supplementary Table 4) and were adjusted for maternal age and BMI at first trimester, weight gain between first and third trimester of pregnancy, gestational age, and newborn gender.

Fig. 1. Correlation between mean LDLR-CpGA DNAm levels, maternal total cholesterol level variation between third and first trimester of pregnancy (Panel A), and relative LDLR mRNA levels in placenta (Panel B). – (a) Adjusted for maternal age, maternal IMC at first trimester of pregnancy, total gestational weight gain, gestational age, and newborn’s gender. (b) Adjusted for gestational age and newborn’s gender.

Fig. 2. Correlation between LRP1-CpGA2 DNAm levels, maternal and fetal characteristics: (A) maternal total cholesterol level variation between third and first trimester of pregnancy; (B) cord blood triglyceride levels; (C) cord blood leptin levels; and (D) relative LRP1 mRNA levels. – (a) Adjusted for maternal age, maternal IMC at first trimester of pregnancy, total gestational weight gain, gestational age, and newborn’s gender. (b) Correlation coefficients (r) and p-values were computed after triglyceride levels were log10-transformed. (c) Adjusted for gestational age and newborn’s gender.

Fig. 3. Correlation between LRP1-CpGC1 DNAm levels and fetal characteristics: (A) cord blood total cholesterol levels; (B) cord blood LDL-cholesterol levels; (C) relative LRP1 mRNA levels. – (a) Adjusted for maternal age, maternal IMC at first trimester of pregnancy, total gestational weight gain, gestational age, and newborn’s gender. (b) Adjusted for gestational age and newborn’s gender.
We then applied stepwise multivariable linear regression analyses to assess which of the identified factors were independent predictors for DNAm levels. Predictors of placental LDLR-CpGA DNAm levels were changes in total cholesterol levels throughout pregnancy (ΔT3-T1), sex of the newborn, and total gestational weight gain, whereas changes in total cholesterol levels throughout pregnancy (ΔT3-T1), maternal age, and BMI at the first trimester of pregnancy predicted LRP1-CpGA2 DNAm levels in the placenta. Overall, these models explained 19.1% and 11.9% of the mean LDLR-CpGA and LRP1-CpGA2 DNAm variations, respectively (Table 4). No maternal characteristic was found to be associated with DNAm levels at LRP1-CpGC1 or LRP1-CpGD1.
Table 4. Multivariable linear regression analyses of LDLR and LRP1 DNA methylation levels predictors in placenta

BMI, body mass index; LDLR, Low-density lipoprotein receptor; LRP1, LDL receptor-related protein 1; NS, not statistically significant.
All β are standardized coefficient values.
Variables with a p-value < 0.10 were retained in the regression model.
Next, we assessed whether DNAm at LDLR and LRP1 gene loci was associated with newborns’ anthropometric characteristics and cord blood leptin and lipid profile (Supplementary Table 5). We observed that higher LRP1-CpGA2 DNAm levels were associated with lower cord blood triglycerides (TGs) and leptin levels (Fig. 2B, 2C). Moreover, LRP1-CpGC1 DNAm levels were positively correlated with cord blood total cholesterol and LDL-C levels (Fig. 3A, 3B).
Mediation analyses were finally applied to assess whether DNAm levels at LDLR and LRP1 gene loci might be part of the mechanisms linking maternal and newborns’ phenotypes. We found that LRP1-CpGA2 DNAm levels significantly mediate the association between changes in maternal total cholesterol levels during pregnancy (ΔT3-T1) and cord blood leptin levels (Fig. 4; p mediation = 0.02). We did not detect significant mediation of DNAm at LDLR gene loci.

