Genomic selection (GS) is a new technology in which selection decisions are based on direct genomic values (DGVs) or genomic enhanced breeding values (GEBVs). During recent years GS has revolutionized dairy cattle breeding and has the potential to double the rate of genetic gain as well as reduce the costs for dairy breeding companies by 92% (Schaeffer, Reference Schaeffer2006). The DGVs are predicted from thousands of genetic markers in the form of single nucleotide polymorphisms (SNP) covering the whole genome (Meuwissen et al. Reference Meuwissen, Hayes and Goddard2001; Goddard & Hayes, Reference Goddard and Hayes2007; Hayes et al. Reference Hayes, Bowman, Chamberlain and Goddard2009; Calus, Reference Calus2010; Mark & Sandoe, Reference Mark and Sandoe2010; Su et al. Reference Su, Guldbrandtsen, Gregersen and Lund2010). This enables all quantitative trait loci (QTL) to be in linkage disequilibrium with at least some of the markers. To estimate DGVs, a reference population consisting of animals with known genotype and phenotypic data is required. By linking these data sets together, estimates of SNP effects for each trait are acquired. For subsequent generations, DGVs can be obtained directly through a DNA sample, thus allowing genetic evaluation and selection early in life (Goddard & Hayes, Reference Goddard and Hayes2007; Hayes et al. Reference Hayes, Bowman, Chamberlain and Goddard2009).
In order for GS to be successfully applied within breeding programs, the accuracy of the prediction of DGVs has to be considered. The accuracy depends on several factors, of which some are the heritability of a trait, the number of animals in the reference population and the number of SNP markers that are used to estimate the SNP effects (Hayes et al. Reference Hayes, Bowman, Chamberlain and Goddard2009; Calus, Reference Calus2010). By increasing the number in the latter factors, the accuracy of GS will be greater (Hayes et al. Reference Hayes, Bowman, Chamberlain and Goddard2009). So far, most studies have based the accuracy of genomic predictions on simulated data. The recent rapid development of genome-wide dense SNP marker maps, that makes it possible to choose makers evenly distributed throughout the genome, has however increased the studies of accuracies based on real cattle populations (Harris et al. Reference Harris, Johnson and Spelman2008; Hayes et al. Reference Hayes, Bowman, Chamberlain and Goddard2009; Luan et al. Reference Luan, Woolliams, Lien, Kent, Svendsen and Meuwissen2009; VanRaden et al. Reference VanRaden, Van Tassell, Wiggans, Sonstegard, Schnabel, Taylor and Schenkel2009; Su et al. Reference Su, Guldbrandtsen, Gregersen and Lund2010). It has been shown that the reliabilities of DGVs are considerably greater than for conventional parent average (Hayes et al. Reference Hayes, Bowman, Chamberlain and Goddard2009; Su et al. 2010).
The composition of cow's milk is of great importance for its nutritional value as well as its processability (Walstra et al. Reference Walstra, Geurts, Noomen, Jellema and van Boekel1999). Several factors influence milk composition, of which some are genetic factors, which shows the importance of powerful genetic selection programs. Lately, the dairy industry has an increasing economic interest of providing milk for specific products, thus making GS an efficient tool in selecting cows early in life for targeted milk production. The aim of this paper is to study DGVs for traditional production traits in relation to several milk traits important for both the nutritional value and processability, including parameters in the protein, fat and mineral profiles, as well as actual technological properties of milk, such as cheese characteristics. The purpose is to investigate correlations between DGVs for traditional production traits included in the present breeding objective and novel production traits. To our knowledge, this is the first time that DGVs for traditional production traits have been studied in relation to bovine milk composition, nutritional value and processability.
Materials and Methods
Animals and milk samples
Morning milk samples and blood samples were collected in September 2008 from 23 Swedish Holstein (SH) cows in the second lactation, belonging to the research farm Nötcenter Viken (Falköping, Sweden). An integrated part of the Swedish breeding occurs at this research farm, which has a unique nucleus breeding herd producing a large number of bull dams. The investigated cows descended from 13 proven bulls. To exclude extremes in the beginning of the lactation, the cows were in lactation week 7–53. All cows in the study were healthy (mean somatic cell count (SCC) below 100 000 cells/ml), fed the same diet and milked three times a day. At the time of sampling, the cows had been fed in the stable for several weeks and been adjusted to the winter feeding regime. Milk yield of the whole sampling day (the total of three milkings) was recorded for each cow. The samples were cooled directly after collection and stored at 4°C overnight. The following day, the samples were thoroughly mixed and subsampled before aliquots of whole and skim milk were either analyzed directly or stored at −20°C until time of analysis. Skim milk was prepared by defatting the samples using centrifugation at 2000 g for 30 min.
