INTRODUCTION
Cryptosporidiosis is a highly prevalent and extremely widespread disease documented by over 1000 reports in humans in 95 countries on all continents except Antarctica (Fayer et al. Reference Fayer, Speer, Dubey and Fayer1997). Considering that cryptosporidiosis is primarily spread by ingestion of contaminated water, was ranked fifth among the 24 most important food-borne parasites in a global ranking by a joint Food and Agriculture Organization (FAO)/World Health Organization (WHO) expert committee in 2012 (http://www.who.int/foodsafety/micro/jemra/meetings/sep12/en/), and can be spread by close proximity to infected humans and animals, the importance of this genus in human and animal health has long been underestimated. Of approximately 8 million worldwide annual deaths of children under 5 years of age, diarrhoea is associated with 10·5% (Liu et al. Reference Liu, Johnson, Cousens, Perin, Scott, Lawn, Rudan, Campbell, Cibulskis, Li, Mathers and Black2012). An epidemiological study of over 22 000 infants and children in Africa and Asia found that Cryptosporidium was one of the four pathogens responsible for most of the severe diarrhoea (Kotloff et al. Reference Kotloff, Nataro, Blackwelder, Nasrin, Farag, Panchalingam, Wu, Sow, Sur, Breiman, Faruque, Zaidi, Saha, Alonso, Tamboura, Sanogo, Onwuchekwa, Manna, Ramamurthy, Kanungo, Ochieng, Omore, Oundo, Hossain, Das, Ahmed, Qureshi, Quadri, Adegbola, Antonio, Hossain, Akinsola, Mandomando, Nhampossa, Acacio, Biswas, O'Reilly, Mintz, Berkeley, Muhsen, Sommerfelt, Robins-Browne and Levine2013) and was considered the second greatest cause of diarrhoea and death in children after rotavirus (Striepen, Reference Striepen2013). In developing countries cryptosporidiosis is reported to account for 20% of all cases of diarrhoea in children (Mosier and Oberst, Reference Mosier and Oberst2000) and, depending on location, at some point in life the percentage of affected persons in a population was estimated to range from 20–90% (Dillingham et al. Reference Dillingham, Lima and Guerrant2002). In the USA, there are approximately 748 000 cases of cryptosporidiosis annually and hospitalizations from cryptosporidiosis cost an estimated $45.8 million (Scallan et al. Reference Scallan, Hoekstra, Angulo, Tauxe and Hoekstra2011). Unfortunately, cryptosporidiosis data are lacking for 11 of the 30 European Union and EEA/EFTA countries and data are underreported for those that did report; in 2007, only 6253 cases, all confirmed, were reported (Anonymous, 2010).
In general, three factors contribute significantly to the success of Cryptosporidium as a parasite. Large numbers of oocysts are excreted into the environment by infected individuals. Oocysts are environmentally hardy and can survive for many months in temperate and moist conditions. And infection can be initiated by a very small number of oocysts; theoretically a single oocyst could cause infection in a susceptible person. In human volunteer studies a median infectious dose for Cryptosporidium hominis ranged from 10 to 83 oocysts and for Cryptosporidium parvum from below 10 to over 1000 oocysts can initiate infection (Okhuysen et al. Reference Okhuysen, Chappell, Crabb, Sterling and DuPont1999; Chappell et al. Reference Chappell, Okhuysen, Langer-Curry, Widmer, Akiyoshi, Tanriverdi and Tzipori2006).
The lack of widespread prophylactic and therapeutic treatment options for cryptosporidiosis in humans and animals also has permitted a high prevalence and widespread distribution of the parasite. Although hundreds of drugs have been tested for prophylaxis and treatment of cryptosporidiosis in animals and humans (Fayer et al. Reference Fayer, Speer, Dubey and Fayer1997), only one has been approved by the US Food and Drug Administration (FDA). Nitazoxanide (Alinia®) liquid has been indicated for treatment of diarrhoea caused by Giardia and Cryptosporidium in patients one year and older and Alinia tablets have been approved for patients 12 years and older. However, Alinia has not been found to be better than a placebo for treatment of cryptosporidiosis in HIV-infected or immunodeficient patients (http://www.romark.com/alinia-product-information). FAO's executive summary of the State of Food Insecurity in the World (http://www.fao.org/docrep/018/i3458e/i3458e.pdf) indicates there are 842 million chronically malnourished persons worldwide, a factor that also significantly contributes to impaired immunity. Furthermore, the epidemiology of diarrhoea in children is thought to overlap with that of pneumonia, perhaps owing to shared risk factors such as insufficient nutrition, suboptimal breastfeeding and zinc deficiency (Walker et al. Reference Walker, Rudan, Liu, Nair, Theodoratou, Bhutta, O'Brien, Campbell and Black2013). Rotavirus, a major cause of diarrhoea in children, is vaccine-preventable, but there is no vaccine for cryptosporidiosis and few published reports of attempts to develop immunoprophylaxis-based modalities (McDonald, Reference McDonald2011).
Because oocysts of Cryptosporidium species from humans and animals are ubiquitous in the environment, cryptosporidiosis can be acquired through multiple routes (reviewed by Robertson and Fayer, Reference Robertson, Fayer, Robertson and Smith2013). Transmission of oocysts is by the faecal–oral route, either directly or indirectly. For humans, direct transmission can be from one person to others primarily from poor hygiene among household members and attendees in day-care centres, nursing facilities, or other institutions, or from animals to persons such as farm-workers, pet owners, veterinarians and farm visitors. Most indirect transmission is from contaminated drinking water or recreational water (especially from faecal accidents in swimming pools). Contaminated food can be a source of transmission, starting at production sites where crops can be contaminated from contact with manure, from contaminated irrigation water, or from the hands of agricultural workers. Food can be contaminated at any point during distribution and preparation by food handlers, washwater, preparation surfaces, equipment or utensils. Although the primary foods implicated in transmission are raw fruits and vegetables, transmission associated with unpasteurized milk and apple cider, raw meat and sauce has also been documented. Although transmission has been reported from contaminated soil virtually nothing is known of the prevalence of infections from camping or gardening activities.
With cryptosporidiosis so widespread and prevalent, and with prophylaxis and therapeutic treatment options so limited, the ability to prevent and control disease appears best served by sanitation. The availability of clean water and toilets, fastidious handling of food and a clear understanding of the sources of Cryptosporidium provide a basis for prevention of transmission. This last topic, prevention, encompasses epidemiology that requires knowledge of the biology and taxonomy of the members of the genus responsible for disease. Because the oocysts of many species are indistinguishable from one another, molecular methods are essential for identification of the species, genotype and subtype of Cryptosporidium to specifically identify the organism responsible for infection and the source and routes of transmission (Xiao and Feng, Reference Xiao and Feng2008).
STANDARDS FOR TAXONOMY OF CRYPTOSPORIDIUM SPP
Clearly described species with stable scientific names are basic and essential for understanding epidemiology. Principals for naming and stability are provided by the International Code of Zoological Nomenclature (ICZN). For names published after 1930 Article 13 of the ICZN states that ‘when describing a new nominal taxon, an author should make clear his or her purpose to differentiate the taxon by including with it a diagnosis, that is to say, a summary of the characters that differentiate the new nominal taxon from related or similar taxa’. The lack of a description of the morphology and other unique features renders the name a nomen nudum, i.e. a naked name, without support, making it non-valid. In such cases the name is available for use at a later time, if desired. Article 13 and recent descriptions that have adopted the guidelines of Xiao et al. (Reference Xiao, Fayer, Ryan and Upton2004, Reference Xiao, Fayer, Ryan and Upton2007) have been applied for accepting or rejecting those species names provided in Tables 1 and 2. The guidelines include the need to: provide morphometric data on oocysts; provide genetic characterization; demonstrate natural, and when feasible, experimental host specificity; and comply with ICZN rules. Those species that meet the guidelines appear in Table 1 and those that do not appear in Table 2 with reasons for making them non-valid species. In addition to these established Cryptosporidium spp., there are also over 40 genotypes of unknown species status. Like most Cryptosporidium species, these genotypes appear largely host-adapted (Feng, Reference Feng2010; Ryan and Power, Reference Ryan and Power2012). Some of them are likely to be named new Cryptosporidium species when biological data become available.