Fig. 4. Mediation analysis between maternal total cholesterol changes in pregnancy, LRP1 DNAm, and cord blood leptin levels. These results show that DNAm level variations at LRP1-CpGA2 induced by mediating the association between maternal total cholesterol changes in pregnancy and cord blood leptin levels in newborns, suggesting a causal relationship between these variables. Path a is the relationship between the ΔT3-T1 maternal total cholesterol levels and LRP1 DNAm levels. Path b is the relationship between LRP1 DNAm levels and cord blood leptin levels. Path c is the direct relationship between ΔT3-T1 maternal total cholesterol levels and cord blood leptin levels, while path c’ is the indirect effect of this relationship when taking into account the LRP1 DNAm profile.
Discussion
Blood lipid levels (TG, LDL-C, and HDL-C) during pregnancy were previously associated with fetal growth and development indices suggesting that maternal dyslipidemia may be involved in fetal metabolic programming.Reference Mudd, Holzman and Evans8, Reference Chatzi, Plana and Daraki34–Reference Ramsay, Ferrell, Crawford, Wallace, Greer and Sattar36 Nevertheless, these associations seemed to be mediated by several other maternal factors including the lipoprotein subtype, maternal BMI (pre- and post-conception), and/or diabetes,Reference Mudd, Holzman and Evans8 which have been supported by a recent Mendelian randomization analysis.Reference Tyrrell, Richmond and Palmer37 In this study, we have decided to focus on maternal blood lipid levels changes throughout pregnancy, which have been overlooked so far. We report that rise in maternal blood lipids levels in pregnancy, in normoglycemic pregnant women, is associated with lower ponderal index and cord blood leptin levels. These associations were not observed with maternal blood lipid levels at first, second, and third trimester of pregnancy, suggesting that changes throughout the pregnancy might be more important than the absolute lipid values at the beginning, middle, or end of pregnancy.
Previous studies suggested that in utero environment perturbations such as maternal dyslipidemia or hyperglycemia contribute to the development of adult chronic diseases possibly through epigenetic adaptations in the placenta and offspring.Reference Clausen, Mathiesen and Hansen38, Reference Van der Graaf, Vissers and Gaudet39 We report that rise in maternal blood lipids levels in pregnancy is associated with DNAm levels variability at LDLR and LRP1 gene loci in placenta (Fig. 5). These results suggest that LDLR and LRP1 epigenetic profile are sensitive to the in utero environment and that DNAm at these loci could explain, to some extent, the link between suboptimal maternal condition and the metabolic health programming in the newborn. In this study, we showed that DNAm variability at LDLR gene locus was associated with functional impact on gene expression. Although the association between LRP1 DNAm and mRNA levels did not reach statistical significance (p < 0.10 but >0.05), the correlation coefficients obtained were modestly strong (r > 0.25) as compared to previous studies showing functional impact of DNAm on gene expression in placenta.Reference Houde, Guay and Desgagne20, Reference Bouchard, Thibault and Guay29 A significant statistical association (p < 0,05) between LRP1 DNAm and mRNA levels could potentially have been reached with larger sample size. Moreover, the gene expression and DNAm studies were done on placental sample collected at delivery. Gene expression in placenta was previously showed to be highly variable according to trimesters.Reference Sitras, Fenton, Paulssen, Vartun and Acharya40 Henceforth, the associations observed between DNAm and mRNA levels at the end of the pregnancy might be even stronger during the first or second trimester. This mechanism may indeed be important for regulation of the lipoprotein metabolism in maternal–placental fetal unit throughout the pregnancy. The functional impact of LRP1 DNAm throughout pregnancy will need to be confirmed in larger cohort.
