Introduction
The trematodes of the genus Echinostoma Rudolphi, 1809 (Echinostomatidae Looss, 1899) are intestinal parasites of birds and mammals with a worldwide distribution. Species bearing 37 spines in the head collar and sharing morphological and biological similarities are considered as belonging to the ‘revolutum’ species complex, referring to the type species of the genus, Echinostoma revolutum (Frölich, 1802) (Kanev, Reference Kanev1994; Kostadinova et al., Reference Kostadinova, Herniou, Barret and Littlewood2003; Fried and Graczyk, Reference Fried and Graczyk2004; Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013, Reference Georgieva, Blasco-Costa and Kostadinova2017; Faltýnková et al., Reference Faltýnková, Georgieva, Soldánová and Kostadinova2015). This speciose group of echinostomes has a controversial taxonomy mostly due to morphological similarities between species, the existence of cryptic diversity and the superficial or inaccurate description of some species (Kostadinova and Gibson, Reference Kostadinova, Gibson, Fried and Graczyk2000; Esteban and Muñoz-Antoli, Reference Esteban, Muñoz-Antoli, Fried and Toledo2009; Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014; Toledo et al., Reference Toledo, Radev, Kanev, Gardner and Fried2014; Chai et al., Reference Chai, Cho, Chang, Jung and Sohn2020).
In order to better understand the interrelationships and actual diversity within the ‘revolutum’ group, more robust analyses based on integrative taxonomy approaches are needed for most species. This approach has shown compelling evidence to support the taxonomic reappraisal of several species that had their affiliation clarified, mainly from Europe and Asia (Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013, Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Faltýnková et al., Reference Faltýnková, Georgieva, Soldánová and Kostadinova2015; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). Among the species that had its taxonomy recently illuminated with new morphological and molecular data is Echinostoma miyagawai Ishii, 1932. This species, originally described from Asia, had its occurrence in Oceania and Europe confirmed, as well as its conspecificity with Echinostoma friedi Toledo et al., 2000, and E. revolutum sensu Morgan and Blair (Reference Morgan and Blair1995, Reference Morgan and Blair1998a, Reference Morgan and Blairb) – both synonyms of E. miyagawai (Kostadinova et al., Reference Kostadinova, Herniou, Barret and Littlewood2003; Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Faltýnková et al., Reference Faltýnková, Georgieva, Soldánová and Kostadinova2015). Recently, some isolates from Bangladesh identified as Echinostoma robustum Yamaguti, 1935 were shown to be molecularly indistinguishable from E. miyagawai (Mohanta et al., Reference Mohanta, Watanabe, Anisuzzaman, Ohari and Itagaki2019; Heneberg, Reference Heneberg2020; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). Based on these findings, the synonymy between E. robustum and E. miyagawai was recently proposed (Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021).
More complexity for this scenario comes with the report of E. robustum in the New World based on larval and adult parasites from the USA and Brazil (Detwiler et al., Reference Detwiler, Bos and Minchella2010). The Brazilian isolate was identified after the molecular study of cercariae found in Biomphalaria glabrata (Say, 1818), for which adult forms are unknown. Although different authors have accepted the occurrence of E. robustum in the Americas mostly relying on the findings of Detwiler et al. (Reference Detwiler, Bos and Minchella2010), there is molecular evidence that these parasites belong to as-of-yet undescribed species of the ‘revolutum’ species complex or known species for which molecular data are not available (Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014; Heneberg, Reference Heneberg2020; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). In the present study, 37-collar-spined echinostomes found in a free-range chicken from Brazil were subject to morphological and molecular studies. Results obtained revealed that the isolate previously identified in the country as E. robustum is a new species herein described.
Materials and methods
Morphological study
The gastrointestinal tracts of 46 free-range chickens slaughtered for consumption were donated by owners from the municipality of Viçosa (20°45′14″S, 42°52′55″W), Minas Gerais, Brazil, in June 2018. Each segment of the gastrointestinal tract was individually opened with scissors in a longitudinal section and washed with tap water over a 100 μm mesh sieve. The helminths recovered were separated in Petri dishes containing physiological saline (0.9% NaCl). Specimens of trematodes were compressed between glass slides and fixed in 10% formalin. Subsequently, they were stained with alum acetocarmine, dehydrated in ethanol series, diaphanized in beechwood creosote and mounted on permanent slides with Canada balsam.
Morphological and morphometric studies of permanent preparations were performed under an Olympus BH2 optical microscope (Olympus, Tokyo, Japan). Measurements were obtained with the aid of a micrometre eyepiece and are expressed in micrometres, unless otherwise indicated. The abbreviations used in the morphological description are as follows: BL, body length; BW, maximum body width; BW/BL (%), body width as a proportion of the body length = shape of the body; FORE, forebody length; FORE (%), forebody as a proportion of body length; CL, head collar length; CW, head collar width; CW/BW (%), head collar as a proportion of body width; ASL, angular spines length; ASW, angular spines width; LSL, lateral spines length; LSW, lateral spines width; DSL, dorsal spines length; DSW, dorsal spines width; OSL, oral sucker length; OSW, oral sucker width; VSL, ventral sucker length; VSW, ventral sucker width; OSW/VSW, oral sucker to ventral sucker width ratio; ESL, esophagus length; ESL/BL (%), esophagus length as a proportion of body length; PL, prepharynx length; PHL, pharynx length; PHW, pharynx width; ATL, anterior testis length; ATW, anterior testis width; PTL, posterior testis length; PTW, posterior testis width; TEND, length of post-testicular region; TEND (%), length of post-testicular field as a proportion of body length; OVL, ovary length; OVW, ovary width; OVAR, distance between ovary and posterior margin of ventral sucker; OVAR(%), distance between ovary and posterior margin of ventral sucker as a proportion of the body length.
