Introduction
Weeds evolving resistance to herbicides are the primary driving force for reduced efficacy of chemical weed control (Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013). Resistance mitigation strategies are needed, therefore, to preserve the usefulness of available herbicide products over time. Because herbicide resistance is an evolutionary outcome, it is necessary to understand the evolutionary forces underpinning the process in order to design effective mitigation strategies.
Herbicide applications are abrupt disturbances to targeted weed populations. The prior existence of a subset of a population that is genetically capable of surviving the perturbation allows the population to persist in the presence of the chemical control; that is, herbicides select for adaptive alleles conferring resistance. The repetition of the selection process over time increases the frequency of the resistance alleles until the population is considered resistant (Heap Reference Heap2014); typically, a field population is considered resistant when more than 20% of the individuals are no longer controlled by the herbicide (Llewellyn and Powles Reference Llewellyn and Powles2001).
As in other evolutionary processes, the capacity of a population to evolve in response to a novel environment requires genetic variation (Mimura et al. Reference Mimura, Ono, Goka and Hara2013; Trucco et al. Reference Trucco, Hager and Tranel2006). Because it is available immediately, the standing genetic variation within the population is thought to be the primary source of resistance alleles and, consequently, the main factor upon which adaptation depends (Délye et al. Reference Délye, Jasieniuk, Le and orre2013b). However, resistance alleles can also be introduced by gene flow—via pollen or seeds from a resistant population (i.e., immigration)—or by de novo mutations—wherein herbicide-resistant mutants arise spontaneously in a given sensitive population (Jasieniuk et al. Reference Jasieniuk, Brûlé-Babel and Morrison1996). Importantly, the relative contribution of each of these three sources influences what strategies should be applied to counteract herbicide-resistance evolution.
The importance of various factors leading to resistance evolution is commonly simulated by models with the purpose of providing insights into herbicide-resistance management (Renton et al. Reference Renton, Busi, Neve, Thornby and Vila-Aiub2014). Because estimations of parameters such as the standing genetic variation or the mutation rate conferring resistance are extremely difficult to calculate empirically, they are often based mostly on assumptions, limiting the reliability of the models. In fact, models are very sensitive to the mutation rate, because it both contributes to the standing genetic variation of the population before selection and causes new genetic variants to arise after the onset of selection (Bagavathiannan et al. Reference Bagavathiannan, Norsworthy, Smith and Neve2013; Neve et al. Reference Neve, Norsworthy, Smith and Zelaya2010). Unfortunately, to our knowledge, there are no peer-reviewed published data on the empirical determination of the spontaneous mutation rate producing herbicide resistance in plants.
In the case of acetolactate synthase (ALS) inhibitors—the group of herbicides with the most cases of resistance reported—an insensitive target site is the main cause of resistance, with multiple mutations conferring resistance (Powles and Yu Reference Powles and Yu2010). At least 29 different amino acid substitutions (Table 1) have been reported in plants, distributed across eight amino acid sites in the ALS protein (Figure 1). The large number of resistance-conferring mutations in the ALS gene likely plays an important role in the high incidence of resistance to ALS inhibitors.

Figure 1. Sites in the acetolactate synthase (ALS) protein at which substitutions providing resistance have been identified in plants. Numbering of amino acids is based on the precursor ALS from Arabidopsis thaliana.
Table 1. Amino acid substitutions conferring resistance to acetolactate synthase inhibitors (Heap Reference Heap2018)

a Abbreviations: SUs, sulfonylureas; IMIs, imidazolinones; S, sensitive; r, weak resistance; R, strong resistance (>10 times the label rate); nd, not determined; multiple listings (e.g., S/r) indicates results varied among species.
It has been suggested that the exposure of weeds to sublethal herbicide treatments could elevate mutation rates, increasing the probability of generating new genetic variants conferring resistance (Gressel and Levy Reference Gressel, Levy, Pareek, Sopory and Bohnert2009). The exposure of weeds to sublethal doses is common in farming situations, for example, at field edges or when weed coverage by the herbicide is reduced due to protection from the crop canopy. There are several reported cases demonstrating that herbicide activity in plants is not only capable of damaging DNA, but can also increase the mutation frequency (Doganlar Reference Doganlar2012; Filkowski et al. Reference Filkowski, Besplug, Burke, Kovalchuk and Kovalchuk2003; Plewa Reference Plewa1985). Moreover, it was also proposed that herbicide-mediated stress may select for mutators (a mutant of a normal gene that increases the mutation rate) and may even induce mutator activity within genomes, leading to adaptive evolution (Gressel Reference Gressel2011).