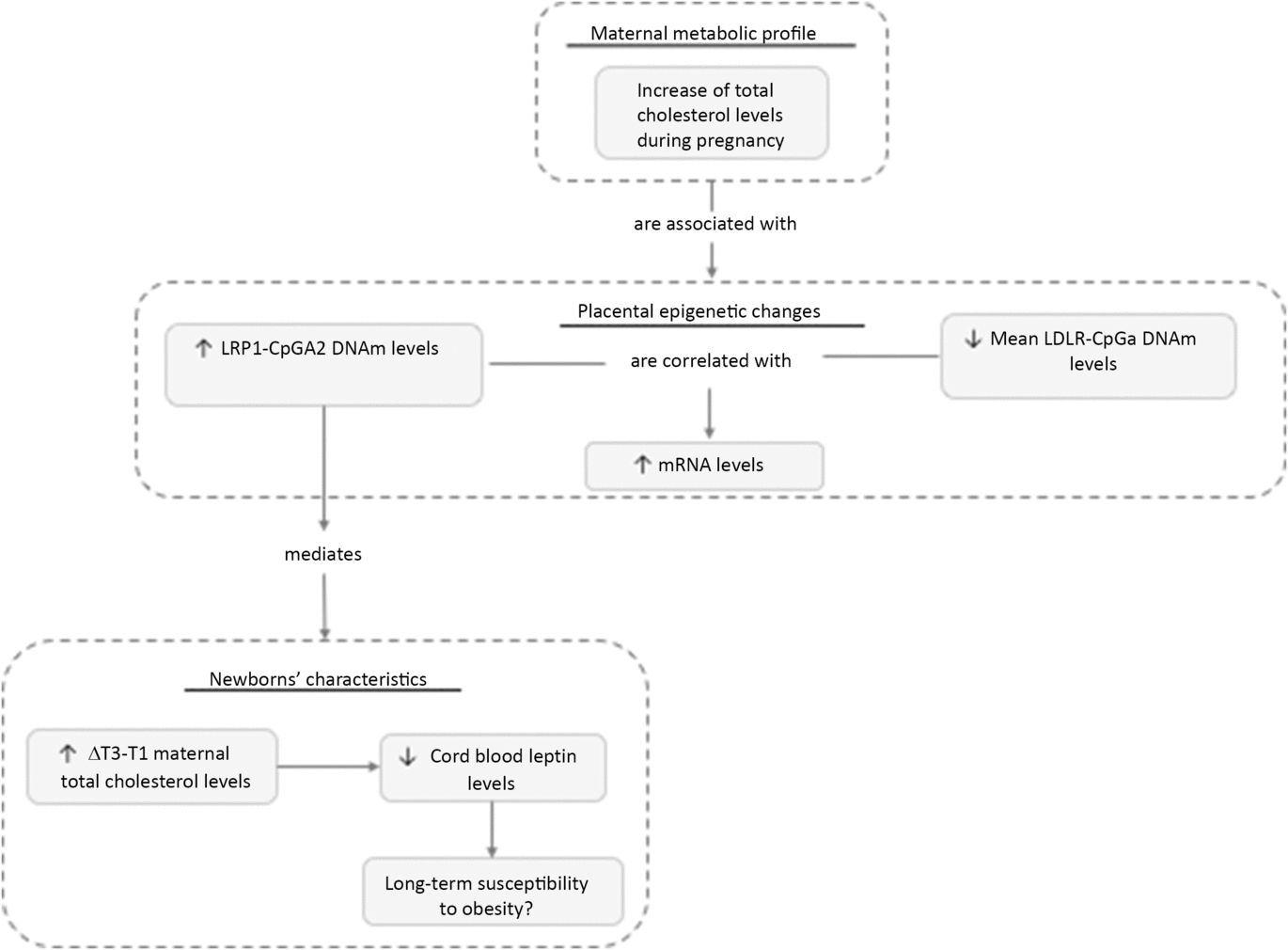
Fig. 5. Maternal metabolic variables associated with placental epigenetics changes and newborns’ characteristics. Maternal blood lipid level variations throughout pregnancy were associated with placental DNAm levels changes at LDLR and LRP1 gene loci. Our results suggest that DNAm at LRP1 gene locus is causally associated with cord blood leptin levels, a biomarker of adiposity.