Table 1. Valid species of Cryptosporidium. Many of these earlier species were originally described based on morphological criteria but have subsequently been validated using molecular data
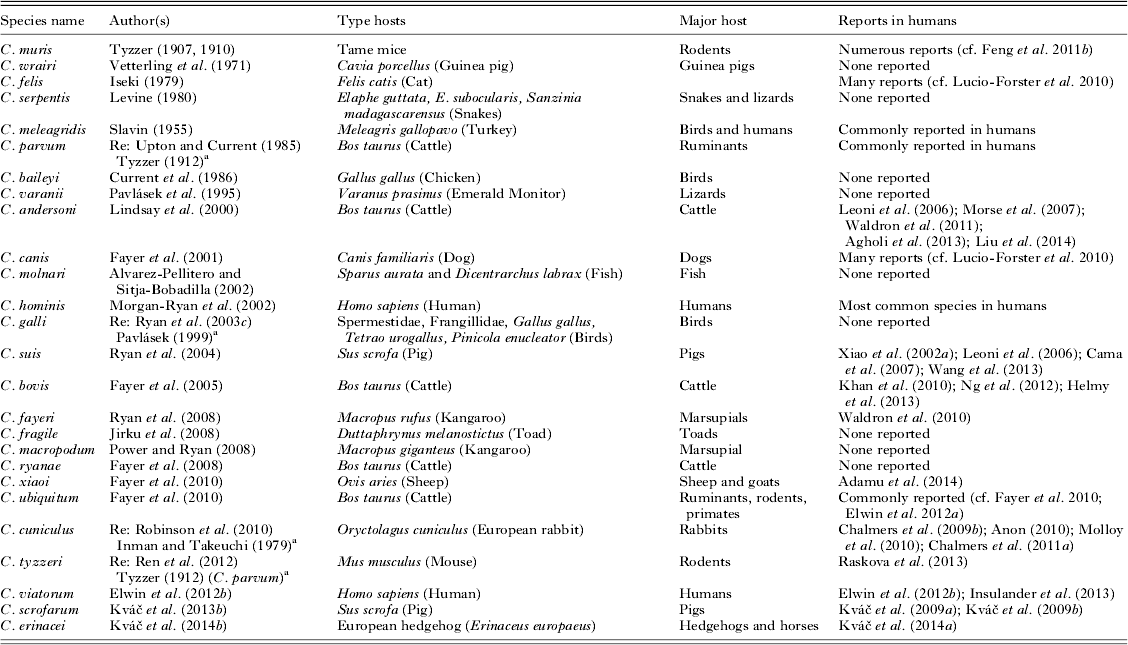
Re, redescription.
a Initial description.
Table 2. Non-valid species names associated with Cryptosporidium

MOLECULAR TYPING TOOLS
After DNA has been extracted from oocysts obtained from environmental, food or biological specimens, different types of molecular diagnostic tools have been used in the identification of Cryptosporidium spp. These tools can be categorized into genotyping, subtyping, multilocus typing/population genetics, and comparative genomics depending on the approaches and usages.
Genotyping
Many small subunit (SSU) rRNA-based tools have been developed for the identification of Cryptosporidium species in humans, animals and water samples. In particular, a PCR-RFLP tool that targets an ∼830-bp fragment of the gene and uses SspI and VspI restrictions for genotyping (Xiao et al. Reference Xiao, Morgan, Limor, Escalante, Arrowood, Shulaw, Thompson, Fayer and Lal1999) is commonly used in the differentiation of Cryptosporidium species in humans, animals and environmental samples. For the analysis of specimens from ruminants, the method has been modified by using SspI and MboII in the RFLP analysis (Feng et al. Reference Feng, Ortega, He, Das, Xu, Zhang, Fayer, Gatei, Cama and Xiao2007). Another format commonly used for genotyping is DNA sequencing of PCR products (Koinari et al. Reference Koinari, Karl, Ng-Hublin, Lymbery and Ryan2013). In recent years, qPCR assays using fluorescent probes and melting curve analysis are increasingly used in Cryptosporidium genotyping (Jothikumar et al. Reference Jothikumar, da Silva, Moura, Qvarnstrom and Hill2008; Hadfield et al. Reference Hadfield, Robinson, Elwin and Chalmers2011; Burnet et al. Reference Burnet, Ogorzaly, Tissier, Penny and Cauchie2012; Lalonde et al. Reference Lalonde, Reyes and Gajadhar2013; Mary et al. Reference Mary, Chapey, Dutoit, Guyot, Hasseine, Jeddi, Menotti, Paraud, Pomares, Rabodonirina, Rieux and Derouin2013; Staggs et al. Reference Staggs, Beckman, Keely, Mackwan, Ware, Moyer, Ferretti, Sayed, Xiao and Villegas2013; Yang et al. Reference Yang, Murphy, Song, Ng-Hublin, Estcourt, Hijjawi, Chalmers, Hadfield, Bath, Gordon and Ryan2013). Although the range of detection or differentiation for Cryptosporidium species is in general narrower, these newer methods are simpler to use and less prone to PCR contamination. The widespread use of the SSU rRNA gene in Cryptosporidium genotyping is largely due to the multi-copy nature of the gene and presence of semi-conserved and hyper-variable regions, which facilitate the design of genus-species primers. In many Cryptosporidium species and genotypes, minor intra-isolate sequence variations are present among different copies of the SSU rRNA gene. Thus, new genotypes should not be named based on one or two nucleotide substitutions or insertions/deletions in the gene (Abeywardena et al. Reference Abeywardena, Jex, Koehler, Rajapakse, Udayawarna, Haydon, Stevens and Gasser2014).
PCR tools based on other genes in general only amplify DNA of C. parvum, C. hominis, Cryptosporidium meleagridis and species/genotypes closely related to them. Thus, studies that have used these tools have usually showed lower Cryptosporidium species diversity (Abd El Kader et al. Reference Abd El Kader, Blanco, Ali-Tammam, Abd El Ghaffar Ael, Osman, El Sheikh, Rubio and de Fuentes2012; Berrilli et al. Reference Berrilli, D'Alfonso, Giangaspero, Marangi, Brandonisio, Kabore, Gle, Cianfanelli, Lauro and Di Cave2012). These tools have limited usefulness in genotyping Cryptosporidium spp. of animals because of their narrow specificity. They, nevertheless, can be used in the identification of mixed infections with C. hominis or C. parvum in humans in developing countries that have been infected with rare Cryptosporidium species based on PCR analysis of the SSU rRNA gene (Cama et al. Reference Cama, Gilman, Vivar, Ticona, Ortega, Bern and Xiao2006). A few other markers such as the 90 kDa heat shock protein and A135 genes have been used in the development of genus-specific PCR-RFLP tools for genotyping Cryptosporidium (Feng et al. Reference Feng, Dearen, Cama and Xiao2009; Tosini et al. Reference Tosini, Drumo, Elwin, Chalmers, Pozio and Caccio2010). Recently, a genus-specific Cryptosporidium qPCR based on the actin gene has been developed. Although it currently can only be used in screening of Cryptosporidium spp., genotyping can be done subsequently using species-specific qPCR (Yang et al. Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014).
Subtyping
Subtyping tools have been used extensively in studies of the transmission of C. hominis in humans, C. parvum in humans and ruminants, and a few other related Cryptosporidium species such as C. meleagridis and Cryptosporidium ubiquitum in both humans and animals (Xiao, Reference Xiao2010; Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014). One of the most common subtyping tools is the DNA sequence analysis of the 60 kDa glycoprotein (gp60, also known as gp40/15) gene. Most of the genetic heterogeneity in this gene is the variation in the number of a tri-nucleotide repeat (TCA, TCG or TCT) in the 5′ end (gp40) of the coding region, although extensive sequence polymorphism is also present in the rest of the gene. The latter is used in defining subtype families within a species, whereas the former is used in identifying subtypes within a subtype family. It should be kept in mind that commonly used gp60 PCR primers do not amplify DNA of C. ubiquitum, Cryptosporidium felis, Cryptosporidium canis and other species distant from C. parvum and C. hominis (Feng et al. Reference Feng, Lal, Li and Xiao2011a ; Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014).
An established subtype nomenclature is used in identifying gp60 subtype family. A subtype name starts with the species and subtype family designation (Ia, Ib, Id, Ie, If, etc. for C. hominis; IIa, IIb, IIc, IId, etc. for C. parvum; IIIa, IIIb, IIIc, IIId, etc. for C. meleagridis; see Table 3 for subtype families of other Cryptosporidium species) followed by the number of TCA (represented by the letter A), TCG (represented by the letter G) or TCT (represented by the letter T) repeats (Sulaiman et al. Reference Sulaiman, Hira, Zhou, Al-Ali, Al-Shelahi, Shweiki, Iqbal, Khalid and Xiao2005; Xiao, Reference Xiao2010; Feng et al. Reference Feng, Lal, Li and Xiao2011a ). Thus, the name IeA11G3T3 indicates that parasite belongs to C. hominis subtype family Ie and has 11 copies of the TCA repeat, three copies of the TCG repeat and three copies of the TCT repeat in the trinucleotide repeat region of the gene.