The objectives of this study were to (1) determine the de novo mutation rate conferring herbicide resistance in a plant population and (2) test whether the mutation rate increases in plants exposed to a sublethal herbicide dose. To achieve these goals, we used grain amaranth (Amaranthus hypochondriacus L.) and resistance to ALS-inhibiting herbicides as a model system, which enabled us to screen millions of plants. We failed to recover any spontaneous mutations among 70.8 million seedlings, suggesting that de novo mutations conferring herbicide resistance might occur less frequently than expected.
Materials and Methods
Model system
Grain amaranth is not known to have herbicide resistance in its genetic background, making it a much better system in which to look for de novo resistance mutations than its weedy counterparts. For example, widespread ALS-inhibitor resistance in waterhemp [Amaranthus tuberculatus (Moq.) J. D. Sauer] (Patzoldt and Tranel Reference Patzoldt and Tranel2007) would make a de novo mutation conferring resistance to ALS inhibitors difficult to distinguish from a contamination event. Additionally, grain amaranth produces pale seeds, which can be used as a contamination checkpoint for black-seeded amaranth weed seeds. Like other Amaranthus species, grain amaranth produces thousands of seeds per mother plant, but it has reduced seed dormancy and higher genetic homogeneity compared with its weedy counterparts. Rapid and uniform seed germination was particularly important, in that it allowed for more uniform exposure of seedlings to soil-applied herbicide selection.
ALS inhibitor–resistant mutations are functionally dominant at typical herbicide use rates, enabling the selection of newly arisen spontaneous mutations, which may be present in only one chromosome during the first generation (heterozygous mode). Furthermore, ALS mutations provide high-level resistance; thus, high rates can be used to strongly select out resistant mutants from a sensitive population, diminishing the occurrence of false positives. In addition, the soil activity of ALS inhibitors permits the selection of plants at the seedling stage, allowing for the screening of millions of individuals in a reduced space.
Experiment size
Our method consisted of producing seeds, planting them at a very high density, screening them at the seedling stage with a PRE herbicide, and looking for surviving resistant individuals. To estimate how many seedlings we should screen, we calculated the probability that at least one resistance-conferring mutation will occur in a single plant as
where each n event is a single-nucleotide substitution known to provide strong resistance (>10 times the field use rate) to imidazolinone herbicides (Table 1) and can occur in grain amaranth based on its ALS gene sequence (GenBank accession EU024568). The per-event probability pn is the probability that the n event will occur in a single generation. We assigned each nucleotide substitution type a probability based on the spontaneous mutation frequencies in Arabidopsis thaliana reported by Ossowski et al. (Reference Ossowski, Schneeberger, Lucas-Lledo, Warthmann, Clark, Shaw, Weigel and Lynch2010), adjusted by the C:G bias in the A. thaliana genome (Arabidopsis Genome Initiative 2000). As a result, the probability of any given plant spontaneously mutating to be resistant is 7.9 × 10−8 (Table 2). We thereby estimated that we should screen at least 38 million plants to have a 95% chance of finding at least one spontaneously resistant plant.
Table 2. Theoretical calculation of the mutation rate conferring acetolactate synthase (ALS)-inhibitor resistance in grain amarantha
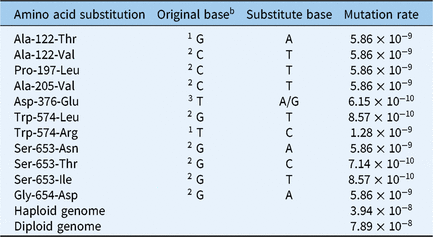
a Mutation rate per haploid genome per generation is determined as the accumulation of the occurrence probability of all the ALS gene base substitutions known to provide resistance to imidazolinones as described in Equation 1. Nucleotide substitution probabilities are based on mutation type frequencies found in Arabidopsis thaliana.
b Numbers indicate base position in DNA codon.
Seed production
Grain amaranth ‘Plainsman’ (Amaranthus hypochondriacus L. × Amaranthus hybridus L.) (Baltensperger et al. Reference Baltensperger, Weber and Nelson1992) was obtained from the North Central Regional Plant Introduction Station (Ames, IA). All plant batches were initiated from sibling seeds from a unique self-pollinated plant. This was done to rule out preexisting mutations conferring resistance to ALS inhibitors. The unique self-pollinated plant was grown during the summer of 2014 in an isolated greenhouse room to avoid contamination from ALS inhibitor–resistant Amaranthus weeds.