Photographs were taken with a Leica ICC50 HD digital camera coupled to the microscope Leica DM500 and analysed in the Leica Application software suite (LAZ EZ), version 2.0 (Leica Microsystems, Wetzlar, Germany). Drawings were made using a camera lucida. The type material is deposited in the Helminthological Collection of the Institute Oswaldo Cruz (CHIOC) and Collection of Trematodes of the Federal University of Minas Gerais (UFMG-TRE).
Molecular study
An ethanol-fixed specimen was used in the molecular study. DNA was extracted using the Wizard® Genomic DNA Purification kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. The concentration of the extracted DNA was estimated using a microvolume spectrophotometer NanoDrop® ND-1000 (Thermo Fisher Scientific, Wilmington, DE, USA). Partial regions of the genes 28S, ITS and Nad-1 were amplified using the primer pairs Dig-12/1500R, BD1/BD2 and NDJ11/NDJ2a, respectively (Luton et al., Reference Luton, Walker and Blair1992; Morgan and Blair, Reference Morgan and Blair1995; Kostadinova et al., Reference Kostadinova, Herniou, Barret and Littlewood2003; Tkach et al., Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003). PCR reactions were performed in a final volume of 25 μL, which included 12.5 μL of GoTaq® G2 Hot Start Polymerase DNA Polymerase (Promega), 1.25 pmol of each primer, about 50 ng of template DNA and sterile ddH2O. PCR products were electrophoresed on a 1.5% agarose gel, and the band of the expected size was purified with polyethylene glycol (PEG) [20% PEG 8000 (Promega) in 2.5 m NaCl solution]. Purified DNA was sequenced in both directions by capillary electrophoresis in an ABI3730 sequencer using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA).
Sequence chromatograms obtained were aligned and edited using the ChromasPro v.2.0.1 (Technelysium Pty Ltd, Australia) and contigs were used for phylogenetic analyses. They were compared with the sequences of species of Echinostoma available in GenBank obtained with Basic Local Alignment Search Tool – BLAST® (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990) searches (https://blast.ncbi.nlm.nih.gov). The sequences were assembled into three alignments with selected echinostomes using the MUSCLE implemented in MEGA X (Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018). Both nuclear and mitochondrial datasets were used according to the main goals of the molecular analyses, as follows: 28S data were used to confirm that our isolate is nested within Echinostoma in the Echinostomatidae, and the ITS and Nad-1 were used to assess its phylogenetic position within the genus. The alignments were trimmed to match the shortest sequence prior to the phylogenetic analyses. The evolutionary models used for the analyses were determined by Bayesian information criterion in MEGA X, corresponding to GTR + G + I for 28S, K2 + G for ITS and HKY + G + I for Nad-1. Outgroups for each dataset were chosen based on previous molecular phylogenetic studies including Echinostoma representatives and related taxa (Tkach et al., Reference Tkach, Kudlai and Kostadinova2016). Maximum Likelihood (ML) and Bayesian Inference (BI) were the methods used to generate phylogenetic hypotheses. ML analyses were performed in MEGA X and the nodal support was measured using the bootstrap test with 1000 replicates. BI analyses were performed with MrBayes v.3.2.6 (Ronquist et al., Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) using Markov chain Monte Carlo (MCMC) for 1 000 000 generations and sampling every 1000 generations. The ‘burn-in’ was established for the first 25% of the trees sampled, and nodal support was estimated with posterior probability.
For testing the taxonomic hypothesis of Echinostoma pseudorobustum sp. nov. as revealed by morphological evidence (see below), besides phylogenetic position and genetic distances, we used three species discovery methods (not requiring prior species assignments), namely the distance-based Assemble Species by Automatic Partitioning (ASAP) (Puillandre et al., Reference Puillandre, Brouillet and Achaz2021), the tree-based Poisson Tree Process (PTP) and multi-rate PTP (mPTP) models (Zhang et al., Reference Zhang, Kapli, Pavlidis and Stamatakis2013; Kapli et al., Reference Kapli, Lutteropp, Zhang, Kobert, Pavlidis, Stamatakis and Flouri2017). These analyses were performed using the Nad-1 dataset as this gene is considered suitable for barcoding species of Echinostoma (Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014). The latter (mPTP) was used especially because it has been shown to be more accurate for species delimitation in datasets with uneven sampling as in our Nad-1 data (Blair and Bryson, Reference Blair and Bryson2017). ASAP analyses were run online (https://bioinfo.mnhn.fr/abi/public/asap/) using the final Nad-1 alignment as input and default parameters for the three-distance metrics available, i.e. Jukes-Cantor (JC69), Kimura (K80) and simple distance (p-distance); only the partition showing the lowest ASAP-score for each metric was considered. PTP analyses were performed in the bPTP and mPTP web servers (https://species.h-its.org/ptp/; https://mptp.h-its.org/#/tree), using the BI tree as input and default parameters, except the MCMC for the bPTP analysis that was run for 500 000 generations; the convergence of chains was checked in the likelihood trace plot. Sequences obtained were deposited in GenBank.
Results
From the total of 46 chickens evaluated during the helminthological study, only one bird (2.2%) was found harbouring three morphologically similar specimens of echinostomes; one individual was used in the molecular analyses and the other two (paragenophores voucher specimens sensu Pleijel et al., Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008) were used in the morphological description. The morphology and molecular data gathered revealed a hitherto unknown species of 37-collar-spined Echinostoma from the ‘revolutum’ group, which is described here as follows.
Family Echinostomatidae Looss, 1899
Genus Echinostoma Rudolphi, 1809
Echinostoma pseudorobustum sp. nov.