Table 3. Major gp60 subtype families of Cryptosporidium spp. and representative sequences
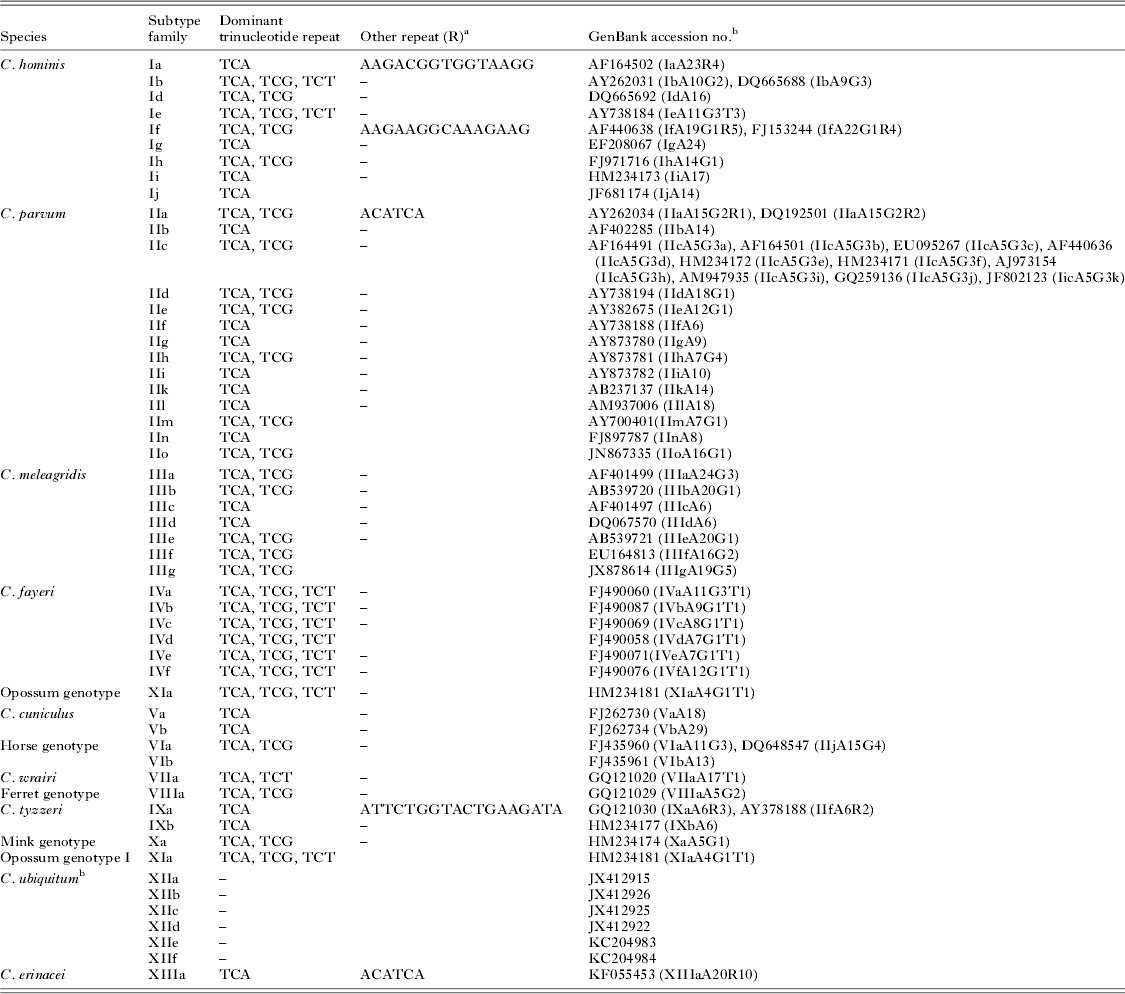
a Consensus repeat; minor sequence variation exists among repeats.
b The gp60 gene of C. ubiquitum subtypes has no trinucleotide repeats, thus cannot be named at the subtype level using the established nomenclature.
In the gp60 gene of a few subtype families, variations in copy numbers of other repeat sequences (designed as R at the end of the subtype name) are also present in the gp40 region. Thus, in the C. parvum IIa subtype family, some subtypes have two or three copies of the ACATCA sequence right after the trinucleotide repeats, which are represented by ‘R2’ or R3 (R1 for most subtypes). Likewise, within the C. hominis subtype family Ia, subtypes are further differentiated by the copy number of a 15-bp repetitive sequence 5′-AA/GGACGGTGGTAAGG-3′ (the last copy is 13-bp: AAA/GACGGTGAAGG) in the gp40 region of the gene. Thus, the name IaA28R4 indicates that parasite belongs to C. hominis subtype family Ia and has 28 copies of the TCA repeat in the trinucleotide region and four copies of the 13–15 bp repeats downstream. Variations in other repeats also occur in other less common C. hominis subtype families such as If. In some subtype families such as C. parvum subtype family IIc, there is no sequence difference in the trinucleotide region (all IIc subtypes have five copies of TCA and three copies of TCG repeats: IIcA5G3). Instead, subtypes differ from each other in the nucleotide sequence of the 3′ region (mostly gp15) of the gene. Subtypes within this subtype families are differentiated by alphabetical extensions, with the original gp60 sequence for the subtype family (GenBank accession number AF164491) assigned as IIcA5G3a (Table 3). Representative sequences of some common subtype families are shown in Table 3.
An advantage of using gp60 for subtyping is the potential association between subtype families and phenotypes of C. parvum and C. hominis. This could be due to the biological importance of the gp60 protein, which is located on the surface of apical region of invasive stages of the parasite, and is one of the dominant targets for neutralizing antibody responses in humans (O'Connor et al. Reference O'Connor, Wanyiri, Cevallos, Priest and Ward2007). Some of the C. parvum subtype families, such as IIa and IId, are found in both humans and ruminants, responsible for zoonotic cryptosporidiosis (Xiao, Reference Xiao2010). In areas with both IIa and IId, such as Spain, IIa subtypes preferentially infect calves whereas IId subtypes preferentially infect lambs and goat kids (Diaz et al. Reference Diaz, Quilez, Chalmers, Panadero, Lopez, Sanchez-Acedo, Morrondo and Diez-Banos2010a ; Quilez et al. Reference Quilez, Vergara-Castiblanco, Monteagudo, Del Cacho and Sanchez-Acedo2013). Some other C. parvum subtype families, especially IIc, have so far only been found in humans (Xiao and Feng, Reference Xiao and Feng2008; Widmer and Lee, Reference Widmer and Lee2010). Host adaptation has recently also been identified in C. ubiquitum based on sequence analysis of its gp60 gene (Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014). There are also significant differences in clinical presentations and virulence among some common C. hominis or C. parvum subtype families in cryptosporidiosis-endemic areas (Cama et al. Reference Cama, Ross, Crawford, Kawai, Chavez-Valdez, Vargas, Vivar, Ticona, Navincopa, Williamson, Ortega, Gilman, Bern and Xiao2007, Reference Cama, Bern, Roberts, Cabrera, Sterling, Ortega, Gilman and Xiao2008; Del Chierico et al. Reference Del Chierico, Onori, Di Bella, Bordi, Petrosillo, Menichella, Caccio, Callea and Putignani2011; Feng et al. Reference Feng, Wang, Duan, Gomez-Puerta, Zhang, Zhao, Hu, Zhang and Xiao2012). Some gp60 subtypes of C. hominis and C. parvum, such as IbA10G2 of C. hominis and IIaA15G2R1 of C. parvum, are widely distributed in the world probably due to their biological fitness (Budu-Amoako et al. Reference Budu-Amoako, Greenwood, Dixon, Sweet, Ang, Barkema and McClure2012a ; Feng et al. Reference Feng, Torres, Li, Wang, Bowman and Xiao2013; Li et al. Reference Li, Xiao, Cama, Ortega, Gilman, Guo and Feng2013).