Subsequently, seeds were produced in independent batches from about 200 grain amaranth plants per batch. Five batches were produced in total from the spring of 2015 to the spring of 2017. To initiate a batch, seeds were planted in 12.3 by 12.3 cm inserts filled with 3:1:1:1 LC1 (soil:peat:torpedo sand; Sun Gro Horticulture, Agawam, MA). Flats were subirrigated overnight before seeds were sowed. Sowing density was 100 seeds per insert. Inserts were placed into 27.4 by 53.9 cm flats with holes that were then placed into display flats for bottom watering. The germination rate of the seeds was between 90% and 95%. Plants were allowed to grow in the 12.3 by 12.3 cm inserts until each plant had its first true leaf that was longer than 0.5 cm. Seedlings then were transplanted into 10.1-cm-diameter pots filled with the same medium described earlier and prewatered before transplanting. Between 400 and 500 seedlings were germinated each time to allow for selection of a uniform group of about 200 plants for each batch. When plants reached 12 to 15 cm in height, they were transplanted into 9.4-L pots filled with a medium that was 1:1:1 (soil:peat:perlite) plus 30 g of 13-13-13 Osmocote® (ICL Specialty Fertilizers, Dublin, OH) and prewatered before transplanting. The pots were placed in a greenhouse at a density between 4 and 10 plants per m2 depending on the batch. The greenhouse room was maintained at 28/22 C day/night temperatures. Natural sunlight was supplemented with mercury-halide lamps to provide a 16:8-h photoperiod during the entire plant cycle. Plants were watered twice daily by an automatic irrigation system that delivered water directly via an independent emitter located in each pot. Liquid fertilizer was applied once a week with a starting dose of 300 ppmv (20-20-20) increasing to 400 ppmv after flowering had started. Flowering started approximately 45 d after planting in all batches. Plants were harvested at least 10 d after it was noted that seeds were easily detached from the plants. Inflorescences were cut manually and stored in paper bags for 1 mo to allow for drying. Seeds were hand threshed and then cleaned using a South Dakota Seed Blower (Seedburo Equipment, Des Plaines, IL). The seed from each parental plant was weighed and stored separately in bottles. Seed bottles were stored in a cold room at 4 C.
Contamination control
Each parental plant (approximately 1,000 total) was tested for the presence of both A. tuberculatus DNA and the most common mutation providing ALS-inhibitor resistance in weedy amaranths, Trp-574-Leu (Patzoldt and Tranel Reference Patzoldt and Tranel2007; Yu and Powles Reference Yu and Powles2014). Amaranthus tuberculatus DNA can be distinguished from that of grain amaranth by the presence of a restriction site recognizable by EcoRV endonuclease in region A of the ALS gene (Foes et al. Reference Foes, Liu, Tranel, Wax and Stoller1998; Trucco et al. Reference Trucco, Jeschke, Rayburn and Tranel2005). Similarly, the Trp-574-Leu mutation is detected by MfeI endonuclease in region B of the ALS gene (Foes et al. Reference Foes, Liu, Vigue, Stoller, Wax and Tranel1999; Patzoldt and Tranel Reference Patzoldt and Tranel2007). DNA was extracted from young leaf samples based on the CTAB procedure indicated by Doyle and Doyle (Reference Doyle and Doyle1990). DNA content of each sample was measured using NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and diluted to 10 ng μl−1. PCR consisted of 12.3 μl H2O, 5 μl 5X GoTaq® green buffer (Promega, Madison, WI), 2 μl dNTP (2.5 mM), 2.5 MgCl2 (25 mM), 1 μl (0.4 μM) of each primer, 0.2 μl GoTaq® DNA polymerase (5 U μl−1; Promega), and 1 μl sample DNA (10 ng μl−1) in a total reaction of 25 μl. Primers used for the amplification of ALS region A were: alsf1, 5′-AGCTCTTGAACGTGAAGGTG; alsr1, 5′-TCAATCAAAAGAGGTCCAGG; and for the ALS region B amplification: alsf2, 5′-TCCCGGTTAAAATCATGCTC; alsr2, 5′-CTAAACGAGAGAACGGCCAG; as described by Foes et al. (Reference Foes, Liu, Tranel, Wax and Stoller1998). The thermocycling program began with 3 min at 95 C; then 35 cycles of 1 min at 95 C, 1 min at 56 C, and 1 min at 72 C; with a final cycle of 5 min at 72 C. DNA amplification was checked with a 1% agarose gel. A volume of 10 μl of product of each PCR reaction was subjected to its respective digestion: 16.5 μl H2O, 3 μl CutSmart™ buffer (10X; New England Biolabs, Ipswich, MA), and 0.5 μl EcoRV (20 U μl−1; New England Biolabs); or 2.2 μl H2O, 2.5 μl CutSmart™ buffer (10X), and 0.3 μl MfeI (20 U μl−1; New England Biolabs); and incubated overnight at 37 C. Digested products were run on a 2% agarose gel. For both digestions, negative samples produced a band at 500 bp, while positive samples produced a band at 400 bp.