[syn. Echinostoma robustum ex Biomphalaria glabrata of Detwiler et al., 2010]
Type host: Gallus gallus domesticus
Type-locality: Viçosa, State of Minas Gerais, Brazil
Type-material: Holotype – CHIOC 39709
Paratype – UFMG-TRE 121
Site in host: caeca
Prevalence: 2.2% (1/46); infected chicken was found harbouring three adult echinostomes.
Representative DNA sequences: 28S OK586835, ITS OK586836, Nad-1: OK564515
ZooBank registration: urn:lsid:zoobank.org:act:273DDBE0-EFF8-4EEA-ACD9-E5915495C382
Etymology: The new species is named based on the previous assignment of its larval stages to Echinostoma robustum by Detwiler et al. (Reference Detwiler, Bos and Minchella2010). The species name derives from the prefix pseudo- (false) added to the epithet robustum.
Description
[Based on two specimens (Figs 1 and 2); measurements are presented in Table 1]. Elongated body, flattened dorsoventrally, narrow, with maximum width at the level of ventral sucker (Figs 1 and 2). Body length/body width ratio 6–7:1. Forebody 19.3% (holotype) and 18.5% (paratype) of body length. Tegument with small spines from head collar to median region of body. Head collar well-developed, reniform, 45.8% and 53.2% of body width, with 37 spines, arranged as follows: 5 angular spines on each edge of head collar, 6 lateral spines on each side, and 15 dorsal spines in a double row. Oral sucker muscular, prepharynx present, small, pharynx ovoid, muscular, oesophagus occupying 9.8 and 10% of total body length, bifurcating in intestinal caeca next to ventral sucker. Blind caeca lateral, almost reaching the posterior end, overlapping vitellaria. Ventral sucker muscular, oval, in the equatorial region of the body. Average ratio between the diameter of oral sucker and ventral sucker 1:2.5. Two testes, rounded or subglobular, tandem, similar in size, in the final quarter of body, at 2 and 2.28 mm from the posterior end, corresponding to 18.4 and 19.8% of total body length. Cirrus-sac oval, between the level of intestinal bifurcation and the anterior margin of ventral sucker, containing seminal vesicle, pars prostatica, ejaculatory duct and cirrus. Genital pore under intestinal bifurcation. Ovary oval, in the third quarter of the body. Mehlis' gland contiguous with ovary and anterior testis. Vitellaria in two lateral fields, from pre-equatorial region to posterior end of the body, overlapping caeca; fields enlarged towards post-testicular region but non-confluent. Uterus intercecal, with many coils, between the posterior margin of ventral sucker and ovary, occupying 38.8% and 44.4% of total body length. Egg oval, yellow, operculated, immature.

Fig. 1. Echinostoma pseudorobustum sp. nov. found in chicken from Brazil: (A) Line drawing of the holotype. (B) Echinonostoma robustum (currently a synonym of Echinostoma miyagawai), a taxon that was previously associated with larval stages of the new species here described (redrawn from Yamaguti, Reference Yamaguti1935). (C) Line drawing of the collar of spines. (D) Detail of the cirrus-sac. Scale bars: A and B: 1 mm; C and D: 100 μm.

Fig. 2. Echinostoma pseudorobustum sp. nov. found in a chicken from Brazil: (A) Micrograph of the paratype. (B) Micrograph of collar spines showing angular (*) and lateral (#) spines. (C) Micrograph of dorsal spines (+). Scale bars: A: 1 mm; B and C: 100 μ m.
Table 1. Morphometric comparisons between Echinostoma pseudorobustum sp. nov. and other morphologically similar species of Echinostoma

Measurements are in μm, unless stated otherwise. Abbreviation of the morphological structures are as presented in the text. Asterisk indicates data taken from original drawings. Hash indicates the range for all collar spines. Measurements not provided or missing drawings are indicated by a dash. Superscripted H and P indicate data for Holotype and Paratype, respectively. Data by Yamaguti (Reference Yamaguti1935) correspond to the original description of Echinosoma robustum, currently a synonymy of Echinostoma miyagawai.
Remarks
The new species can be readily distinguished from the remaining Echinostoma of the ‘revolutum’ group by having the posterior testis located at the shortest distance to the posterior end, representing <20% of the body length (TEND 18.4 and 19.8%). Echinostoma pseudorobustum sp. nov. further differs from its congeners described in South America, namely Echinostoma barbosai Lie and Basch, 1966; Echinostoma chloephagae Sutton and Lunaschi, 1980; Echinostoma erraticum Lutz, 1924; Echinostoma luisreyi Maldonado et al., 2003; Echinostoma mendax Dietz, 1910; Echinostoma microrchis Lutz, 1924; Echinostoma neglectum Lutz, 1924; Echinostoma nephrocystis Lutz, 1924, and Echinostoma rodriguesi Hsu et al., 1968, by its larger body size (BL 10.8 and 11.5 mm vs <10.8 mm) and longer forebody (FORE representing at least 18.5% of the body length, instead of <18% in the above-mentioned species) (see Table 2). The other species with a similar body size is Echinostoma paraensei Lie and Basch, 1967, which can also be easily differenced by its smaller forebody (FORE 18.5 and 19.3% vs 11.3%), oesophagus length as a proportion of body length (ESL/BW 9.8 and 10% vs 4.3%), head collar as a proportion of body width (CW/BW 45.8 and 53.2% vs 36%) and uterine field (OVAR 38.8 and 44.4% vs 31.3%) but more importantly by the size of dorsal collar spines. Regarding Echinostoma lindoense Sandground and Bonne, 1940, a species described in Asia and reported in Brazil by Lie (Reference Lie1968), the new species is differentiated in having a smaller body size (BL 10.8 and 11.5 mm vs 12.4–17.2 mm). This species is very similar to E. miyagawai, but considered distinct from it due to the limits for the body length and width, head collar and ventral sucker width and size of cirrus-sac, ovary and testes (Kostadinova et al., Reference Kostadinova, Gibson, Biserkov and Chipev2000).