Multilocus typing and population genetics
The whole genome sequencing (WGS) of C. parvum, C. hominis and Cryptosporidium muris has allowed the identification of microsatellite and minisatellite sequences in Cryptosporidium genomes and other targets that are highly polymorphic between C. parvum and C. hominis (Robinson and Chalmers, Reference Robinson and Chalmers2012; Li et al. Reference Li, Xiao, Cama, Ortega, Gilman, Guo and Feng2013). They are frequently used in either multilocus fragment typing (MLFT) or multilocus sequence typing (MLST) to increase the subtyping resolution offered by gp60 sequence analysis (Quilez et al. Reference Quilez, Vergara-Castiblanco, Monteagudo, Del Cacho and Sanchez-Acedo2011, Reference Quilez, Vergara-Castiblanco, Monteagudo, Del Cacho and Sanchez-Acedo2013; Diaz et al. Reference Diaz, Hadfield, Quilez, Soilan, Lopez, Panadero, Diez-Banos, Morrondo and Chalmers2012; Herges et al. Reference Herges, Widmer, Clark, Khan, Giddings, Brewer and McEvoy2012; De Waele et al. Reference De Waele, Van den Broeck, Huyse, McGrath, Higgins, Speybroeck, Berzano, Raleigh, Mulcahy and Murphy2013; Feng et al. Reference Feng, Torres, Li, Wang, Bowman and Xiao2013, Reference Feng, Tiao, Li, Hlavsa and Xiao2014). Recently, a MLST tool has been developed for subtyping C. muris and Cryptosporidium andersoni (Feng et al. Reference Feng, Yang, Ryan, Zhang, Kvac, Koudela, Modry, Li, Fayer and Xiao2011b ). In addition to subtyping, MLFT and MLST data can be analysed for linkage disequilibrium and recombination rates with various population genetic tools such as DnaSP (http://www.ub.es/dnasp/), LIAN (http://pubmlst.org/perl/mlstanalyse/mlstanalyse.pl?site=pubmlst&page=lian&referer=pubmlst.org) and Arlequin 3.1 (http://cmpg.unibe.ch/software/arlequin3/). These data can also be analysed for population differentiation using the Bayesian analysis tool STRUCTURE (http://pritch.bsd.uchicago.edu/structure.html) and network analysis software eBURST (http://eburst.mlst.net/). Any population differentiation can be confirmed by the calculation of the Wright's fixation index (F ST). These tools have been used widely in population genetics studies and geographic tracking of C. parvum, C. hominis, C. muris and C. andersoni (Quilez et al. Reference Quilez, Vergara-Castiblanco, Monteagudo, Del Cacho and Sanchez-Acedo2011, Reference Quilez, Vergara-Castiblanco, Monteagudo, Del Cacho and Sanchez-Acedo2013; Herges et al. Reference Herges, Widmer, Clark, Khan, Giddings, Brewer and McEvoy2012; Diaz et al. Reference Diaz, Hadfield, Quilez, Soilan, Lopez, Panadero, Diez-Banos, Morrondo and Chalmers2012; Wang et al. Reference Wang, Jian, Zhang, Ning, Liu, Zhao, Feng, Qi, Wang, Lv, Zhao and Xiao2012; De Waele et al. Reference De Waele, Van den Broeck, Huyse, McGrath, Higgins, Speybroeck, Berzano, Raleigh, Mulcahy and Murphy2013; Feng et al. Reference Feng, Torres, Li, Wang, Bowman and Xiao2013, Reference Feng, Tiao, Li, Hlavsa and Xiao2014; Zhao et al. Reference Zhao, Ren, Gao, Bian, Hu, Cong, Lin, Wang, Qi, Qi, Zhu and Zhang2013). Some of these studies have shown a panmictic population structure with frequent recombination in C. parvum (Herges et al. Reference Herges, Widmer, Clark, Khan, Giddings, Brewer and McEvoy2012; De Waele et al. Reference De Waele, Van den Broeck, Huyse, McGrath, Higgins, Speybroeck, Berzano, Raleigh, Mulcahy and Murphy2013), whereas others have demonstrated the existence of a flexible reproductive strategy (co-occurrence of panmictic, clonal or epidemic structure) in this species (Tanriverdi et al. Reference Tanriverdi, Grinberg, Chalmers, Hunter, Petrovic, Akiyoshi, London, Zhang, Tzipori, Tumwine and Widmer2008; Drumo et al. Reference Drumo, Widmer, Morrison, Tait, Grelloni, D'Avino, Pozio and Caccio2012). Similarly, some genetic studies conducted on C. hominis identified largely a clonal population structure (Mallon et al. Reference Mallon, MacLeod, Wastling, Smith, Reilly and Tait2003; Gatei et al. Reference Gatei, Das, Dutta, Sen, Cama, Lal and Xiao2007; Li et al. Reference Li, Xiao, Cama, Ortega, Gilman, Guo and Feng2013; Feng et al. Reference Feng, Tiao, Li, Hlavsa and Xiao2014), whereas others showed the common occurrence of genetic recombination in C. hominis in developing countries (Widmer and Sullivan, Reference Widmer and Sullivan2012). In areas with an overall clonal population structure of C. hominis, genetic recombination has been shown to be a driving force for the emergence of virulent subtypes such as IbA10G2 and IaA28R4 (Li et al. Reference Li, Xiao, Cama, Ortega, Gilman, Guo and Feng2013; Feng et al. Reference Feng, Tiao, Li, Hlavsa and Xiao2014). Genetic recombination appears also to play an important role in the emergence of the virulent C. parvum subtype IIaA15G2R1 (Feng et al. Reference Feng, Torres, Li, Wang, Bowman and Xiao2013).
Comparative genomics
The WGS of C. parvum, C. hominis and C. muris and recent advances in next-generation sequencing (NGS) techniques have made the comparative genomic analysis of Cryptosporidium spp. possible. It is expected that comparative genomics of Cryptosporidium spp. will significantly improve our understanding of the transmission of cryptosporidiosis, especially in the investigation of outbreaks and monitoring of emerging and virulent subtypes (Widmer and Sullivan, Reference Widmer and Sullivan2012). Thus far, WGS analysis of Cryptosporidium spp. using NGS has only been done on one isolate each of the C. parvum anthroponotic subtype family IIc (Widmer et al. Reference Widmer, Lee, Hunt, Martinelli, Tolkoff and Bodi2012) and C. ubiquitum (Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014), although efforts are underway to sequence more isolates of Cryptosporidium species of public health and veterinary importance. NGS analysis of HSP70 and gp60 PCR products from two C. parvum isolates showed the presence of two HSP70 and 10 gp60 subtype in these isolates in spite of the initial detection of one subtype per locus using the traditional Sanger sequencing (Grinberg et al. Reference Grinberg, Biggs, Dukkipati and George2013). If verified, this finding of high intra-isolate heterogeneity would have important implications in our understanding of the epidemiology and population genetics of Cryptosporidium spp.
CRYPTOSPORIDIUM SPECIES IN HUMANS
Currently, nearly 20 Cryptosporidium species and genotypes have been reported in humans, including C. hominis, C. parvum, C. meleagridis, C. felis, C. canis, Cryptosporidium cuniculus, C. ubiquitum, Cryptosporidium viatorum, C. muris, Cryptosporidium suis, Cryptosporidium fayeri, C. andersoni, Cryptosporidium bovis, Cryptosporidium scrofarum, Cryptosporidium tyzzeri, Cryptosporidium erinacei and Cryptosporidium horse, skunk and chipmunk I genotypes (Xiao, Reference Xiao2010; Waldron et al. Reference Waldron, Dimeski, Beggs, Ferrari and Power2011; Elwin et al. Reference Elwin, Hadfield, Robinson and Chalmers2012a ; Ng et al. Reference Ng, Eastwood, Walker, Durrheim, Massey, Porigneaux, Kemp, McKinnon, Laurie, Miller, Bramley and Ryan2012; Kváč et al. Reference Kváč, Sakova, Kvetonova, Kicia, Wesolowska, McEvoy and Sak2013a ; Raskova et al. Reference Raskova, Kvetonova, Sak, McEvoy, Edwinson, Stenger and Kvac2013; Liu et al. Reference Liu, Shen, Yin, Yuan, Jiang, Xu, Pan, Hu and Cao2014). Humans are most frequently infected with C. hominis and C. parvum. Other species, such as C. meleagridis, C. felis, C. canis, C. cuniculus, C. ubiquitum and C. viatorum are less common. The remaining Cryptosporidium species and genotypes have been found in only a few human cases (Xiao, Reference Xiao2010; Elwin et al. Reference Elwin, Hadfield, Robinson and Chalmers2012a ). These Cryptosporidium spp. infect both immunocompetent and immunocompromised persons.
The distribution of these species in humans is different among geographic areas and socioeconomic conditions. In European countries and New Zealand, both C. hominis and C. parvum are commonly detected in humans. In contrast, C. parvum is the dominant species in humans in Middle Eastern countries, whereas C. hominis is the dominant species in other industrialized nations and developing countries (Xiao and Feng, Reference Xiao and Feng2008; Xiao, Reference Xiao2010; Nazemalhosseini-Mojarad et al. Reference Nazemalhosseini-Mojarad, Feng and Xiao2012). Likewise, human infections with C. canis and C. felis are reported in studies conducted in developing countries, C. ubiquitum mostly in industrialized nations and C. cuniculus mostly in the UK. Differences also exist in the distribution of C. parvum and C. hominis between urban and rural areas, with the former more commonly detected in rural and the latter in urban areas (Learmonth et al. Reference Learmonth, Ionas, Ebbett and Kwan2004; Llorente et al. Reference Llorente, Clavel, Goni, Varea, Seral, Becerril, Suarez and Gomez-Lus2007; Zintl et al. Reference Zintl, Proctor, Read, Dewaal, Shanaghy, Fanning and Mulcahy2009; Chalmers et al. Reference Chalmers, Smith, Elwin, Clifton-Hadley and Giles2011a ). This difference in Cryptosporidium species distribution is probably the results of differences in infection sources and transmission routes (Xiao, Reference Xiao2010).
There are also temporal and age-associated variations in the disease burdens between C. parvum and C. hominis (Chalmers et al. Reference Chalmers, Elwin, Thomas, Guy and Mason2009a , Reference Chalmers, Smith, Elwin, Clifton-Hadley and Giles2011a ). Like earlier observations in the UK and New Zealand, C. hominis was more prevalent in autumn, and C. parvum was more prevalent in spring in some more recent studies conducted in Canada, Ireland and the Netherlands (Wielinga et al. Reference Wielinga, de Vries, van der Goot, Mank, Mars, Kortbeek and van der Giessen2008; Zintl et al. Reference Zintl, Proctor, Read, Dewaal, Shanaghy, Fanning and Mulcahy2009; Budu-Amoako et al. Reference Budu-Amoako, Greenwood, Dixon, Barkema and McClure2012b ). In the Netherlands, C. hominis was more commonly found in children and C. parvum more in adults (Wielinga et al. Reference Wielinga, de Vries, van der Goot, Mank, Mars, Kortbeek and van der Giessen2008). In the UK, C. hominis was more prevalent in infants less than one year, females aged 15–44 years and international travellers, and there has been a decline in C. parvum cases since 2001 (Chalmers et al. Reference Chalmers, Elwin, Thomas, Guy and Mason2009a , Reference Chalmers, Smith, Elwin, Clifton-Hadley and Giles2011a ). In some studies, rare Cryptosporidium species were more commonly detected in immunocompromised persons than immunocompetent persons (ANOFEL Cryptosporidium National Network, 2010 ), while in others, there were no significant differences in the distribution of Cryptosporidium species between children and HIV-positive persons (Cama et al. Reference Cama, Ross, Crawford, Kawai, Chavez-Valdez, Vargas, Vivar, Ticona, Navincopa, Williamson, Ortega, Gilman, Bern and Xiao2007, Reference Cama, Bern, Roberts, Cabrera, Sterling, Ortega, Gilman and Xiao2008).