Because hybridization between different Amaranthus species is common (Trucco et al. Reference Trucco, Jeschke, Rayburn and Tranel2005), batches were grown in a greenhouse room, and planting dates were managed to avoid flowering during the time of the year when weedy amaranth (particularly A. tuberculatus) pollen is most prevalent in the air in central Illinois (from June to September). We also ensured that amaranth plants were not present in any of the adjacent greenhouse rooms.
Resistance screening
The seed obtained from the batches was planted in 12.3 by 12.3 cm inserts filled with the same medium described earlier (3:1:1:1; LC1: soil:peat:torpedo sand) and covered with a layer of 2 mm of the same growth medium. Seed was kept separated by parental plant. Planting density did not exceed 23,000 seeds per insert, in order to respect the upper limit of 160 seeds cm−2, determined from preliminary experiments as not detrimental for the germination rate. Heterozygous A. tuberculatus plants (approximately 100 seeds planted) with the Trp-574-Leu mutation were included in each screening experiment to verify effective resistance selection. Inserts were placed into 27.4 by 53.9 cm flats with holes that were then placed on display flats for bottom watering. To moisten the growth medium before seeds were sowed, flats were subirrigated overnight. Right after sowing, flats received an application of imazethapyr (Pursuit®, BASF, Florham Park, NJ) at a 10X rate (700 g ae ha−1), which was previously determined to effectively discriminate between A. tuberculatus heterozygous resistant mutants (carrying the Trp-574-Leu substitution) and sensitive grain amaranth plants. Herbicide was applied using a research spray chamber (De Vries Manufacturing, Hollandale, MN) calibrated to deliver 185 L ha−1 at 276 kPa. The flats were located 45 cm below the nozzle (80015 EVS, TeeJet Technologies, Wheaton, IL). After herbicide spraying, water was sprayed over the flats at a 1-mm depth to incorporate the herbicide. Afterward, flats were watered from the top daily. Seeds started to germinate on the same day. Most seedlings emerged from the second to the seventh day after treatment (DAT). Sensitive seedlings stopped their growth about 10 DAT. At 15 DAT, surviving plants were evident and were transplanted to 10.1-cm-diameter pots filled with the same medium described earlier (3:1:1:1; LC1: soil:peat:torpedo sand).
Germination rate
Germination rate was calculated in separate 7.7 by 5.5 cm inserts filled with the same medium described earlier (3:1:1:1; LC1: soil:peat:torpedo sand) to estimate the germination rate of the seed bulk. From each parental plant, 160 seeds were sowed in 1 cm2 of soil to simulate the density used for resistance screening. This density was maintained by sowing the seeds with a tube that had a constant measured diameter. These flats were not treated with herbicide. After germination, seedlings were counted manually.