Table 2. Comparative data between body length, forebody (FORE%) and post-testicular (TEND%) distance proportional to the body between Echinostoma pseudorobustum sp. nov. and Echinostoma spp. belonging to the ‘revolutum’ group reported in South America

Asterisk indicates data taken from original drawings. Measurement not provided or missing drawings are indicated by a dash.
Even though molecularly characterized cercariae of the new species was identified as E. robustum by Detwiler et al. (Reference Detwiler, Bos and Minchella2010) (see below), currently accepted as E. miyagawai, adult morphology clearly shows the distinctiveness of E. pseudorobustum sp. nov. Comparing with the original description of E. miyagawai by Ishii (Reference Ishii1932), our specimens are much smaller (BL 10.8 and 11.5 mm vs 21–26 mm), more robust (BW/BL 14.3 and 16.1% vs 11.7%), have rounded testes instead of lobulated ones, longer oesophagus (ESL/BL 9.8 and 10% vs 5.4%) and uterine field (OVAR 38.8 and 44.4% vs 30.5%). In relation to the most recent reports of E. miyagawai from Eurasia, E. pseudorobustum sp. nov. also shows differences in the morphology of testes (smooth vs irregularly lobed), (Kostadinova et al., Reference Kostadinova, Gibson, Biserkov and Chipev2000; Toledo et al., Reference Toledo, Muñoz-Antoli and Esteban2000; Faltýnková et al., Reference Faltýnková, Georgieva, Soldánová and Kostadinova2015; Mohanta et al., Reference Mohanta, Watanabe, Anisuzzaman, Ohari and Itagaki2019; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021), while comparing to the original description of E. robustum of Yamaguti (Reference Yamaguti1935) (Fig. 1B), our specimens were larger and narrower (BW/BL 14.3 and 16.1% vs 23.2%), possess larger forebody (FORE 18.5 and 19.3% vs 16%), head collar width (CW/BW 45.8 and 53.2% vs 43.3%), and uterine field (OVAR 38.8 and 44.4% vs 28%), and smooth testes, unlike the irregularly lobulated ones.
The description of E. pseudorobustum sp. nov. led us to propose a slight modification in the diagnosis of Echinostoma. It remains as provided by Kostadinova (Reference Kostadinova, Jones, Gibson and Bray2005) with the following modification: Testes tandem, elongated-oval, smooth, indented or lobed, contiguous or separated, just post-equatorial; occasionally, anterior testis equatorial (TEND = 18–35%).
Molecular study
Molecular sequences of 28S (1063 bp), ITS (960 bp) and Nad-1 (442 bp) genes were successfully obtained for a specimen E. pseudorobustum sp. nov. obtained in the same host. After alignment and trimming, datasets composed of 1063 bp of 28S (56 sequences from 55 species/species-level lineages); 947 bp of ITS (12 sequences from 11 species/species-level lineages) and 442 bp of Nad-1 (156 sequences from 21 species/species-level lineages) were used in the subsequent analyses. Regarding 28S, >99% of similarity was verified between the Brazilian isolate and all 13 species of Echinostoma included in the analysis. Despite being of limited use for tackling the intrageneric relationships, the phylogenetic trees based on this gene clearly show that our isolate is nested within the representatives of Echinostoma (Fig. 3). However, the nodal supports for the different clades verified in the 28S tree were most of the time low. On the other hand, phylogenetic trees resulting from ML and BI analyses of both ITS and Nad-1 genes showed the newly sequenced Echinostoma clustering in a clade together with a Brazilian isolate identified as E. robustum lineage D by Detwiler et al. (Reference Detwiler, Bos and Minchella2010) and also confirmed that the new species belongs to the ‘revolutum’ group of Echinostoma (Figs 4 and 5).

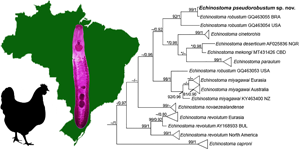
Fig. 3. Phylogenetic relationship between Echinostoma pseudorobustum sp. nov. (in bold) and other 37-collar-spined Echinostoma species inferred from sequences of 28S rDNA (1063 bp) based on Maximum Likelihood (ML) and Bayesian Inference (BI) analyses. Nodal support is indicated as ML/BI; values <0.70 (BI) and <70 (ML) are indicated by a dash. BRA, Brazil; UKR, Ukraine; KOR, South Korea; CZE, Czech Republic; USA, United States; GBR, United Kingdom; ISL, Iceland; NZL, New Zealand; KEN, Kenya; SVK, Slovakia; IND, India; LTU, Lithuania; PER, Peru.