Milk composition
For all the sampled cows, fresh milk samples were analyzed for contents of protein, fat, lactose and urea as well as for the freezing point by using an infrared technique and for somatic cells by using flow cytometry. This was performed at a certified dairy analysis laboratory (Eurofins Steins Laboratory, Jönköping, Sweden), all according to Glantz et al. (Reference Glantz, Lindmark, ånsson, Stålhammar, Bårström, Fröjelin, Knutsson, Teluk and Paulsson2009). Yields per cow per day were calculated by multiplying each percentage by the milk yield of the sampling day. Additionally, data were obtained on milk, protein and fat yield as well as contents of protein and fat for the sampled cows’ 305-days lactation during the second lactation from the national cow database.
In addition, a more comprehensive study of milk composition traits was made on nine of the analyzed SH cows. The nine cows with the lowest SCC out of the 23 cows were selected for the comprehensive study. Milk samples stored at −20°C were analyzed for contents of casein, whey proteins and non-protein nitrogen using the Kjeldahl method, contents of free fatty acids using a colorimetric technique, fatty acid composition using gas chromatography and contents of total Ca and P using inductively coupled plasma mass spectrometry, as described previously (Glantz et al. Reference Glantz, Lindmark, ånsson, Stålhammar, Bårström, Fröjelin, Knutsson, Teluk and Paulsson2009). The components were analyzed at another certified dairy analysis laboratory (Eurofins Steins Laboratory, Holstebro, Denmark). In addition, pH was measured on fresh milk samples. Duplicate measurements were made on each sample. Yields per cow per day were calculated as described above.
Free Ca2+ concentration was measured at 25 and 32°C in skim milk samples using an Orion 97–20 Ionplus Calcium Electrode (Thermo Electron Corporation, Beverly, MA, USA), all according to Glantz et al. (Reference Glantz, Lindmark Månsson, Stålhammar and Paulsson2011). Each sample was analyzed in triplicate.
Technological properties
Milk samples from the nine selected cows described above were also analyzed for technological properties, such as cheese characteristics and size determination of casein micelles and fat globules.
Rheological measurements
Using the method described by Glantz et al. (Reference Glantz, Lindmark Månsson, Stålhammar and Paulsson2011), rennet-induced gelation was carried out at 32°C on fresh skim milk samples for 40 min with low-amplitude oscillation measurements (Stresstech rheometer; Reologica Instruments AB, Lund, Sweden) using chymosin (0·90 ml Chy-Max Plus/l, 200 international milk clotting U/ml; Christian Hansen A/S, Hørsholm, Denmark) to obtain gel strength and gelation time. After 40 min gel formation, stress sweeps were run to study the stress dependence of the viscosity to obtain the yield stress. The samples were subjected to an increasing shear stress in 100 intervals from 0·1 to 300 Pa. The yield stress was defined as the shear stress at the first local maximum of the viscosity. All samples were analyzed at least in duplicate.
Model cheeses
Model cheeses were produced from skim milk as described earlier (Glantz et al. Reference Glantz, Lindmark Månsson, Stålhammar and Paulsson2011) using glucono-δ-lactone (3·76 g/kg Roquette, Lestrem Cedrix, France) (Lucey et al. Reference Lucey, van Vliet, Grolle, Geurts and Walstra1997) and chymosin (1·25 ml Chy-Max Plus/l, 200 International Milk Clotting Units/ml, Christian Hansen A/S, Denmark) to determine cheese yield (expressed as g cheese/100 g milk), pH and cheese hardness, all according to (Glantz et al. Reference Glantz, Lindmark Månsson, Stålhammar and Paulsson2011). One batch of cheese was made for each sample and for each batch of cheese, triplicate measurements were made to obtain cheese hardness.
Casein micelle size determination
Photon correlation spectroscopy was used to determine the z-average hydrodynamic diameter of casein micelles at 90° scattering angle at 25°C on skim milks diluted in simulated milk ultrafiltrate (Jenness & Koops, Reference Jenness and Koops1962) as described previously (Glantz et al. Reference Glantz, Devold, Vegarud, Lindmark Månsson, Stålhammar and Paulsson2010). Each sample was analyzed in triplicate.