Using mediation analyses, we then explored the causality chain between maternal blood lipid changes, placental DNAm variations, and newborns’ anthropometric and metabolic profiles (Figs. 4 and 5). We report that the associations between maternal total cholesterol changes in pregnancy and cord blood leptin levels are mediated, at least partially, by placental epigenetic adaptations at the LRP1 gene locus. This suggests that placental DNAm responds to maternal lipid changes in pregnancy, which in turn impacts fetal growth. As we are studying complex traits, there are probably other factors contributing to interindividual variability in cord blood leptin levels, such as genetic, diet, or other environmental risk factors.Reference Rakyan, Down, Balding and Beck41 However, the fact that only one CpG site can explain that much of this variability is highly interesting. Moreover, even if the observed DNAm variations in our study are relatively modest, we and other groups already shown these can affect gene expression and hence could obviously have physiological consequences over a long period of time.Reference Houde, Guay and Desgagne20, Reference Toperoff, Aran and Kark42
Interestingly, lower placental LRP1 mRNA levels have already been reported in pregnancies complicated by preeclampsia and characterized by a suboptimal fetal development.Reference Hentschke, Poli-de-Figueiredo, da Costa, Kurlak, Williams and Mistry10, Reference Wadsack, Tabano and Maier11, Reference Stepan, Faber and Walther43 In addition, hypercholesterolemia in pregnant rabbit has been associated with placental collagen deposition and atherosclerotic lesions that compromised placental nutrients transfer from the maternal circulation to the fetus.Reference Frantz, Menezes and Lange44In utero exposure to increased maternal cholesterol levels may thus impair placental function and restrict materno-fetal nutrients transport leading to developmental restriction, possibly manifested as lower cord blood leptin levels in our study. Henceforth, we hypothesized that epigenetic modifications at LRP1 locus in response to risen maternal blood lipids are an attempt to improve materno-placental lipids transport in functionally impaired placenta. Nevertheless, this may also have triggered a compensatory response in the newborns (increased cord blood lipids), which might increase their long-term susceptibility to CVD and adult metabolic diseases.
LRP1 encodes for a large cell surface receptor ubiquitously expressed in a variety of organs including, placenta, adipose tissue, liver, and brain.Reference Hussain, Strickland and Bakillah45 It is also highly expressed in neurons: a lack of LRP1 expression caused motor and behavioral impairment (i.e., hyperactivity, tremor, dystonia) on transgenic mice, suggesting altered neurotransmission.Reference May, Rohlmann and Bock46 Interestingly, LRP1 expression in the brain could also affect body weight and adiposity through the regulation of food intake and energy expenditure: LRP1-KO mice showed significantly accelerated body weight gain, insulin resistance, hyperlipidemia, and an increase in plasma leptin concentration suggesting the development of leptin resistance.Reference Liu, Zhang and Zerbinatti47 Similarly, we report that decreased LRP1 DNAm in placenta and corresponding mRNA levels are associated with higher cord blood leptin concentration. However, some mechanisms behind LRP1 function are still unclear, with at least one other study reporting that the inactivation of LRP1 in white and brown adipose tissues decreased fat storage, and had protective effects against diet-induced obesity.Reference Hofmann, Zhou and Perez-Tilve48LRP1 inactivation also protected the mice from glucose intolerance and diabetes potentially because of their increased metabolic rate and energy expenditure, which supports that LRP1 is probably an important regulator of adipocyte energy homeostasis.Reference Hofmann, Zhou and Perez-Tilve48 Interestingly, our study suggested that LRP1 epigenetic profile in placenta is sensitive to the maternal blood lipid levels variability during the pregnancy. Previous studies showed that DNAm variability observed in a specific tissue might also be reflected in other tissues and have tissue-specific impact on gene expression regulation.Reference Guay, Brisson and Lamarche32 Moreover, recent researches suggest that newborns’ methylome in different tissues, not only in placenta, might also be sensitive to suboptimal maternal condition.Reference Cardenas, Koestler and Houseman49, Reference Banik, Kandilya, Ramya, Stunkel, Chong and Dheen50 Therefore, it possible that LRP1 DNAm profile in newborns’ brain or adipose tissue cells might have been affected by maternal blood lipid variability during pregnancy and could explain, to some extent, the link between suboptimal maternal condition, cord blood leptin levels, and newborns’ anthropometric measurements. Nevertheless, this remains a hypothesis that will need to be investigated.