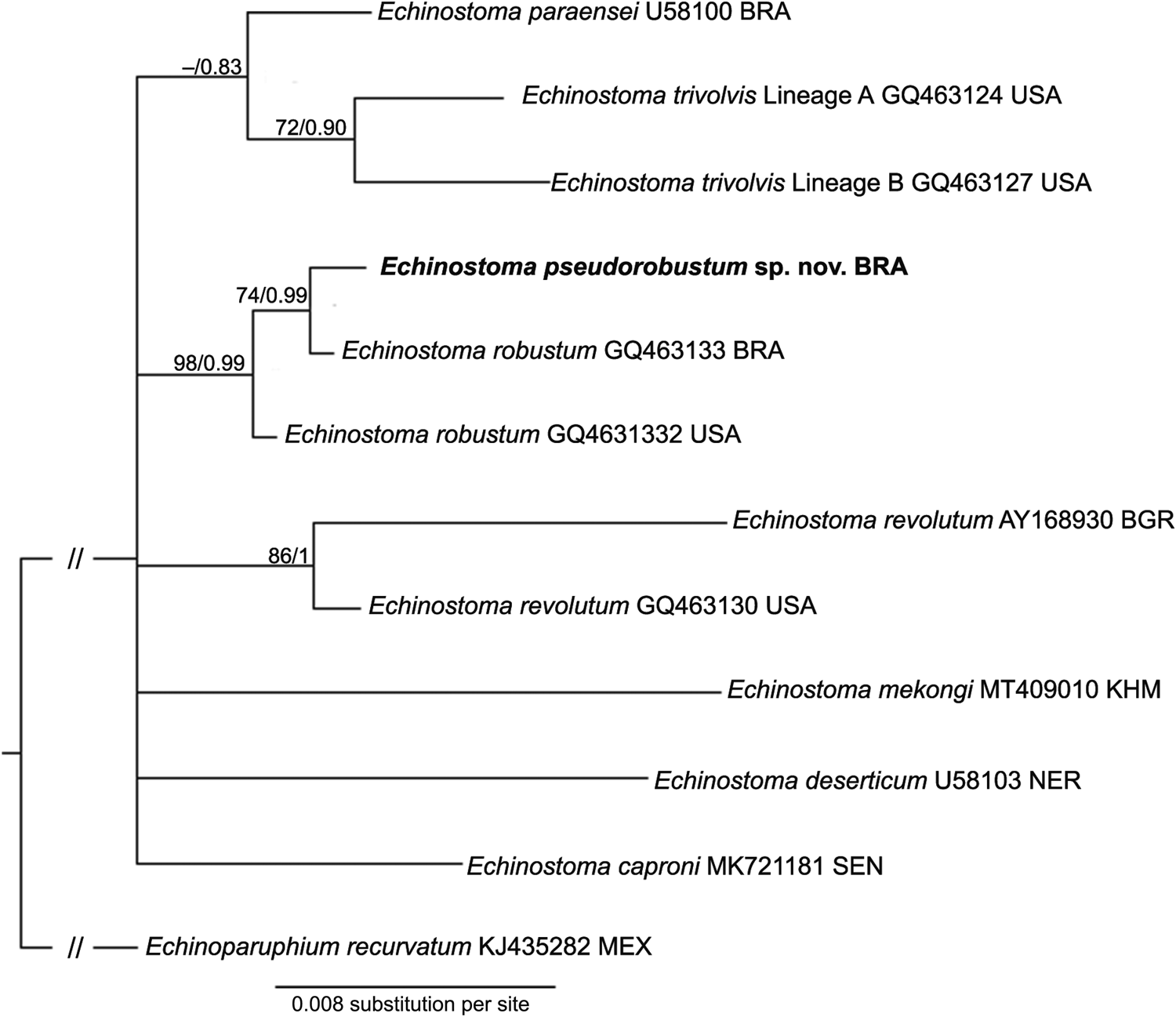
Fig. 4. Phylogenetic relationship between Echinostoma pseudorobustum sp. nov. (in bold) and other 37-collar-spined Echinostoma species inferred from sequences of internal transcribed spacer (947 bp) based on Maximum Likelihood (ML) and Bayesian Inference (BI) analyses. Nodal support is indicated as ML/BI; values <0.70 (BI) and <70 (ML) are indicated by a dash. BRA, Brazil; USA, United States; BGR, Bulgaria; KHM, Cambodia; NER, Niger; SEN, Senegal; MEX, Mexico.

Fig. 5. Phylogenetic relationship between Echinostoma pseudorobustum sp. nov. (in bold) and other 37-collar-spined Echinostoma species inferred from sequences of mitochondrial Nad-1 sequences (442 bp) based on Maximum Likelihood (ML) and Bayesian Inference (BI) analyses. Nodal support is indicated as ML/BI; values <0.80 (BI) and <80 (ML) are indicated by a dash. BRA, Brazil; USA, United States; RUS, Russia; NER, Niger; KHM, Cambodia; NZL, New Zealand; BGR, Bulgaria; SVK, Slovakia; RUS, Russia; CZE, Czech Republic.
Considering the ITS sequences, E. pseudorobustum sp. nov. differed 0.11–2.27% from other 9 species belonging to the genus Echinostoma included in the analyses. The newly-generated sequence falls into a clade containing two isolates [one from Brazil (GQ463133 – 0.11% of divergence – 1 nucleotide) and another from the USA (GQ463132 – 0.21% of divergence – 2 nucleotides)] identified as E. robustum by Detwiler et al. (Reference Detwiler, Bos and Minchella2010) (Fig. 4).