Fat globule size determination
The volume-weighted droplet diameter, d(4,3), of the fat globules was determined in fresh milk samples at 20°C by using light diffraction, all according to Glantz et al. (Reference Glantz, Lindmark, ånsson, Stålhammar, Bårström, Fröjelin, Knutsson, Teluk and Paulsson2009). All samples were analyzed in duplicate.
Analysis and estimation of direct genomic values
All 23 cows were genotyped with the Illumina BovineSNP50 BeadChip (Illumina, Inc., San Diego, CA, USA). Analysis and estimation of DGVs were conducted at Århus University, Denmark. The reference population consisted of approximately 16 000 progeny tested bulls from the cooperation within EuroGenomics (David et al. Reference David, de Vries, Feddersen and Borchersen2010). Most of the bulls in the reference population were typed with the above-mentioned Illumina BovineSNP50 BeadChip. In one of the participating countries, some of the bulls had also been typed with a customized chip. Information from the customized chip was imputed, making the same information available for all the bulls. Genotype information was exchanged between the participating partners within EuroGenomics and phenotypes were available through Interbull. Standard editing procedures were applied concerning calling rate and minor allele frequencies. Used model of estimation of DGV was GBLUP and response variables were deregressed proofs weighted according to expected daughter contribution. DGVs were calculated for each cow for milk (DGVmilk), fat (DGVfat) and protein yield (DGVprotein), milk index (−0·25·DGVmilk+0·25·DGVfat+ DGVprotein+100; DGVmilk index), Nordic total merit (NTM; DGVNTM), which is a combined value of production, functional and conformation traits, and udder health (mastitis resistance; DGVudder health). The estimated DGVs for udder health include information from three different sources, that is SCC, treatment of mastitis and udder conformation (udder depth and fore udder attachment). In addition, a quota was calculated between DGVfat and DGVmilk as well as DGVprotein and DGVmilk to obtain DGVs for fat (DGVfat%) and protein content (DGVprotein%).
Statistical analyses
Linear correlations between DGVs and milk traits were estimated with Pearson correlation combined with the corresponding P-values. The level of significance was set at P<0·10. Statistical analyses were performed using Minitab (version 14, Minitab Ltd., Coventry, UK).
Results
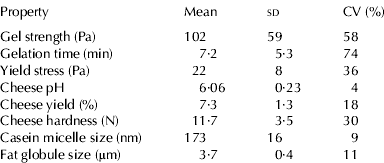
An extensive study of milk parameters and technological properties was performed on morning milk samples from individual SH cows, in order to study relations between DGVs and milk traits. Means, sd and coefficients of variation of the analyzed parameters and technological properties are summarized in Tables 1 & 2. The values obtained for milk components are in the range found previously in milk from individual SH cows (Wedholm et al. Reference Wedholm, Hallén, Larsen, Lindmark, ånsson, Karlsson and Allmere2006a; Hallén et al. Reference Hallén, Wedholm, Andrén and Lundén2008; Näslund et al. Reference Näslund, Fikse, Pielberg and Lundén2008) and SH bulk milk (Glantz et al. Reference Glantz, Lindmark, ånsson, Stålhammar, Bårström, Fröjelin, Knutsson, Teluk and Paulsson2009). The values obtained for technological properties are in the range reported earlier in SH bulk milk (Glantz et al. Reference Glantz, Lindmark, ånsson, Stålhammar, Bårström, Fröjelin, Knutsson, Teluk and Paulsson2009, Reference Glantz, Devold, Vegarud, Lindmark Månsson, Stålhammar and Paulsson2010). Despite the high amount of milk produced by the cows, the contents of milk components are equal to or somewhat higher compared with average contents of protein and fat for the SH breed (Swedish Dairy Association, 2009). The coefficients of variation for milk components range from 6% for lactose content to 30% for fat content. Moderate coefficients of variation are observed for milk proteins and minerals (13–20%), whereas the coefficients of variation for yield parameters are higher (16–54%). For most of the technological properties, the coefficients of variation are moderate or high (>18%).