Resistance confirmation
Once a surviving seedling was detected, it was first examined visually for potentially distinguishing phenotypic traits (e.g., stem pubescence and leaf shape) and tested for A. tuberculatus DNA contamination by PCR as described earlier. Similarly, Palmer amaranth (Amaranthus palmeri S. Watson) contamination was checked by PCR that consisted of the mix described earlier with the following primers: forward, 5′-GCGAACATGTTTATCATACCTGG-3′; reverse, 5′-CTCAATACTGGGTGCATCCAC-3′ (Murphy et al. Reference Murphy, Plewa, Phillippi, Bissonnette and Tranel2017). Thermocycling started with 5 min at 95 C; then 27 cycles of 1.5 min at 95 C, 1 min at 59 C, and 2 min at 72 C; with a final cycle of 5 min at 72 C. DNA amplification was checked on a 1% agarose gel for direct confirmation of any A. palmeri DNA presence. When a plant was verified to be none of these weed species, a POST application of imazethapyr at 1X rate (70 g ae ha−1) was sprayed when the plant was 10-cm tall. If the plant survived, it was allowed to produce seed. DNA was extracted to sequence the ALS gene to look for the presence of mutations. Sanger sequencing was performed by the University of Illinois Core Sequencing Facility (334 Edward R. Madigan Laboratory, 1201 W. Gregory, Urbana, IL). When the plants finished their cycle, the seed was cleaned manually and stored in the cold room at 4 C. At least 10 offspring from each resistant plant were planted. When offspring plants were 10-cm tall, imazethapyr at 1X rate was applied to check for resistance inheritance.
Sublethal herbicide treatment
The fifth batch of plants was treated with a sublethal dose of herbicide. The selected herbicide treatment aimed to increase the mutation rate without significantly reducing the seed output. Before flower initiation, a mutation would likely remain in the vegetative tissue without reaching the reproductive structures, while after embryo formation, embryo cells are not dividing actively, decreasing the number of DNA replication events susceptible to mutation. Consequently, we targeted the herbicide application at 5 d before appearance of the inflorescence. At that time, many meiotic events should have already started; thus, the mutation rate could be significantly increased, but the reproductive tissue would not be directly exposed, decreasing the chance of dramatically reducing seed output. Plant height at the time of herbicide treatment ranged from 15 to 25 cm.
For the herbicide stress, we used atrazine (AAtrex®, Syngenta Crop Protection, Greensboro, NC) applied POST. Like other contact herbicides, atrazine leads to the accumulation of free radicals that damage DNA, but unlike most other contact herbicides that are nonsystemic, atrazine moves through the xylem, increasing its opportunity to affect the meristem. We did not use a truly systemic herbicide, because they strongly accumulate in the meristem, radically decreasing seed output. Additionally, given grain amaranth’s indeterminate growth, atrazine’s residual activity was advantageous; any atrazine landing on the soil potentially could prolong plant exposure to herbicide stress.
Dose level was determined from preliminary experiments to identify a rate that was injurious but did not decrease the seed output by more than 10% compared with a nontreated plant. Two rates were selected, and plants were divided into two treatments: 110 plants treated at 0.2 kg ha−1 and 110 plants treated at 0.1 kg ha−1. Also, an untreated group of 20 plants was included as a control. Herbicide applications were performed in the research spray chamber calibrated with the same parameters as described earlier. The nozzle was maintained 45 cm above the plants.
Results and Discussion
Seed production
The five batches of grain amaranth produced more than 87 million seeds in total (Table 3). Seed production between batches was mostly homogeneous, with the exception of one batch that had about triple the yield of the others. The seed produced per parental plant was highly variable, ranging from less than 1,000 to more than 300,000 seeds, with an average of 77,000 seeds per plant (SD = 59,500). The average seed weight was similar across batches and plants, with a mean of 0.06 g per 100 seeds (SD = 0.004).
Table 3. Calculated number of screened grain amaranth plants per seed batch based on their respective germination rates

a Means are grouped based on the Tukey’s studentized range (HSD). Comparisons significant at the 0.05 level are indicated by different letters.
For the atrazine-treated batch (Batch E in Table 3), although visually affected (Figure 2), treated plants recovered well from atrazine treatment, with growth and development similar to those of the nontreated group. The yield per plant did not differ significantly between atrazine treatments (unpublished data). Moreover, yield parameters of the whole batch of plants did not differ significantly from those of the untreated batches. More than 14 million seeds were obtained from the atrazine-treated batch.

Figure 2. Grain amaranth plants at 15 d after atrazine treatment showed clear symptoms of herbicide injury. Plants treated at 0.2 kg ha−1 were visually more affected on average than plants treated at 0.1 kg ha−1.