Cryptosporidium parvum and C. hominis are responsible for most cryptosporidiosis outbreaks, with C. hominis responsible for more outbreaks than C. parvum (Xiao, Reference Xiao2010). This is even the case for the UK, where C. parvum and C. hominis are both common in the general population. Recently, there was one drinking water associated cryptosporidiosis outbreak caused by C. cuniculus (Chalmers et al. Reference Chalmers, Robinson, Elwin, Hadfield, Xiao, Ryan, Modha and Mallaghan2009b ). An outbreak of C. meleagridis also occurred in a high school dormitory in Japan, although the infection source for the outbreak was not clear (Asano et al. Reference Asano, Karasudani, Okuyama, Takami, Oseto, Inouye, Yamamoto, Aokage, Saiki, Fujiwara, Shiraishi, Uchida, Saiki, Suzuki, Yamamoto, Udaka, Kan, Matsuura and Kimura2006).
Subtyping studies based on gp60 have shown that many C. parvum infections in humans are not results of zoonotic transmission (Xiao, Reference Xiao2010). Among several C. parvum subtype families identified, IIa and IIc are the two most common families in humans. In developing countries, most C. parvum infections in children and HIV-positive persons are caused by the subtype family IIc, with IIa largely absent, indicating that anthroponotic transmission of C. parvum is common in these areas. In contrast, both IIa and IIc subtype families are seen in humans in developed countries (Xiao and Feng, Reference Xiao and Feng2008; Xiao, Reference Xiao2010), whereas IId is the dominant C. parvum subtype family in humans in Middle Eastern countries (Nazemalhosseini-Mojarad et al. Reference Nazemalhosseini-Mojarad, Feng and Xiao2012). In a recent study conducted in Sweden, all IIc infections were acquired when travelling in developing countries, whereas almost all IIa and IId infections were acquired locally or in other European countries (Insulander et al. Reference Insulander, Silverlas, Lebbad, Karlsson, Mattsson and Svenungsson2013). As expected infections with subtype family IIa in the UK are frequently associated with farm visits (Chalmers et al. Reference Chalmers, Smith, Hadfield, Elwin and Giles2011b ).
CRYPTOSPORIDIUM SPECIES IN ANIMALS
Cryptosporidium in cattle, sheep and goats
Molecular studies have identified a wide range of Cryptosporidium species and genotypes in animals, many of which are not commonly found in humans. Livestock however, particularly cattle and sheep, are important reservoirs for C. parvum. In case–control studies, contact with cattle was implicated as a risk factor for human cryptosporidiosis in the USA, UK, Ireland and Australia (Robertson et al. Reference Robertson, Sinclair, Forbes, Veitch, Cunliffe, Willis and Fairley2002; Goh et al. Reference Goh, Reacher, Casemore, Verlander, Chalmers, Knowles, Williams, Osborn and Richards2004; Hunter et al. Reference Hunter, Hughes, Woodhouse, Syed, Verlander, Chalmers, Morgan, Nichols, Beeching and Osborn2004; Roy et al. Reference Roy, DeLong, Stenzel, Shiferaw, Roberts, Khalakdina, Marcus, Segler, Shah, Thomas, Vugia, Zansky, Dietz and Beach2004).
Studies worldwide suggest that cattle are infected with four major Cryptosporidium species: C. parvum, C. bovis, C. andersoni and C. ryanae (Xiao and Feng, Reference Xiao and Feng2008; Xiao, Reference Xiao2010). Studies of dairy cattle in industrialized nations have shown a dominance of C. parvum, especially its IIa subtypes in pre-weaned calves (Xiao, Reference Xiao2010; Amer et al. Reference Amer, Zidan, Adamu, Ye, Roellig, Xiao and Feng2013). Subtype IIaA15G2R1 is especially common and is overwhelmingly the dominant subtype in most areas (Xiao, Reference Xiao2010). However, the C. parvum IId subtype has been found in low frequencies in cattle in European countries. Intensive farming practices may facilitate the persistent transmission of C. parvum IIa subtypes on most dairy farms in industrialized nations (Xiao, Reference Xiao2010; Amer et al. Reference Amer, Zidan, Adamu, Ye, Roellig, Xiao and Feng2013; Santín, Reference Santín2013).
In developing countries C. bovis is the dominant species in pre-weaned calves, in addition to C. parvum, C. ryanae and C. andersoni (Silverlås et al. Reference Silverlås, de Verdier, Emanuelson, Mattsson and Björkman2010; Meireles et al. Reference Meireles, de Oliveira, Teixeira, Coelho and Mendes2011; Muhid et al. Reference Muhid, Robertson, Ng and Ryan2011; Wang et al. Reference Wang, Wang, Sun, Zhang, Jian, Qi, Ning and Xiao2011a ; Budu-Amoako et al. Reference Budu-Amoako, Greenwood, Dixon, Barkema and McClure2012b ; Venu et al. Reference Venu, Latha, Basith, Raj, Sreekumar and Raman2012; Amer et al. Reference Amer, Zidan, Adamu, Ye, Roellig, Xiao and Feng2013; Silva et al. Reference Silva, Richtzenhain, Barros, Gomes, Silva, Kozerski, de Araújo Ceranto, Keid and Soares2013; Silverlås and Blanco-Penedo, Reference Silverlås and Blanco-Penedo2013; Zhang et al. Reference Zhang, Wang, Yang, Zhang, Cao, Zhang, Ling, Liu and Shen2013a ). A small number of these studies subtyped C. parvum (Amer et al. Reference Amer, Harfoush and He2010; Imre et al. Reference Imre, Lobo, Matos, Popescu, Genchi and Dărăbuş2011; Meireles et al. Reference Meireles, de Oliveira, Teixeira, Coelho and Mendes2011; Muhid et al. Reference Muhid, Robertson, Ng and Ryan2011; Wang et al. Reference Wang, Wang, Sun, Zhang, Jian, Qi, Ning and Xiao2011a ; Silva et al. Reference Silva, Richtzenhain, Barros, Gomes, Silva, Kozerski, de Araújo Ceranto, Keid and Soares2013), with C. parvum IId subtypes identified as the dominant C. parvum in China and Malaysia (Muhid et al. Reference Muhid, Robertson, Ng and Ryan2011; Wang et al. Reference Wang, Wang, Sun, Zhang, Jian, Qi, Ning and Xiao2011a ; Zhang et al. Reference Zhang, Wang, Yang, Zhang, Cao, Zhang, Ling, Liu and Shen2013a ) and both IIa and IId dominant in Egypt (Amer et al. Reference Amer, Harfoush and He2010; Reference Amer, Zidan, Adamu, Ye, Roellig, Xiao and Feng2013). Thus, the prevalence and consequently the potential for transmission of Cryptosporidium infecting dairy cattle differ in developing countries vs industrialized nations at both species and subtype levels.
Cryptosporidium bovis (Fayer et al. Reference Fayer, Santin and Xiao2005) primarily infects young post-weaned cattle and has a wide geographic distribution (Feng et al. Reference Feng, Ortega, He, Das, Xu, Zhang, Fayer, Gatei, Cama and Xiao2007). To date there have been only three reports of humans infected with C. bovis: a dairy farm worker in India, a farm hand in Australia, and a 5-year-old village boy with animal contact in Egypt; the infection was asymptomatic in the first two cases and symptomatic in the third case (Khan et al. Reference Khan, Debnath, Pramanik, Xiao, Nozaki and Ganguly2010; Ng et al. Reference Ng, Eastwood, Walker, Durrheim, Massey, Porigneaux, Kemp, McKinnon, Laurie, Miller, Bramley and Ryan2012; Helmy et al. Reference Helmy, Krücken, Nöckler, von Samson-Himmelstjerna and Zessin2013). There have been a few reports of C. andersoni in humans in the UK, Australia, Iran and Malawi (Leoni et al. Reference Leoni, Amar, Nichols, Pedraza-Díaz and McLauchlin2006; Morse et al. Reference Morse, Nichols, Grimason, Campbell, Tembo and Smith2007; Waldron et al. Reference Waldron, Dimeski, Beggs, Ferrari and Power2011; Agholi et al. Reference Agholi, Hatam and Motazedian2013). A recent study in China identified C. andersoni in 34/252 diarrhoea patients in Shanghai (Liu et al. Reference Liu, Shen, Yin, Yuan, Jiang, Xu, Pan, Hu and Cao2014), but this together with the observation of common occurrence of Giardia duodenalis assemblage C in these patients needs confirmation by other studies. One study in New Zealand identified C. hominis (subtype IbA10G2) in cattle (Abeywardena et al. Reference Abeywardena, Jex, Nolan, Haydon, Stevens, McAnulty and Gasser2012).