Table 1. Means, sd and coefficients of variation (CV) of analyzed milk composition traits from individual Swedish Holstein cows

† n=number of cows
‡ Mean fatty acid (FA) as a proportion (wt/wt) of the total fat fraction of 100%
Table 2. Means, sd and coefficients of variation (CV) of analyzed technological properties of milk from individual Swedish Holstein cows (n=9)
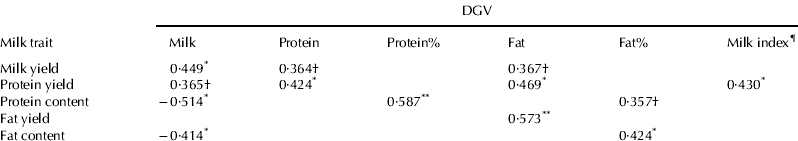
Linear correlations between DGVs and milk traits based on one morning milk sampling are presented in Table 3. Results are shown only for components or technological properties correlated significantly (P<0·10) with at least one of the studied DGVs. Both DGVmilk and DGVprotein correlate positively with milk yield (P=0·001 and 0·077, respectively) and yields in the protein (P=0·006–0·093) and mineral profiles (P=0·041–0·064), whereas DGVprotein% and DGVfat% have positive correlations with contents of these traits (P=0·002–0·097). Negative correlations exist between DGVmilk and protein profile contents (P=0·013–0·072). DGVfat on the other hand, shows no correlation with yields or contents of milk components, except for a positive correlation with free fatty acids (P=0·040). Negative correlations exist between DGVudder health and yields of the protein profile (P=0·054–0·084) as well as contents of fat (P=0·030) and minerals (P=0·099). However, no significant correlation exists between DGVudderhealth and SCC. Furthermore, DGVNTM correlates negatively with casein (P=0·094) and fat content (P=0·055) as well as freezing point (P=0·099). Additionally, correlations also exist between the studied DGVs and technological properties of milk. DGVmilk and DGVNTM show a negative correlation with cheese yield (P=0·011 and 0·065, respectively), in contrast to DGVprotein% and DGVfat% having positive correlations (P=0·030 and 0·033, respectively). DGVprotein% and DGVudder health both correlate with gelation properties (P=0·006–0·043). One may also consider non-linear relationships, such as exponential relationships, that may exist between DGVs and milk traits, however, no such correlations were found in this data set.
Table 3. The Pearson correlation coefficient between direct genomic values (DGV) and milk composition and technological properties based on data from one morning milk sampling‡

† P<0·10; *P<0·05; **P<0·01; ***P<0·001
‡ Results are shown only for components correlated significantly (P<0·10) with at least 1 DGV
§ −0·25·DGVmilk+0·25·DGVfat+DGVprotein+100
¶ Mastitis resistance
†† NTM=Nordic total merit. Combined value of production, functional and conformation traits
‡‡ n=number of cows
The correlations presented above are based on data from one morning milk sampling. To ensure that these samples represent the entire lactation, available data for the cows’ 305-days lactation were correlated with both the data from one morning milk sampling and the studied DGVs. The results from these analyses are presented in Tables 4 & 5. Significant linear correlations are found between one morning milk sampling and data from 305-days lactation regarding milk (P=0·002), protein (P=0·000) and fat yield (P=0·023) as well as protein (P=0·004) and fat content (P=0·047; Table 4). Similar to the one morning milk sample, DGVmilk and DGVprotein correlate positively with 305-days milk (P=0·032 and 0·088, respectively) and protein yield (P=0·087 and 0·044, respectively). DGVfat also correlates positively with 305-days milk and protein yield (P=0·085 and 0·024, respectively). However, DGVfat also has a positive correlation with fat yield (P=0·004). Additionally, DGVmilk correlates negatively and DGVfat% positively with protein (P=0·012 and 0·094, respectively) and fat content (P=0·049 and 0·044, respectively). Thus, similar correlations for DGVs are obtained using one morning milk sample and data on 305-days lactation, thus confirming the usefulness of one milk sampling.
Table 4. The Pearson correlation coefficient (r) between data from one morning milk sampling and data from 305-days lactation on milk, protein and fat yield as well as protein and fat content (n=23)

* P<0·05; **P<0·01; ***P<0·001
Table 5. The Pearson correlation coefficient between direct genomic values (DGV) and milk composition‡ based on data from 305-days lactation (n=23)§
† P<0·10; *P<0·05; **P<0·01; ***P<0·001
‡ Milk, protein and fat yield as well as protein and fat content
§ Results are shown only for components correlated significantly (P<0·10) with at least 1 DGV
¶ −0·25·DGVmilk+0·25·DGVfat+DGVprotein+100
Discussion
With the limited number of animals in this study, estimation of DGVs for novel traits of milk composition and technological properties based on observation from the present investigation was not possible. However, the DGVs for production traits included in the traditional evaluations have a reliability that is considerably larger than corresponding pedigree index, due to the connection to a large reference population. This has made it interesting to investigate the correlations between proofs for traditional production traits and the novel traits. Ongoing research will increase the number of genotyped cows with records to enhance milk composition and technological properties to a level exceeding this study. The ultimate goal is, however, in the future to have sufficient direct or most likely indirect records of the novel traits to be able to calculate DGVs for bulls in the reference population based on large daughter groups.