The analyses of the Nad-1 dataset showed high sequence similarity of E. pseudorobustum sp. nov. with E. robustum lineage D (GQ463055) (0.23% of divergence – 1 nucleotide), confirming their conspecificity. These Brazilian sequences clustered in a well-supported clade, sister to E. robustum lineage C (GQ463054) from the USA (Fig. 5). The latter most likely represent either an undescribed species or a known taxon yet not characterized molecularly as well as E. robustum lineage A (GQ463053), also from the USA, which appeared in a distantly related clade, sister to a clade composed by 35 sequences previously assigned to different taxa: E. friedi (Marcilla et al., unpublished), E. revolutum by Morgan and Blair (Reference Morgan and Blair1998a, Reference Morgan and Blairb), E. miyagawai by Georgieva et al. (Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017), Nagataki et al. (Reference Nagataki, Tantrawatpan, Agatsuma, Sugiura, Duenngai, Sithithaworn, Andrews, Petney and Saijuntha2015), Li et al. (Reference Li, Qiu, Zeng, Diao, Chang, Gao, Zhang and Wang2019), Fu et al. (Reference Fu, Jin, Li and Liu2019), and E. robustum by Mohanta et al. (Reference Mohanta, Watanabe, Anisuzzaman, Ohari and Itagaki2019). Currently, these isolates are treated under a single species with Eurasian + Oceanian distribution, E. miyagawai, given the rough similarity in the Nad-1 sequences (0.23–2.94%), which falls in the intraspecific range of variability (Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014; Nagataki et al., Reference Nagataki, Tantrawatpan, Agatsuma, Sugiura, Duenngai, Sithithaworn, Andrews, Petney and Saijuntha2015; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021).
The distance- and tree-based methods of species discovery based on Nad-1 yielded similar results (Supplementary Fig. 1), recognizing the conspecificity between the Brazilian isolate (GQ463055) of Detwiler et al. (Reference Detwiler, Bos and Minchella2010) with our newly sequenced specimen and their independent status as E. pseudorobustum sp. nov, yet the mPTP analysis also recognized in this new taxon the isolate GQ463054 of the same authors (see below). All distance metrics of the ASAP method recognized 19 species of Echinostoma, whereas the bPTP and mPTP methods informed us 20 and 18 taxa, respectively; this difference is due to the disputed affiliation of isolates assigned to Echinostoma caproni Richard, 1964 and those potentially belonging to the new species. While the ASAP and mPTP recognized a single entity for all isolates of E. caproni, the bPTP admitted two discrete entities. In turn, the bPTP and ASAP support the conspecificity of our isolate only with GQ463055, which further includes the North American isolate GQ463054 from Ladislavella elodes (Say, 1821) ( = Lymnaea elodes) considering the mPTP method. Agreeing with the phylogenetic analyses, all methods indicate that the American isolates assigned to E. robustum by Detwiler et al. (Reference Detwiler, Bos and Minchella2010) belong to at least two species other than E. miyagawai, including E. pseudorobustum sp. nov. and at least one undescribed or non-sequenced species. All methods confirmed that only isolates of Eurasian distribution can be assigned to E. revolutum (sensu stricto), setting them apart from the North American E. revolutum (sensu lato) (Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013), and that the large clade composed of 35 sequences represents a single species, E. miyagawai, with a wide geographic distribution (Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021).
Discussion
The actual diversity, host-associations, phylogenetic relationships and distribution of the ‘revolutum’ species complex within Echinostoma have been obscured partially because, in most contributions, authors have relied only on a single line of evidence (e.g. morphology or DNA) to support their species hypotheses, which ultimately rendered a dubious taxonomic status for several species (Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Faltýnková et al., Reference Faltýnková, Georgieva, Soldánová and Kostadinova2015; Chai et al., Reference Chai, Cho, Chang, Jung and Sohn2020). In the present study, thorough morphological evaluation along with phylogenetic reconstructions based on three loci (nuclear 28S and ITS, and mitochondrial Nad-1 markers; the latter also used in the species discovery analyses) made it possible to describe E. pseudorobustum sp. nov. from a chicken in Brazil and to confirm its conspecificity with cercariae previously found in B. glabrata from the same country identified as E. robustum (Detwiler et al., Reference Detwiler, Bos and Minchella2010). The results obtained reinforce, as already advocated by others (Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Nagataki et al., Reference Nagataki, Tantrawatpan, Agatsuma, Sugiura, Duenngai, Sithithaworn, Andrews, Petney and Saijuntha2015; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021), that isolates identified as E. robustum from the Americas, in fact, belongs to yet undescribed or molecularly non-characterized species, one of them here described as new. The morphological data for E. pseudorobustum sp. nov. clearly separate this species from E. robustum sensu Yamaguti (Reference Yamaguti1935), currently considered a synonym of E. miyagawai (Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). Moreover, all but one molecular analysis indicates that the other two American isolates identified as E. robustum represent two distinct species that need further scrutiny for a better characterization. These two North American isolates were identified after morphological study of experimentally obtained adults (Detwiler et al., Reference Detwiler, Bos and Minchella2010), but a detailed morphological description or deposition of vouchers was not provided, hampering the advance in this taxonomic issue.
In the last years, new species belonging to the ‘revolutum’ species complex, some of them with overlapping morphological characteristics, have been described after molecular studies, revealing a greater diversity than previously thought for this group (Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013, Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Faltýnková et al., Reference Faltýnková, Georgieva, Soldánová and Kostadinova2015; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). In South America, 13 species of Echinostoma have already been described with 37 head collar spines, but molecular studies are extremely scarce. In fact, E. pseudorobustum sp. nov. is the second species of the genus for which molecular data were generated (the other one is E. paraensei). This fact greatly hinders the advance in taxonomic aspects and life cycle, especially considering morphologically similar species. Despite these difficulties, the new species described here can be separated from other South American 37-collar-spined echinostomes by the longest forebody (highest FORE%) and the shortest post-testicular distance (testes in the last quarter of the body, i.e. lowest TEND%).