The longitudinal follow-up of our women from the first trimester of pregnancy to delivery is one of the strengths of this study. It has allowed us to accurately assess changes in maternal fasting blood lipids throughout pregnancy and its impact on the newborn methylome, anthropometric characteristics, and cord blood lipid and leptin levels. However, we cannot exclude that DNAm levels at LDLR and LRP1 loci may have caused fluctuations in maternal lipemia during pregnancy because placenta samples were only analyzed at birth. The relatively small sample size (n = 69) and the common ethnicity of the population studied are also limiting our study, as our results may not be representative of a more diversified population. Moreover, the risk of false positive due to multiple testing cannot be excluded, as we tested a number of 4 CpG loci. Further studies are thus needed to confirm our result and to assess the functional impacts of the maternal lipid level variation on placental methylome, transcriptome, and newborns’ characteristics.
Conclusion
Our study showed that maternal blood lipids levels variability in pregnancy, in normoglycemic pregnant women, is associated with placental DNAm levels changes and newborn’s anthropometric measurements and blood marker of adiposity, such as cord blood leptin levels. Our findings support that placental DNAm at LRP1 gene loci mediates the relationship between maternal blood lipid changes during pregnancy and cord blood leptin levels. This is of great interest, as we know that lipid transfer from the mother through the placenta is an important factor for adequate fetal growth and development. Moreover, these findings support that epigenetic is a crucial element in human development, although more studies are needed to fully elucidate the long-term impacts of fetal epigenetic programming. As obesity and diabetes are of great health concern these days, a better understanding of how maternal metabolic factors can influence children metabolism could be helping in the worldwide effort to prevent such epidemics.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S204017441900076X
Acknowledgments
The authors acknowledge the contribution of Cécilia Légaré, MSc, Sébastien Claveau, MSc, Nadia Mior, Jeanine Landry, RN, and Chantal Aubut, RN from the ECOGENE-21 Biocluster for their dedicated work in this study. The authors also express their gratitude to Céline Bélanger, Chicoutimi Hospital, for her thoughtful language revision of the manuscript. LB is a junior research scholar from the Fonds de la recherche du Québec en santé (FRQS).
Authors’ Contributions
AAH and SPG contributed to the study design, participated to data collection analysis/interpretation, and wrote the manuscript. EB critically reviewed the literature and wrote the manuscript. MFH, JPB, and PP participated in study design conception and revised the manuscript. DG contributed to the study design and revised the manuscript. DB participated to data collection and analysis/interpretation and revised the manuscript. LB conceived the study design, participated to data collection and analysis/interpretation, and revised the manuscript.
Financial Support
MFH is a recipient of an American Diabetes Association Pathways Accelerator Award (#1-15-ACE-26). LB, MFH, PP, and JPB are members of the FRQS-funded Centre de recherche du Centre hospitalier universitaire de Sherbrooke. During this research, SPG was the recipient of a doctoral research award from the Canadian Institutes for Health and Research (CIHR), whereas AAH was a recipient of a FRQS doctoral training award and was supported by Diabète Québec. This project was supported by ECOGENE-21, the Canadian Institutes of Health Research (CIHR team in community genetics (grant #CTP–82941)), FRSQ, and Diabète Québec.
Conflicts of Interest
None.
Ethical Standards
A written informed consent was obtained for all women before their inclusion in the study, and all clinical data were de-identified. This project received the approval by the Chicoutimi Hospital Ethics Committee, in accordance with the Declaration of Helsinki.
Availability of Data and Material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.