Spontaneous mutation rate leading to herbicide resistance
Based on each batch’s germination rate, we determined that at least 70.8 million plants were screened (Table 3). Our high-throughput screening procedure was demonstrated to be sufficiently robust to identify resistant individuals within a sensitive population (Figure 3). In total, about 150 plants were recovered from the herbicide screening. Of these, 25 were confirmed resistant and the remaining were determined (using procedures described earlier) to be sensitive escapes. The recovery of a small number of sensitive escapes indicated that we used an appropriate herbicide rate: a higher rate would not have allowed marginally resistant plants to survive, whereas a lower rate would have resulted in too many sensitive escapes, requiring extensive follow-up research. The 25 recovered resistant individuals were all determined to be contaminations from ALS–inhibitor resistant Amaranthus weeds: A. tuberculatus and A. palmeri. Of these 25 individuals, 18 were assumed to arise from seed contamination and 7 from pollen contamination, based on homozygosity and heterozygosity, respectively, at the ALS locus (unpublished data). No spontaneously resistant genotypes were detected among 70.8 million plants, suggesting the probability of finding a spontaneous ALS–inhibitor resistant mutant in a given sensitive plant population is less than 1.4 × 10−8.

Figure 3. An amaranth plant resistant to acetolactate synthase inhibitors was easily distinguished among thousands of sensitive amaranth plants at 15 d after treatment with imazethapyr at 10X rate (700 g ha−1).
In our theoretical approach to this study, we initially calculated the probability of a mutation giving rise to a grain amaranth plant resistant to imidazolinone herbicides to be 7.9 × 10−8 (Table 2). Thus, from 70.8 million seedlings, the chance of detecting at least one resistant plant was 99.6% (Table 4). Our failure to identify a resistant mutant indicates (P = 0.003) that the mutation rate was lower than expected. It should be considered, though, that some C:G→T:A mutations found in A. thaliana were produced in C:T sites that are known to be methylated. Spontaneous deamination of methylated cytosine leads to thymine substitution being an important source of mutation (Schmitz et al. Reference Schmitz, Schultz, Lewsey, Malley, Urich, Libiger, Schork and Ecker2011). However, we do not know the methylation status of C:T sites in the ALS gene of grain amaranth. Assuming C:T sites are not methylated in the grain amaranth ALS gene, the expected theoretical probability of finding an ALS–inhibitor resistant plant diminishes to 5.8 × 10−8, being still significantly higher (P = 0.015) than the observed results. More importantly, the mutation rate between genic and intergenic regions varies substantially in A. thaliana (Ossowski et al. Reference Ossowski, Schneeberger, Lucas-Lledo, Warthmann, Clark, Shaw, Weigel and Lynch2010). This was attributed to a higher mutation rate in pericentromeric regions, where gene density is lower than that found farther away from the centromere. Although we do not know the exact location of the ALS gene in relation to the centromere, we may speculate that it is situated away from the centromere, as is generally the case for most genes. Considering the mutation rate within genic regions only and subtracting the mutations originated in mobile elements, the theoretical probability for selecting an ALS–inhibitor resistant mutant in a sensitive population decreases to 1.38 × 10−8. In that case, the chance of getting at least one resistant mutant in 70.8 million plants is reduced to 62.4%. That estimation is not significantly different (P = 0.376) than the observed mutation rate. If this calculation is close to reality, we should have screened at least 218 million plants to have a 95% chance of finding at least one resistant individual (Table 4).
Table 4. Theoretical calculation approaches for determining the ALS-inhibitor resistance mutation rate in grain amarantha

a Mutation rate is the probability that one or more resistant plants arise in one generation. Nucleotide substitution probabilities are based on mutation type frequencies found in Arabidopsis thaliana.
b Reciprocal = 1/mutation rate per plant per generation.
c Probability of finding at least one spontaneous ALS-resistant mutant in 70,883,089 screened plants based on the theoretical mutation rate.
d Probability that the observed number of mutants (<1.4 × 10−8) was different from the expected number based on the binomial exact test.
e Probability that any one of all nucleotide substitutions known to provide resistance to imidazolinones occurs.
f Nucleotide substitution probability adjusted by the proportion of C:G methylated sites in the Arabidopsis thaliana genome.
g Nucleotide substitution probability adjusted by the proportion of mutations in genic regions of the Arabidopsis thaliana genome.
h Probability that any one of the three mutations, Ala-122-Thr, Trp-574-Leu, and Ser-653-Asn, occurs.
i Probability that the mutation Trp-574-Leu occurs.