We recognized that the type and only known definitive host of E. pseudorobustum sp. nov., the chicken, a non-native domestic bird, probably is not the main final host for this echinostome species. In fact, the low prevalence of infection and worm burden, ultimately, suggests we are dealing with an accidental host. The process involving spillover of wild hosted echinostomes to chicken was previously reported in the Americas by Mello (Reference Mello1933), even though the identification as E. revolutum requires molecular confirmation. Moreover, the experimental susceptibility of G. g. domesticus to some South American species of Echinostoma (e.g. E. barbosai, E. erraticum, E. lindoense and E. rodriguesi) was shown by different authors (Lie and Basch, Reference Lie and Basch1966; Hsu et al., Reference Hsu, Lie and Basch1968; Lie, Reference Lie1968; Komma, Reference Komma1972). Information on the exact environment in which the infected chicken was established is not available. We speculate that it became infected in the rural area of the city, in which the birds are reared in free-ranging system and with access to an aquatic environment containing molluscs or amphibians harbouring echinostome metacercariae. Probably, a wild bird is the natural definitive host of E. pseudorobustum sp. nov. Given the natural host of the new species here described is likely distinct from the type-host here described, the existence of some level of host-induced phenotypic plasticity cannot be ruled out. However, clear morphological differences that set the studied specimens apart from the other Echinostoma spp. of the ‘revolutum’ group (see remarks) hardly could be associated with some host-induced variability. Moreover, a high level of morphological variation has not been reported to other species of Echinostoma known from experimental hosts (including chicken) compared to natural hosts (e.g. E. trivolvis, E. caproni and E. miyagawai) (Beaver, Reference Beaver1937; Jeyarasasingam et al., Reference Jeyarasasingam, Heyneman, Lim and Mansoul1972; Fried and Huffman, Reference Fried and Huffman1996; Kostadinova et al., Reference Kostadinova, Gibson, Biserkov and Chipev2000). It is also worth mentioning that, yet the validity of the new species is supported by morphological and molecular data, further morphological studies based on a larger number of specimens may evidence a wider breadth of intraspecific variability in the taxon but still not overlapping key diagnostic features with other species.
The incorrect assignment to some sequences of Echinostoma (‘revolutum’ group) that are available in the GenBank database probably stems from an inaccurate morphological identification of the sequenced isolates. It is also likely true that a finer morphological study combined with molecular findings may reveal that those so-called cryptic species, e.g. the three independent lineages under the same name in the GenBank, E. trivolvis, are in fact discrete entities, even morphologically, that each deserves their own name. Therefore, integrated approaches are extremely important to unravel the actual species diversity in Echinostoma and make it more reliable information available in the genetic repositories. In the present study, the levels of morphological variation, combined with phylogenetic support, confirm that the clades in which the new species here described as E. pseudorobustum sp. nov. nested and the one that grouped the Australasian lineage of E. miyagawai represent genetically distinct species.
In general, molecular data have not supported the occurrence of the same species of Echinostoma in the New and Old World. For instance, a widely known issue is related to the distribution of E. revolutum in the Americas, and even in Brazil (Kanev, Reference Kanev1994; Sorensen et al., Reference Sorensen, Curtis and Minchella1998; Kostadinova et al., Reference Kostadinova, Gibson, Biserkov and Chipev2000, 2003; Detwiler et al., Reference Detwiler, Bos and Minchella2010, Reference Detwiler, Zajac, Minchella and Belden2012; Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013; Nagataki et al., Reference Nagataki, Tantrawatpan, Agatsuma, Sugiura, Duenngai, Sithithaworn, Andrews, Petney and Saijuntha2015). Phylogenetic and species delineation analyses, as well as the nucleotide divergence verified among the E. revolutum (sensu stricto) and the North American isolates, do not support the occurrence of this species in the Americas (Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013, Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Nagataki et al., Reference Nagataki, Tantrawatpan, Agatsuma, Sugiura, Duenngai, Sithithaworn, Andrews, Petney and Saijuntha2015; Mohanta et al., Reference Mohanta, Watanabe, Anisuzzaman, Ohari and Itagaki2019; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021; present study). Moreover, data obtained in this study also do not support the occurrence of E. miyagawai (reported as E. robustum) in the Americas, as previously suggested by phylogenetic analysis carried out by different authors (Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013, Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014; Nagataki et al., Reference Nagataki, Tantrawatpan, Agatsuma, Sugiura, Duenngai, Sithithaworn, Andrews, Petney and Saijuntha2015; Mohanta et al., Reference Mohanta, Watanabe, Anisuzzaman, Ohari and Itagaki2019; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). An exception to this geographical pattern is the curious finding of a Brazilian species (E. paraensei) molecularly identified in Australia (0.3% of divergence in Nad-1 in relation to an isolate from the type locality) (Morgan and Blair, Reference Morgan and Blair1998a, Reference Morgan and Blairb). The occurrence of Asian E. lindoense in Brazil (Lie, Reference Lie1968), identified by morphological approach, needs verification, especially including molecular evidence.
Phylogenetic analyses based on the mitochondrial Nad-1 gene were found to be effective in exploring the interrelationships and intraspecific variability among 37-collar-spined echinostomes, also revealing congruence with available morphological data (Morgan and Blair, Reference Morgan and Blair1998b; Georgieva et al., Reference Georgieva, Blasco-Costa and Kostadinova2017). Owing to its high evolutionary rate, the probability of detecting diagnostic characters among closely related species increases (Vilas et al., Reference Vilas, Criscione and Blouin2005), which seems the case for the unambiguous differentiation between the new species and its sister taxon ‘E. robustum’ lineage C of Detwiler et al. (Reference Detwiler, Bos and Minchella2010), a result not achieved using the ITS dataset alone. In a recent study that included comprehensive multigene phylogenetic analyses for the Echinostomatidae, Izrailskaia et al. (Reference Izrailskaia, Besprozvannykh and Tatonova2021) suggested that the Cox-1 gene might be more promising for tackling taxonomic issues in this family due to its uniform accumulation of mutations across lineages, unlike the variable rates of mutation of the Nad-1 gene. They draw this conclusion chiefly based on the lack of significant genetic variation (using Nad-1) between isolates of E. miyagawai from East Asia and Europe that exhibit differences in the cercariae morphology and time to maturation, suggesting that Nad-1 does not provide enough phylogenetic signal for the distinction between these closely related lineages. However, considering the substantial body of evidence for the utility of the Nad-1 in unravelling the interrelations of Echinostoma spp. (Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013, Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021, present study), which in many cases corroborates the morphological data, further studies based on a wider taxon coverage for Cox-1 data, obviously including isolates of E. miyagawai from Europe, should be performed before accepting such assumption. Ideally, gathering complete mitogenomes may indicate more precisely which genes are more reliable and informative (individually or in concatenated analyses); currently, complete mitogenome is available only to four species, i.e. E. caproni, E. miyagawai, E. paraensei and E. revolutum, and mostly used to assess higher-level phylogenetic relationships (Fu et al., Reference Fu, Jin, Li and Liu2019; Le et al., Reference Le, Pham, Doan, Le, Saijuntha, Rajapakse and Lawton2020).