The results from this investigation show that linear correlations exist between the studied DGVs and milk composition traits and technological properties. To our knowledge, this is the first time DGVs for traditional production traits have been evaluated in relation to bovine milk composition and processability data. Earlier studies on genetic correlations show a negative correlation between milk yield and protein (−0·39), casein (−0·38) (Ikonen et al. Reference Ikonen, Morri, Tyrisevä, Ruottinen and Ojala2004) and fat content (−0·58) (Schennink et al. Reference Schennink, Heck, Bovenhuis, Visker, van Valenberg and van Arendonk2008), which is in agreement with the results obtained in our study. The positive linear correlations existing for DGVmilk, DGVprotein and DGVfat with milk and protein yield as well as for DGVprotein% and DGVfat% with protein content are expected and have been shown for genetic and phenotypic correlations previously (Stoop et al. Reference Stoop, Bovenhuis and van Arendonk2007; Schennink et al. Reference Schennink, Heck, Bovenhuis, Visker, van Valenberg and van Arendonk2008; Nixon et al. Reference Nixon, Bohmanova, Jamrozik, Schaeffer, Hand and Miglior2009). Both a positive genetic (0·86) and phenotypic correlation (0·91) between protein and lactose yield has been reported by Stoop et al. (Reference Stoop, Bovenhuis and van Arendonk2007), which is in accordance with the results found in this study. A high SCC has been shown to be associated with a high fat content and protein yield (Carlén et al. Reference Carlén, Strandberg and Roth2004; Stoop et al. Reference Stoop, Bovenhuis and van Arendonk2007; Cunha et al. Reference Cunha, Molina, Carvalho, Facury Filho, Ferreira and Gentilini2008). This is in accordance with the negative correlations found between DGVudder health and protein yield and fat content in our study. No significant correlation was found between DGVudder health and SCC in this study, which may be explained by the fact that DGVudder health includes information not only on SCC but also on treatment of mastitis and udder conformation. Thus, the linear correlations obtained between the studied DGVs and milk components are apparently reasonable and valid. The same holds for the technological properties of the milk. This study indicated a negative correlation between DGVudder health and gelation time, which is in accordance with earlier studies that report on an association between a high SCC and longer gelation time (Amenu & Deeth, Reference Amenu and Deeth2007; Cassandro et al. Reference Cassandro, Comin, Ojala, Dal Zotto, De Marchi, Gallo, Carnier and Bittante2008; Barowska et al. Reference Barowska, Litwinczuk, Wolanciuk and Brodziak2009). However, Wedholm et al. (Reference Wedholm, Larsen, Lindmark, ånsson, Karlsson and Andrén2006b) found no effect of SCC on milk clotting properties. The negative correlation between DGVmilk and cheese yield seems reasonable, given that high contents of protein and fat result in a high cheese yield (Lucey & Kelly, Reference Lucey and Kelly1994; Wedholm et al. Reference Wedholm, Larsen, Lindmark, ånsson, Karlsson and Andrén2006b). This also confirms the positive correlations existing between DGVprotein% and DGVfat% and cheese yield. In this study, model cheeses were produced without protein standardisation, pH adjustment or salt addition. Therefore cheese yield as defined in this study is highly dependent on the protein concentration in the milk and the results from this study could be compared with other research studies on model cheeses, such as the study of Wedholm et al. (Reference Wedholm, Larsen, Lindmark, ånsson, Karlsson and Andrén2006b). Hence, the results show the possibilities of obtaining valuable information on both detailed milk composition and technological properties for individual animals through DGVs for traditional production traits. This gives opportunities to select the most interesting heifers based on their DNA profile. By including new properties in the GS analyses, such as polyunsaturated fatty acids and gelation time, heifers can in the future be selected early in life to produce milk for specific dairy products and bulls with desired DGVs for the traits can be selected for breeding purpose.