In addition, it must be pointed out that in many of the published ALS–inhibitor resistant cases, mutations reported to confer resistance to a 10X rate (strong resistance) were in the homozygous state. However, newly arisen spontaneous mutations are expected to be mostly in one chromosome only (heterozygous) during the first generation, likely conferring a lower level of resistance than in homozygotes. Therefore, screening at a 10X rate may have eliminated individuals containing some of the mutations known to confer resistance. To examine this possibility, we first confirmed that this herbicide rate, applied in the same manner as used for our resistance screening, effectively selected heterozygous A. tuberculatus plants containing the Trp-574-Leu mutation from sensitive plants. We also confirmed that heterozygous smooth pigweed (Amaranthus hybridus L.) plants containing the Ala-122-Thr mutation survived a 10X rate of imazethapyr (unpublished data). Moreover, we determined that A. tuberculatus plants homozygous for the Ser-653-Asn mutation survived a 20X rate (unpublished data). Assuming that we were selecting for these three mutations only, the probability of finding at least one ALS–inhibitor resistant plant was 2.51 × 10−8. In this scenario, the chance of finding at least one resistant mutant in this experiment was 83.12%, and we should have screened 120 million plants to have a 95% chance to identify an individual with any of these three mutations. The probability of finding any of these mutants decreases if adjusted by methylation and genic regions as described earlier (Table 4).
Mutation rate in herbicide-treated plants
According to the germination rate (0.84), more than 11 million seedlings derived from atrazine-treated parental plants were screened for ALS-inhibitor resistance (Table 3). No resistant mutants were found. Many reasons may account for the nonidentification of resistant mutants. First of all, we do not know the lower limit of the mutation rate conferring ALS-inhibitor resistance, so we do not know exactly how many screened individuals are needed to identify a resistant mutant. Furthermore, as just described, the treatment may have generated ALS–inhibitor resistant mutants that our screening procedure did not select. Alternatively, it may be that the herbicide treatment used in this experiment is not effective in generating mutant offspring or did not generate plant stress in the right tissue or at the right time. Because we were not able to establish a baseline mutation rate, we were not able to rigorously test the hypothesis that herbicide stress increases the mutation rate. All that we can conclude is that we found no evidence that herbicide stress increased the mutation rate.
Implications for weed management
To our knowledge, these are the first peer-reviewed published data on the empirical determination of the spontaneous mutation rate providing herbicide resistance in plants. Stannard and Fay (Reference Stannard and Fay1987) reported the selection of 15 ALS–inhibitor resistant individuals by screening 20 million individuals of alfalfa (Medicago sativa L.) seedlings with chlorsulfuron. However, those data were not published beyond an abstract format, and the origin of the plant material was not well characterized (which is required to rule out resistance arising from standing genetic variation). In the present study, an empirically determined probability of 1.4 × 10−8 per individual is established as a higher limit for the occurrence of spontaneous ALS–inhibitor resistant mutants in a sensitive Amaranthus population. Although this probability is lower than expected, it is still sufficient to give rise to herbicide resistance in weeds, given their large populations and high reproductive output.
It is also possible that the plant population used in this study has a lower mutation rate than weed populations. Domesticated populations are a product of genetic bottlenecking and selection for genome stability, to the detriment of genetic variation and mutator alleles. Moreover, high heterozygosity as well as intraspecific hybridization events in outcrossing species (e.g., A. tuberculatus and A. palmeri) may favor a higher mutation frequency than in self-pollinated species such as grain amaranth (Bashir et al. Reference Bashir, Sailer, Gerber, Loganathan, Bhoopalan, Eichenberger, Grossniklaus and Baskar2014; Yang et al. Reference Yang, Wang, Huang, Zhang, Yuan, Chen, Hurst and Tian2015). In addition, under field conditions, plants are commonly exposed to stresses that may increase the mutation rate. Different studies demonstrated that natural stresses, such as drought, flood, cold, salinity, and UV light may cause severe DNA damage in plants (Filkowski et al. Reference Filkowski, Kovalchuk and Kovalchuk2004; Jiang et al. Reference Jiang, Mithani, Belfield, Mott, Hurst and Harberd2014; Yao and Kovalchuk Reference Yao and Kovalchuk2011). Consequently, weed populations in the field may have higher mutation rates than reported in this study, in which, being performed in the greenhouse, most environmental variables were controlled. For example, UV rays were partially blocked by glass, potentially decreasing their mutagenic effects. Moreover, recently it has been proposed that nonlethal herbicide applications may increase the mutation rate in surviving plants (Gressel and Levy Reference Gressel, Levy, Pareek, Sopory and Bohnert2009); although we did not find empirical evidence supporting this, the hypothetical stress-mediated increase of mutation rates leading to herbicide resistance remains biologically possible. Furthermore, while the mutation rate may remain low, the potential occurrence of spontaneously resistant mutants ultimately depends on the number of viable offspring produced per season. Therefore, population parameters such as the number of escapes, number of seeds produced per plant, and the survivability of escapes will determine the emergence of new resistant genotypes, regardless of the mutation rate. In this sense, spontaneous mutations could potentially play an important role in generating new resistance alleles depending on environmental variables, farming conditions, and weed population parameters. Nonetheless, the low mutation rate we observed in this study suggests that de novo mutations may contribute less than previously thought to resistance evolution.