The species delimitation methods handle one of the most problematic issues in describing the biological diversity, which is identifying taxa at the species level (Carstens et al., Reference Carstens, Pelletier, Reid and Satler2013). Even though these approaches cannot be considered the panacea to the species problem, a bulk of methods have been recently employed to strengthen the hypotheses delineated from more traditional pieces of evidence (e.g. morphology, ecology and behaviour), and often unravelling cryptic diversity (Carstens et al., Reference Carstens, Pelletier, Reid and Satler2013). Two categories of species delimitation methods can be recognized: the species discovery, such as the GMYC, PTP and ASAP, which do not require prior assignments for the putative species, and the species validation ones, which demand the discrimination of the lineages a priori (e.g. *BEAST, BPP, spedeTEM) (Carstens et al., Reference Carstens, Pelletier, Reid and Satler2013; da Cruz and Weksler, Reference da Cruz and Weksler2018). Both strategies have been recently used for parasitic helminths such as trematodes (Pérez-Ponce de León et al., Reference Pérez-Ponce de León, García-Varela, Pinacho-Pinacho, Sereno-Uribe and Poulin2016), acanthocephalans (Pinacho-Pinacho et al., Reference Pinacho-Pinacho, García-Varela, Sereno-Uribe and Pérez-Ponce de León2018), nematodes (Ailán-Choke et al., Reference Ailán-Choke, Tavares, Luque and Pereira2020) and cestodes (Alves et al., Reference Alves, de Chambrier, Luque and Scholz2021), and their usefulness is acknowledged in these works. In the present study, the species discovery methods were useful to confirm the conspecificity between our newly sequenced specimen of E. pseudorobustum sp. nov. with the Brazilian isolate previously found in the first intermediate host (GQ463055), and to refute the occurrence of E. miyagawai in the Americas, as previously suggested by other authors (Georgieva et al., Reference Georgieva, Selbach, Faltýnková, Soldánová, Sures, Skirnisson and Kostadinova2013, Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). Even though Izrailskaia et al. (Reference Izrailskaia, Besprozvannykh and Tatonova2021) suggested that isolates identified as E. revolutum from North America and Eurasia represent a single species, the results obtained reported them as independent lineages similarly as found by other workers (Georgieva et al., Reference Georgieva, Faltýnková, Brown, Blasco-Costa, Soldánová, Sitko, Scholz and Kostadinova2014, Reference Georgieva, Blasco-Costa and Kostadinova2017; Chai et al., Reference Chai, Jung, Chang, Shin, Cho, Ryu, Kim, Park, Jeong, Hoang and Abullah2021). The differences observed in the number of species recognized by the mPTP (18), ASAP (19) and bPTP (20) are expected as each method has its own limitations, e.g. overlumping or oversplitting (Dellicour and Flot, Reference Dellicour and Flot2018; Puillandre et al., Reference Puillandre, Brouillet and Achaz2021), therefore incongruencies among them are common (Carstens et al., Reference Carstens, Pelletier, Reid and Satler2013). Considering the amount of genetic divergence between the two Brazilian isolates, including the newly generated one, and the poorly known North American isolate GQ463054, we do not consider the latter isolated in our concept of E. pseudorobustum as suggested by the mPTP analysis, yet this decision needs verification based on adult specimens for allowing morphological comparisons, and higher taxon sampling (accounting for the intraspecific variability) to be more useful for certain species delimitation methods.
Integrative taxonomy approach in the study of species of Echinostoma from South America is in its infancy. New broader approach studies are necessary to advance the knowledge of life history and host–parasite relationship involving this complex group of trematodes. Besides the description of a new species, data presented here suggest that E. miyagawai ( = E. robustum) must be considered a species with distribution restricted to Eurasia and Oceania, not occurring in New World.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182021001864.
Author contribution
M.C.V., P.V.A. and H.A.P. conceived and designed the study. M.C.V. conducted fieldwork. M.C.V., D.L.H. and P.V.A. performed molecular and phylogenetic analyses. M.C.V., D.L.H., P.V.A. and H.A.P. wrote the article. H.A.P. supervising, reviewing and editing the article.
Financial support
This work was also supported by the National Council for Scientific and Technological Development (CNPq), Brazil (research scholarship to H.A.P.) and the National Council for the Improvement of Higher Education (CAPES), Brazil (student scholarship to M.C.V. and D.L.H. and post-doctoral scholarship to P.V.A.).
Conflict of interest
None.
Ethical standards
The use of parasites from chickens was in accordance with the local animal research ethics committees of Universidade Federal de Viçosa (protocol CEUA/UFV 69/2017).