The coefficients of variation for milk components are in the range to be expected and what has been reported earlier or somewhat higher (Schennink et al. Reference Schennink, Stoop, Visker, Heck, Bovenhuis, van der Poel, van Valenberg and van Arendonk2007; Cassandro et al. Reference Cassandro, Comin, Ojala, Dal Zotto, De Marchi, Gallo, Carnier and Bittante2008; Stoop et al. Reference Stoop, van Arendonk, Heck, van Valenberg and Bovenhuis2008). For gelation properties, however, the coefficients of variation are higher than those reported by Cassandro et al. (Reference Cassandro, Comin, Ojala, Dal Zotto, De Marchi, Gallo, Carnier and Bittante2008). Studies have shown high heritability for contents of fat (0·52), protein (0·60) and lactose (0·64), whereas the heritability for yields of these components are moderate (0·34–0·47) (Stoop et al. Reference Stoop, Bovenhuis and van Arendonk2007, Reference Stoop, van Arendonk, Heck, van Valenberg and Bovenhuis2008). Also for Ca and P the heritabilities are high (0·57 and 0·62, respectively), which indicates that these minerals may be changed by genetic selection (van Hulzen et al. Reference van Hulzen, Sprong, van der Meer and van Arendonk2009). The heritability for saturated fatty acids (0·19–0·59) is higher than for mono- and polyunsaturated fatty acids (0·09–0·21) (Schennink et al. Reference Schennink, Stoop, Visker, Heck, Bovenhuis, van der Poel, van Valenberg and van Arendonk2007; Stoop et al. Reference Stoop, van Arendonk, Heck, van Valenberg and Bovenhuis2008). The moderate coefficients of variation for the fat fraction (Table 1) thus imply that milk fat composition may be altered by genetic selection. More interestingly, however, are the opportunities arising for technological properties of milk. It has been shown that moderate heritability exists for gelation time (approximately 0·25) and gel strength (between 0·15 and 0·40) (Ikonen et al. Reference Ikonen, Ahlfors, Kempe, Ojala and Ruottinen1999; Cassandro et al. Reference Cassandro, Comin, Ojala, Dal Zotto, De Marchi, Gallo, Carnier and Bittante2008). Hence, the results indicate that these properties can be changed by genetic selection, which is an unused variation within the Nordic breeding work. This opens up for new challenges for the breeding and dairy industry.
It is often not possible for farmers to optimize every specific trait or character and usually farmers will focus on combined values. DGVNTM is a total merit index that is a combination of production, functional and conformation traits, which makes it an important economic tool for farmers. The results in this study show a tendency for lower casein and fat content as well as cheese yield when breeding for a high NTM. Therefore, it is of the outmost importance to not only consider the DGVs related to production traits when studying the impact of DGVs on milk traits, but also other DGVs that are essential to the farmers, such as DGVNTM. If these DGVs are not considered, there is a risk that milk traits may be adversely altered. This may result in contents and yields of milk components that are unfavourable from a processing and nutritional perspective.
We have demonstrated the potential of using DGVs of traits included in the breeding objective of today to obtain information on detailed milk composition and technological properties early in the heifers’ life. However, further studies using more animals, larger reference population and more SNP markers will reveal relationships with greater accuracy. Through the use of GS, new and interesting approaches are arising when a blood sample can be used to predict composition and technological properties of milk, thus making it possible for breeding and dairy companies to select cows for targeted milk production. This, in turn, will certainly increase the economic output for the industry as well as for the farmers, given that the industry changes the payment system for additional components to the farmers. The results obtained in this study are a first evaluation of the relations between DGVs for traditional production traits and novel milk traits. Today, these results can be used for indirect selection on traits that are not registered in the milk recording scheme, such as casein and minerals or technological properties. The future challenge will be to enhance the recording of new important traits in a reference population, making direct selection available for traits and properties that improve milk quality and are of economical importance.
The Genomic Selection project at Århus University, Denmark is acknowledged for the contribution of DGVs for the cows. Laure Bourgeois, Mia Davidsson and Johanna Ryssnäs, all M.Sc. at the Department of Food Technology, Engineering and Nutrition, Lund University, Sweden are greatly acknowledged for their help with the experimental work. We also wish to thank the staff at Nötcenter Viken for invaluable help during milking and the collection of data. This work was supported by grants from the Swedish Farmer's Foundation for Agricultural Research (SLF), Stockholm, Sweden.