In contrast, the standing genetic variation of weed populations appears to contribute substantially to the number of resistant individuals in every season. Preston and Powles (Reference Preston and Powles2002) found the frequency of ALS-inhibitor resistance in unselected populations of rigid ryegrass (Lolium rigidum Gaudin) in Australia to be on the order of 10−5 to 10−4. Similarly, Délye et al. (Reference Délye, Deulvot and Chauvel2013a), by DNA analysis of herbarium specimens collected before the use of herbicides, calculated the frequency of one mutation providing resistance to acetyl-CoA carboxylase inhibitors in unselected French populations of blackgrass (Alopecurus myosuroides Huds.) to be on the order of 10−4. In addition to the frequency of resistance alleles in the standing genetic variation of the population, the number of emerged resistant plants in the field will depend on the number of seeds present in the soil seedbank, which may vary from hundreds to millions, and their emergence rate, which will vary according to several variables such as temperature, soil humidity, seed depth, seed dormancy and viability, and tillage method. For example, assuming a resistance frequency of 5 × 10−5 (from the standing genetic variation) in the population, 1,000 seeds m−2 in the soil, and a 5% emergence rate, at least 25 resistant plants should emerge per hectare per season. In contrast, even in a poorly managed field, wherein there is 1 escape m−2 and each plant produces 25,000 seeds with 10% survivability, a mutation rate of 1.4 × 10−8 is expected to produce not even one resistant mutant per hectare. From this comparison, it can be concluded that resistance mitigation strategies should be designed considering the standing genetic variation as the main source of resistance alleles in weed populations.
If alleles come from the standing genetic variation of the population, these genetic variants have been conserved by natural selection for years. Therefore, adaptive alleles must have little or no fitness cost, in accordance with several publications reporting a negligible fitness penalty for many mutations conferring herbicide resistance (Powles and Yu Reference Powles and Yu2010). As a consequence, these mutations are expected to remain in the population in the absence of selection. In addition, when adaptive alleles are derived from the standing genetic variation, adaptive evolution is determined by the amount of variation present before selection (Jasieniuk et al. Reference Jasieniuk, Brûlé-Babel and Morrison1996). Stated differently, the evolutionary outcome of a current selection process would depend on how previous selection events constrained genetic variants in the population. For example, recurrent selection by only one herbicide—by bottlenecking genetic diversity—will reduce the probability of the target weed population evolving resistance to another herbicide (assuming no cross-resistance to the two herbicides exists). In contrast, if alleles providing resistance are mainly a product of new mutations, selection by one herbicide will not reduce the likelihood for herbicide resistance evolving to subsequent herbicides.
The main source of resistance alleles in weed populations determines which management practices (Norsworthy et al. Reference Norsworthy, Ward, Shaw, Llewellyn, Nichols, Webster, Bradley, Frisvold, Powles, Burgos, Witt and Barrett2012) are more suitable to counteract resistance evolution. For example, if resistance primarily comes from new mutations, preventing weeds from producing seed would be paramount, because it would be from these newly produced seeds that resistance would arise. In contrast, if resistance primarily arises from standing genetic variation, new seed production from weed escapes (assuming escapes are not a result of herbicide resistance) is not as problematic in terms of herbicide-resistance evolution. However, before acting on such conclusions, more research is needed to determine the spontaneous mutation rate conferring herbicide resistance, and what factors might alter this rate. The model system described herein, which uses grain amaranth and resistance to ALS inhibitors, could be scaled up to further investigate herbicide-resistance mutation rates.
Author ORCID
Patrick J. Tranel, https://orcid.org/0000-0003-0666-4564.
Acknowledgments
The USDA National Institute of Food and Agriculture (AFRI Project 2015-67013-22965) provided partial funding of this research. No conflicts of interest have been declared.