Globally, the prevalence of Cryptosporidium spp. in sheep can vary drastically from <5 to >70% (Robertson, Reference Robertson2009). At least eight Cryptosporidium species have been identified in sheep faeces including C. parvum, C. hominis, C. andersoni, C. suis, Cryptosporidium xiaoi, C. fayeri, C. ubiquitum and C. scrofarum, with C. xiaoi, C. ubiquitum and C. parvum most prevalent (Ryan et al. Reference Ryan, Bath, Robertson, Read, Elliot, McInnes, Traub and Besier2005; Santín et al. Reference Santín, Trout and Fayer2007; Fayer and Santín, Reference Fayer and Santín2009; Giles et al. Reference Giles, Chalmers, Pritchard, Elwin, Mueller-Doblies and Clifton-Hadley2009; Yang et al. Reference Yang, Jacobson, Gordon and Ryan2009, Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014; Robertson, Reference Robertson2009; Díaz et al. Reference Diaz, Quilez, Chalmers, Panadero, Lopez, Sanchez-Acedo, Morrondo and Diez-Banos2010a ; Wang et al. Reference Wang, Feng, Cui, Jian, Ning, Wang, Zhang and Xiao2010a ; Fiuza et al. Reference Fiuza, Cosendey, Frazão-Teixeira, Santín, Fayer and de Oliveira2011a ; Sweeny et al. Reference Sweeny, Ryan, Robertson, Yang, Bell and Jacobson2011; Cacciò et al. Reference Cacciò, Sannella, Mariano, Valentini, Berti, Tosini and Pozio2013; Connelly et al. Reference Connelly, Craig, Jones and Alexander2013; Koinari et al. 2014). There are geographic differences in the distribution of the three dominant species in sheep; C. parvum is the dominant species in Europe, C. xiaoi is the dominant species in Australia, whereas C. ubiquitum appears to dominate in the Americas and Asia (see Table 2 in Ye et al. Reference Ye, Xiao, Wang, Wang, Amer, Roellig, Guo and Feng2013). As with cattle, there are probably age-associated differences in the distribution of Cryptosporidium spp. in sheep (Sweeny et al. Reference Sweeny, Ryan, Robertson, Yang, Bell and Jacobson2011; Ye et al. Reference Ye, Xiao, Wang, Wang, Amer, Roellig, Guo and Feng2013), although the picture remains unclear due to a lack of longitudinal studies. In one longitudinal study in China, periparturient shedding of oocysts in ewes was identified as the source of C. xiaoi infection in newborn lambs (Ye et al. Reference Ye, Xiao, Wang, Wang, Amer, Roellig, Guo and Feng2013).
In locations with common occurrence of C. parvum in sheep, the subtype IIaA15G2R1 is also dominant, although IId subtypes are also commonly reported (Xiao, Reference Xiao2010; Yang et al. Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014). The IIaA15G2R1 subtype was reported in three lambs linked to a human infection in the UK (Chalmers et al. Reference Chalmers, Ferguson, Cacciò, Gasser, Abs EL-Osta, Heijnen, Xiao, Elwin, Hadfield, Sinclair and Stevens2005). Recently, sheep have also been implicated as a potential source for human C. ubiquitum infections in the UK by gp60 subtyping (Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014). Furthermore, two studies reported the occurrence of the C. hominis IbA10G2 subtype in lambs in the UK (Giles et al. Reference Giles, Chalmers, Pritchard, Elwin, Mueller-Doblies and Clifton-Hadley2009; Connelly et al. Reference Connelly, Craig, Jones and Alexander2013).
Although fewer epidemiological studies have examined Cryptosporidium spp. in goats, C. parvum, C. hominis, C. ubiquitum and C. xiaoi have also been identified in goats (Park et al. Reference Park, Guk, Han, Shin, Kim and Chai2006; Goma et al. Reference Goma, Geurden, Siwila, Phiri, Claerebout and Vercruysse2007; Geurden et al. Reference Geurden, Thomas, Casaert, Vercruysse and Claerebout2008; Quílez et al. Reference Quílez, Torres, Chalmers, Hadfield, Del Cacho and Sánchez-Acedo2008; Giles et al. Reference Giles, Chalmers, Pritchard, Elwin, Mueller-Doblies and Clifton-Hadley2009; Robertson, Reference Robertson2009; Diaz et al. Reference Diaz, Quilez, Robinson, Chalmers, Diez-Banos and Morrondo2010b ; Fayer et al. Reference Fayer, Santin and Macarisin2010; Koinari et al. 2014). The C. parvum IId subtype predominates in goats (Xiao, Reference Xiao2010).
Cryptosporidium in pigs
The main Cryptosporidium species identified in pigs worldwide are C. suis and C. scrofarum (formally pig genotype II), although C. muris, C. tyzzeri and C. parvum have been reported (Ryan et al. Reference Ryan, Samarasinghe, Read, Buddle, Robertson and Thompson2003a ; Xiao et al. Reference Xiao, Moore, Ukoh, Gatei, Lowery, Murphy, Dooley, Millar, Rooney and Rao2006; Zintl et al. Reference Zintl, Neville, Maguire, Fanning, Mulcahy, Smith and De Waal2007; Johnson et al. Reference Johnson, Buddle, Reid, Armson and Ryan2008; Kváč et al. Reference Kváč, Hanzlíková, Sak and Kvetonová2009a , Reference Kváč, Kestřánová, Pinková, Květoňová, Kalinová, Wagnerová, Kotková, Vítovec, Ditrich, McEvoy, Stenger and Sak2013b ; Jeníková et al. Reference Jeníková, Němejc, Sak, Květoňová and Kváč2010; Jenkins et al. Reference Jenkins, Liotta, Lucio-Forster and Bowman2010; Sevá et al. Reference Sevá Ada, Funada, Souza Sde, Nava, Richtzenhain and Soares2010; Xiao, Reference Xiao2010; Wang et al. Reference Wang, Qiu, Jian, Zhang, Shen, Zhang, Ning, Cao, Qi and Xiao2010b ; Budu-Amoako et al. Reference Budu-Amoako, Greenwood, Dixon, Barkema, Hurnik, Estey and McClure2011; Chen et al. Reference Chen, Mi, Yu, Shi, Huang, Chen, Zhou, Cai and Lin2011; Farzan et al. Reference Farzan, Parrington, Coklin, Cook, Pintar, Pollari, Friendship, Farber and Dixon2011; Fiuza et al. Reference Fiuza, Gallo, Frazão-Teixeira, Santín, Fayer and Oliveira2011b ; Yin et al. Reference Yin, Shen, Yuan, Lu, Xu and Cao2011; Němejc et al. Reference Němejc, Sak, Květoňová, Kernerová, Rost, Cama and Kváč2013; Yui et al. Reference Yui, Nakajima, Yamamoto, Kon, Abe, Matsubayashi and Shibahara2014). As in dairy cattle, there is also an age-associated distribution of C. suis and C. scrofarum in pigs, with the former more commonly seen in pre-weaned piglets and the latter more commonly seen in older pigs (Němejc et al. Reference Němejc, Sak, Květoňová, Kernerová, Rost, Cama and Kváč2013; Zhang et al. Reference Zhang, Yang, Liu, Wang, Zhang, Shen, Cao and Ling2013b ; Yui et al. Reference Yui, Nakajima, Yamamoto, Kon, Abe, Matsubayashi and Shibahara2014). Cryptosporidium suis has been reported in humans (Xiao et al. Reference Xiao, Bern, Arrowood, Sulaiman, Zhou, Kawai, Vivar, Lal and Gilman2002a ; Leoni et al. Reference Leoni, Amar, Nichols, Pedraza-Díaz and McLauchlin2006; Cama et al. Reference Cama, Ross, Crawford, Kawai, Chavez-Valdez, Vargas, Vivar, Ticona, Navincopa, Williamson, Ortega, Gilman, Bern and Xiao2007; Wang et al. Reference Wang, Zhang, Zhao, Zhang, Zhang, Guo, Liu, Feng and Xiao2013) and has been frequently recovered from water samples (Feng et al. Reference Feng, Zhao, Chen, Jin, Zhou, Li, Wang and Xiao2011c ). Cryptosporidium scrofarum has also been reported in an immunocompetent human (Kváč et al. Reference Kváč, Kestřánová, Pinková, Květoňová, Kalinová, Wagnerová, Kotková, Vítovec, Ditrich, McEvoy, Stenger and Sak2013b ).
Cryptosporidium parvum has been reported at least six times in pigs: (1) in four 19-day-old pre-weaned piglets with diarrhoea from an indoor farm in Western Australia (Morgan et al. Reference Morgan, Buddle, Armson and Thompson1999a ); (2) in asymptomatic sows from intensive commercial pig production units in Ireland (Zintl et al. Reference Zintl, Neville, Maguire, Fanning, Mulcahy, Smith and De Waal2007); (3) in two piglets from Prince Edward Island, Canada (Budu-Amoako et al. Reference Budu-Amoako, Greenwood, Dixon, Barkema, Hurnik, Estey and McClure2011); (4) in piglets in Ontario, where it was the most prevalent species detected (55·4%) (Farzan et al. Reference Farzan, Parrington, Coklin, Cook, Pintar, Pollari, Friendship, Farber and Dixon2011); (5) in pig lagoons in the USA (Jenkins et al. Reference Jenkins, Liotta, Lucio-Forster and Bowman2010); and (6) in one pig isolate from the Czech Republic (IIa A16G1R1b) (Němejc et al. Reference Němejc, Sak, Květoňová, Kernerová, Rost, Cama and Kváč2013). This suggests that pigs may play a potential role in the transmission of zoonotic Cryptosporidium. However, further research is required to understand the prevalence of Cryptosporidium species in wild pigs. Recently, C. suis and C. scrofarum have also been detected in wild boars in the Czech Republic (Němejc et al. Reference Němejc, Sak, Květoňová, Hanzal, Jeníková and Kváč2012) and Spain (García-Presedo et al. Reference García-Presedo, Pedraza-Díaz, González-Warleta, Mezo, Gómez-Bautista, Ortega-Mora and Castro-Hermida2013a ). The latter also reported the occurrence of C. parvum subtypes IIaA16G2R1 and IIaA13G1R1, two common subtypes in humans in the country.
Cryptosporidium in cats and dogs
Genotyping studies of Cryptosporidium oocysts in faeces of dogs and cats have demonstrated that most infections in these animals are caused by C. canis and C. felis, respectively. Cryptosporidium muris and C. parvum have also occasionally been reported in dogs and cats (cf. Lucio-Forster et al. Reference Lucio-Forster, Griffiths, Cama, Xiao and Bowman2010). Cryptosporidium muris has a wide host range and has also been identified in a few humans in developing countries (Gatei et al. Reference Gatei, Wamae, Mbae, Waruru, Mulinge, Waithera, Gatika, Kamwati, Revathi and Hart2006; Muthusamy et al. Reference Muthusamy, Rao, Ramani, Monica, Banerjee, Abraham, Mathai, Primrose, Muliyil, Wanke, Ward and Kang2006; Palmer et al. Reference Palmer, Traub, Robertson, Devlin, Rees and Thompson2008). Cryptosporidium felis has a much more restricted host range and, using molecular techniques, has been confirmed to infect cats, immunocompetent and immunocompromised humans and a cow (Bornay-Llinares et al. Reference Bornay-Llinares, da Silva, Mourna, Myjap, Pietkiewicz, Kruminis-Lozowska, Graczak and Pieniazek1999; Lucio-Forster et al. Reference Lucio-Forster, Griffiths, Cama, Xiao and Bowman2010). Similarly, using molecular techniques, C. canis has been confirmed to infect dogs, foxes, wolves and immunocompetent and immunocompromised humans (Lucio-Forster et al. Reference Lucio-Forster, Griffiths, Cama, Xiao and Bowman2010). In children in developing countries, C. felis and C. canis are responsible for as much as 3·3 and 4·4%, respectively, of overall cryptosporidiosis cases (Lucio-Forster et al. Reference Lucio-Forster, Griffiths, Cama, Xiao and Bowman2010). However, most human cases of cryptosporidiosis, worldwide, are associated with C. hominis and C. parvum (Xiao, Reference Xiao2010) and therefore C. muris, C. canis and C. felis are of low zoonotic risk to humans.
Cryptosporidium in wild mammals
Cryptosporidium cuniculus (previously rabbit genotype) was first identified genetically in rabbits from the Czech Republic (Ryan et al. Reference Ryan, Xiao, Read, Zhou, Lal and Pavlasek2003b ), then formally re-described as a species, based on biological and genetic data in 2010 (Robinson et al. Reference Robinson, Wright, Elwin, Hadfield, Katzer, Bartley, Hunter, Nath, Innes and Chalmers2010). Cryptosporidium cuniculus was initially thought to be host-specific until the discovery that C. cuniculus was linked to a human cryptosporidiosis outbreak in the UK (Chalmers et al. Reference Chalmers, Robinson, Elwin, Hadfield, Xiao, Ryan, Modha and Mallaghan2009b ), which raised considerable awareness about the importance of investigating rabbits as a source of Cryptosporidium transmissible to humans. Subsequently, a study reported that C. cuniculus was the third most commonly identified Cryptosporidium species in patients with diarrhoea in the UK (Chalmers et al. Reference Chalmers, Smith, Elwin, Clifton-Hadley and Giles2011a ). It has also been identified in a few human patients in France, South Australia and Nigeria (Anon, 2010; unpublished; Molloy et al. Reference Molloy, Smith, Kirwan, Nichols, Asaolu, Connelly and Holland2010).
Few studies have characterized Cryptosporidium spp. in wild deer, with C. ubiquitum, C. bovis, C. ryanae, the deer genotype and C. parvum and a C. hominis-like genotype reported (cf. Cinque et al. Reference Cinque, Stevens, Haydon, Jex, Gasser and Campbell2008; Amer et al. Reference Amer, Honma, Ikarashi, Oishi, Endo, Otawa and Nakai2009; Feng, Reference Feng2010; Ng et al. Reference Ng, Yang, Whiffin, Cox and Ryan2011; Nolan et al. Reference Nolan, Jex, Koehler, Haydon, Stevens and Gasser2013; García-Presedo et al. Reference García-Presedo, Pedraza-Díaz, González-Warleta, Mezo, Gómez-Bautista, Ortega-Mora and Castro-Hermida2013b ). In captive cervids C. parvum and C. ubiquitum predominated (cf. Feng, Reference Feng2010).
House mice are commonly infected with C. muris and C. tyzzeri (formerly mouse genotype I), and occasionally with the mouse genotype II (Morgan et al. Reference Morgan, Sturdee, Singleton, Gomez, Gracenea, Torres, Hamilton, Woodside and Thompson1999b ; Foo et al. Reference Foo, Farrell, Boxell, Robertson and Ryan2007; Silva et al. Reference Silva, Richtzenhain, Barros, Gomes, Silva, Kozerski, de Araújo Ceranto, Keid and Soares2013). There have been numerous reports of C. muris in humans (cf. Feng et al. Reference Feng, Yang, Ryan, Zhang, Kvac, Koudela, Modry, Li, Fayer and Xiao2011b ) and a co-infection of C. tyzzeri and C. parvum in a young woman (Raskova et al. Reference Raskova, Kvetonova, Sak, McEvoy, Edwinson, Stenger and Kvac2013). Confirmed C. parvum infections have been reported in relatively few rodents (Morgan et al. Reference Morgan, Sturdee, Singleton, Gomez, Gracenea, Torres, Hamilton, Woodside and Thompson1999b ; Lv et al. Reference Lv, Zhang, Wang, Jian, Zhang, Ning, Wang, Feng, Wang, Ren, Qi and Xiao2009; Feng, Reference Feng2010). Recently C. erinacei (formerly the hedgehog genotype) has been described biologically and genetically from the European hedgehog (Erinaceus europaeus) (Kváč et al. Reference Kváč, Hofmannová, Hlásková, Květoňová, Vítovec, McEvoy and Sak2014b ). It has previously been described in horses and an immunocompetent human (Kváč et al. Reference Kváč, Saková, Kvĕtoňová, Kicia, Wesołowska, McEvoy and Sak2014a ).
Of the few genotyping studies conducted with foxes, the Cryptosporidium fox genotype, C. canis fox subtype (a variant of C. canis) and a Cryptosporidium macropodum-like genotype have been reported (Xiao et al. Reference Xiao, Sulaiman, Ryan, Zhou, Atwill, Tischler, Zhang, Fayer and Lal2002b ; Ng et al. Reference Ng, Yang, Whiffin, Cox and Ryan2011; Ruecker et al. Reference Ruecker, Matsune, Wilkes, Lapen, Topp, Edge, Sensen, Xiao and Neumann2012, Reference Ruecker, Matsune, Lapen, Topp, Edge and Neumann2013; Nolan et al. Reference Nolan, Jex, Koehler, Haydon, Stevens and Gasser2013).
Cryptosporidium fayeri (previously marsupial genotype I) and C. macropodum (previously marsupial genotype II) infect marsupials (Ryan et al. Reference Ryan, Power and Xiao2008; Power and Ryan, Reference Power and Ryan2008; Power, Reference Power2010; Ryan and Power, Reference Ryan and Power2012). There has been one report of C. fayeri in a 29-year-old immunocompetent woman who suffered prolonged gastrointestinal illness in Sydney (Waldron et al. Reference Waldron, Cheung-Kwok-Sang and Power2010). Identical gp60 subtypes were found in marsupials in the area, indicating potential zoonotic transmission (Waldron et al. Reference Waldron, Cheung-Kwok-Sang and Power2010).
Cryptosporidium in birds
Currently only three avian Cryptosporidium spp. are recognized in birds: C. meleagridis, Cryptosporidium baileyi and Cryptosporidium galli (Table 1). Cryptosporidium meleagridis has a wide host range and is the third most prevalent species infecting humans (Leoni et al. Reference Leoni, Amar, Nichols, Pedraza-Díaz and McLauchlin2006; Xiao, Reference Xiao2010; Elwin et al. Reference Elwin, Hadfield, Robinson and Chalmers2012a ). The ability of C. meleagridis to infect humans and other mammals, and its close phylogenetic relationship to C. parvum and C. hominis, has led to the suggestion that mammals actually were the original hosts, and that the species has later adapted to birds (Xiao et al. Reference Xiao, Sulaiman, Ryan, Zhou, Atwill, Tischler, Zhang, Fayer and Lal2002b , Reference Xiao, Fayer, Ryan and Upton2004). Recently, sequence analysis of the SSU gene and HSP70 loci has been used to provide evidence of zoonotic transmission of C. meleagridis from chickens to a human on a Swedish farm (Silverlås et al. Reference Silverlås, Mattsson, Insulander and Lebbad2012). In addition to the three recognized species of Cryptosporidium, 11 genotypes: the avian genotypes I–V, the black duck genotype, the Eurasian woodcock genotype and goose genotypes I–IV have been reported (Ryan and Xiao, Reference Ryan, Xiao, Cacciò and Widmer2014), none of which are considered human pathogens. In addition, C. hominis, C. parvum, Cryptosporidium serpentis, C. muris, C. andersoni and muskrat genotype I have also been identified in a small number of birds, most of which were probably the results of accidental ingestion of oocysts by these organisms (cf. Ryan, Reference Ryan2010; Ryan and Xiao, Reference Ryan, Xiao, Cacciò and Widmer2014).
Cryptosporidium in marine mammals, reptiles, amphibians and fish
Very little is known about the prevalence and genetic diversity of species of Cryptosporidium in marine environments and the role that marine animals play in transmission of these parasites to humans. Molecular research on marine mammals has identified C. muris, seal genotypes 1–4 and a genotype similar to the skunk genotype in seals (Santín et al. Reference Santín, Dixon and Fayer2005; Rengifo-Herrera et al. Reference Rengifo-Herrera, Ortega-Mora, Gómez-Bautista, García-Moreno, García-Párraga, Castro-Urda and Pedraza-Díaz2011; Bass et al. Reference Bass, Wallace, Yund and Ford2012; Rengifo-Herrera et al. Reference Rengifo-Herrera, Ortega-Mora, Gómez-Bautista, García-Peña, García-Párraga and Pedraza-Díaz2013).
Cryptosporidium serpentis (Levine, Reference Levine1980) and Cryptosporidium varanii (syn. Cryptosporidium saurophilum) Pavlásek et al. Reference Pavlásek, Lávicková, Horák, Král and Král1995; Pavlasek and Ryan, Reference Pavlasek and Ryan2008) are the only valid species in reptiles. Several genotypes including tortoise genotype I and snake genotypes I and II have also been identified (cf. Ryan and Xiao, Reference Ryan, Xiao, Cacciò and Widmer2014), which have not been reported in humans. A new intestinal species, Cryptosporidium ducismarci has been reported in several species of tortoises, snakes and lizards (Traversa, Reference Traversa2010). Because only molecular data are presented, this species has to be regarded as a nomen nudum, pending the support of morphological and biological data. Relatively little is known about Cryptosporidium in amphibians and currently the only accepted species is Cryptosporidium fragile, which was described from the stomach of naturally infected black-spined toads (Duttaphrynus melanostictus) from the Malay peninsula (Jirku et al. Reference Jirku, Valigurova, Koudela, Krzek, Modry and Slapeta2008).
Cryptosporidium has been described in both fresh and marine water piscine species, with Cryptosporidium molnari (Alvarez-Pellitero and Sitja-Bobadilla, Reference Alvarez-Pellitero and Sitja-Bobadilla2002), the only currently recognized species. Cryptosporidium scophthalmi was described in 2004 in turbot (Psetta maxima, syn. Scophthalmus maximus) (Alvarez-Pellitero et al. Reference Alvarez-Pellitero, Quiroga, Sitja-Bobadilla, Redondo, Palenzuela, Padros, Vazquez and Nieto2004), but is considered a nomen nudum until genetic data are provided, as several morphologically identical species can be found in one fish species. A total of 13 additional species/genotypes have been identified in fish using molecular tools; piscine genotypes 1–8, rat genotype III, C. parvum, C. hominis C. xiaoi and C. scrofarum (cf. Koinari et al. Reference Koinari, Karl, Ng-Hublin, Lymbery and Ryan2013; Ryan and Xiao, Reference Ryan, Xiao, Cacciò and Widmer2014). The very low prevalence of C. parvum and C. hominis in fish (<1%) suggests that fish are not an important reservoir for human infection.
The taxonomic status of Cryptosporidium in fish presents a problem based on the location of the oocyst in the basal part of the stomach epithelium and the retained residual parasitophorous sac with the retained remnants of the attachment organelle, features not found in species of Cryptosporidium from homoeothermic hosts. Recognition of these features prompted Paperna and Vilenkin (Reference Paperna and Vilenkin1996) to propose the new genus Piscicryptosporidium for two new species – Piscicryptosporidium reichenbachklinkei and Piscicryptosporidium cichlidis from cichlid fishes of the genus Oreochromis. Other forms of divergence however have been accepted for Cryptosporidium species found in hosts within the same major host taxon, such as the large oocysts of Cryptosporidium species that develop in the stomach vs the small oocysts of species that develop in the small intestine. Genetic data also suggest that Cryptosporidium in fish are genetically very distinct (Palenzuela et al. Reference Palenzuela, Alvarez-Pellitero and Sitjà-Bobadilla2010; Koinari et al. Reference Koinari, Karl, Ng-Hublin, Lymbery and Ryan2013), however additional genetic and biological data are required to better understand the taxonomic status of Piscicryptosporidium.
RESEARCH GAPS AND PERSPECTIVES
Despite the considerable progress in the last 20 years on the taxonomy and molecular epidemiology of Cryptosporidium, significant research gaps remain. Differences still exist on the interpretation of ICZN rules and opinions on validity of some Cryptosporidium species (Slapeta, Reference Slapeta2013). Morphological and biological data are not yet available for some common Cryptosporidium genotypes with public health and veterinary importance, such as the horse and skunk genotypes. Taxonomic and molecular epidemiological studies on Cryptosporidium spp. in wildlife, especially those in watershed are still scarce. Resolutions to those issues should greatly improve our understanding of the species structure and transmission of Cryptosporidium in humans and animals.
Cryptosporidium was the aetiological agent in 60·3% (120) of the waterborne protozoan parasitic outbreaks that have been reported worldwide between 2004 and 2010, (Baldursson and Karanis, Reference Baldursson and Karanis2011). Yet the public health significance of various zoonotic Cryptosporidium species detected in animals in water catchments and in drinking water remains unclear. For example, do different species of Cryptosporidium have different sensitivities (inactivation rates) to drinking water treatments and environmental conditions such as temperature, UV and solar inactivation? Are there differences in infectious dose for different zoonotic Cryptosporidium species? There is also a need to confirm if molecular detection of zoonotic Cryptosporidium species in wildlife is commonly associated with actual infections or mechanical transmission. Cryptosporidium cuniculus is the only species besides C. hominis and C. parvum known to be associated with a waterborne outbreak of human cryptosporidiosis, yet little is known about the prevalence and oocyst shedding rates of C. cuniculus in rabbits. Measuring the infectivity of different Cryptosporidium species under different climatic conditions is also crucial for accurate risk assessment of public health implications, particularly as more extreme precipitation is predicted globally (IPCC, 2013).
Studies on the transmission of Cryptosporidium in humans and domesticated animals are currently hampered by the lack of suitable subtyping tools for Cryptosporidium species that are genetically distant from C. parvum and C. hominis. The ability to subtype all major Cryptosporidium species at the gp60 is also important for understanding the transmission dynamics, particularly as recent research suggests that gp60 plays an active and essential role in the life cycle of the parasite and that genetic variation at this locus might be essential for the parasite's long-term success (Abal-Fabeiro et al. Reference Abal-Fabeiro, Maside, Bello, Llovo and Bartolomé2013; Feng et al. Reference Feng, Torres, Li, Wang, Bowman and Xiao2013, Reference Feng, Tiao, Li, Hlavsa and Xiao2014; Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014). Currently only C. parvum, C. hominis, C. meleagridis, C. tyzzeri, C. cuniculus, C. fayeri and C. ubiquitum can be subtyped at this locus (Chalmers et al. Reference Chalmers, Robinson, Elwin, Hadfield, Xiao, Ryan, Modha and Mallaghan2009b ; Lv et al. Reference Lv, Zhang, Wang, Jian, Zhang, Ning, Wang, Feng, Wang, Ren, Qi and Xiao2009; Power et al. Reference Power, Cheung-Kwok-Sang, Slade and Williamson2009; Xiao, Reference Xiao2010; Feng et al. Reference Feng, Lal, Li and Xiao2011a ; Kváč et al. Reference Kváč, McEvoy, Loudová, Stenger, Sak, Květoňová, Ditrich, Rašková, Moriarty, Rost, Macholán and Piálek2013c ; Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014). The most recent study, which used NGS to identify and develop a gp60-based typing tool for C. ubiquitum, revealed that the gp60 gene of C. ubiquitum has extensive sequence differences from the gp60 gene of other Cryptosporidium spp. (Li et al. Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers, Santin, Fayer, Kvac, Ryan, Sak, Stanko, Guo, Wang, Zhang, Cai, Roellig and Feng2014). These findings highlight the need for WGS analysis of diverse Cryptosporidium species to develop subtyping tools for other common Cryptosporidium species in humans and domestic animals. Extensive WGS of Cryptosporidium spp. will likely lead to improved understanding of virulence factors in C. parvum and C. hominis and the genetic basis for host specificity and human infectivity of various Cryptosporidium species. This in turn will promote the development of vaccines and new therapies to help control the spread of Cryptosporidium (Striepen, Reference Striepen2013).
CONCLUSIONS
As molecular tools improve and data accumulate, our understanding of the role of zoonotic transmission in epidemiology and clinical manifestations is becoming clearer. In the future, these tools and advanced techniques developed from WGS of diverse Cryptosporidium species will be essential in understanding intricate associations between wildlife, domestic animals and humans in the context of health and climate changes to enable management of the zoonotic risk of Cryptosporidium.






