Introduction
Eriophyoid mites are microscopic, highly specialized, phytophagous mites known to colonize over 2500 plant species (Amrine & Stansy, Reference Amrine and Stansy1994). As a crop pest, they not only have the potential to cause physical damage to their host, but they also have the ability to transmit plant viruses (Slykhuis, Reference Slykhuis1953; Slykhuis, Reference Slykhuis1955; Slykhuis, Reference Slykhuis1956; Oldfield, Reference Oldfield1970; Slykhuis, Reference Slykhuis1976; Oldfield & Proeseler, Reference Oldfield, Proeseler, Lindquist, Sabelis and Bruin1996; Styer & Nault, Reference Styer, Nault, Lindquist, Sabelis and Bruin1996; Seifers et al., Reference Seifers, Harvey, Martin and Jensen1997). The viruses transmitted by eriophyoid mites are quite species-specific, with a particular virus often only transmitted by a single mite species (Krantz & Lindquist, Reference Krantz and Lindquist1979). This means correctly identifying a mite species is an important factor for understanding the spread of eriophyoid transmitted viruses in agriculture.
The wheat curl mite (WCM), Aceria tosichellaKeifer (Reference Keifer1969), is an eriophyoid mite commonly associated with cereals worldwide (Amrine & Stansy, Reference Amrine and Stansy1994; Frost & Ridland, Reference Frost, Ridland, Lindquist, Sabelis and Bruin1996). The mite itself usually causes little damage in the field other than the characteristic leaf rolling and occasional trapping of the flag leaf. In isolated cases, mite feeding can result in reduced grain yields, but only when mites are found in very high numbers (Harvey et al., Reference Harvey, Martin and Seifers2002). The agricultural significance of A. tosichella lies in its ability to spread the damaging plant disease, wheat streak mosaic virus (WSMV). This virus was first recorded in 1929 from cropping areas of Kansas, USA (McKinney, Reference McKinney1937). The virus began to cause serious economic problems via reductions in wheat grain yield from 1949 onwards (Oldfield, Reference Oldfield1970), and the virus is now known to be widely distributed throughout the cropping regions of the USA and Europe. In a severe year, the virus can cause up to 100% loss of yield (Tosic, Reference Tosic1971; Slykhuis, Reference Slykhuis1976; Sanchez-Sanchez et al., Reference Sanchez-Sanchez, Henry, Cardenas-Soriano and Alvizo-Villasana2001). Although wheat is the primary host of A. tosichella, this mite has been recorded on at least 60 alternate plant hosts (Slykhuis, Reference Slykhuis1955; Connin, Reference Connin1956; Somsen & Sill Jr., Reference Somsen and Sill1970; Harvey et al., Reference Harvey, Seifers and Martin2001; Skoracka & Kuczyński, Reference Skoracka, Kuczyński, Naukowa and Ignatowicz2006).
Due to their small size and the existence of numerous morphologically similar species, taxonomic work on eriophyoid mites is difficult. This is illustrated by long-standing confusion and misidentification of A. tosichella. Aceria tosichella was described in 1969, and its hosts were listed as wheat and barley (Keifer, Reference Keifer1969). Previously, A. tosichella was misidentified as A. tulipae, a species believed to occur on wheat, onions, garlic and tulips (Shevtchenko et al., Reference Shevtchenko, De-Millo and Razvyazhina1996). However, Shevtchenko et al. (Reference Shevtchenko, De-Millo, Razvyazkina and Kapkova1970) were able to show that the species occurring on bulbs (A. tulipae) was not the same species as that found on wheat. They described the species occurring on wheat and named it Aceria tritici, but this name is considered a synonym for A. tosichella.
There is evidence that there may be multiple species or strains of WCM. The presence of different strains of WCM was first indicated in work by Slykhuis (Reference Slykhuis1955) based on performance in transplants between plant hosts. Shevtchenko et al. (Reference Shevtchenko, De-Millo, Razvyazkina and Kapkova1970) published a description of an eriophyoid form, similar to that of A. tosichella but differing in body measurements. Sukhareva (Reference Sukhareva1981) reported a wide variation in morphological characters for A. tosichella. Skoracka & Kuczyński (Reference Skoracka, Kuczyński, Naukowa and Ignatowicz2006) also described intraspecific morphological variation in A. tosichella and suggested these differences, along with inconsistencies in host preference, could be due to the presence of different strains or species. Frost (Reference Frost1995) identified two distinct forms of Aceria sp. (presumably Aceria tosichella) on wheat in Australia. Many of these specimens were later confirmed as being A. tosichella, but some collections that were taken from wheat differed slightly from published descriptions of A. tosichella (Halliday & Knihinicki, Reference Halliday and Knihinicki2004) and included the alternate form identified by Frost (Reference Frost1995).
Molecular approaches using DNA-based techniques could provide a powerful tool to help resolve the taxonomic issues surrounding A. tosichella. Many studies have successfully applied molecular approaches to small organisms, like mites, to examine their population structure, evolutionary history, life history and for identification purposes (Fenton et al., Reference Fenton, Malloch, Jones, Amrine, Gordon, Ahara, McGavin and Birch1995, Reference Fenton, Jones, Malloch and Thomas1996; Osakabe et al., Reference Osakabe, Goka, Toda, Shintaku and Amano2005; Carbonnelle et al., Reference Carbonnelle, Hance, Migeon, Baret, Cros-Arteil and Navajas2007; Laumann et al., Reference Laumann, Norton, Weigmann, Scheu, Maraun and Heethoff2007; Ros & Breeuwer, Reference Ros and Breeuwer2007). Some molecular studies have also distinguished closely related species of eriophyoid mites. Variation in the nuclear ribosomal internal transcribed spacer 1 (ITS1) has been used to differentiate morphologically cryptic pest species of grapevine mites, Colomerus species (Carew et al., Reference Carew, Goodisman and Hoffmann2004), and to identify Cecidophyopsis species from around the world (Kumar et al., Reference Kumar, Fenton and Jones1999).
Here, we examine DNA variation in WCM in Australia using mitochondrial and nuclear molecular markers. We collected WCM throughout south-eastern and Western Australia and characterized representative samples for nuclear and mitochondrial genes. For internal transcribed spacer 1 (ITS1), we show that the mites fall into two discrete lineages. A restriction fragment length polymorphism (RFLP) test based on ITS1 sequence was developed to separate these two lineages. Sequence data from the adenine nucleotide translocase (ANT) and the mitochondrial 16S rRNA gene also supported the separation of the ITS1 lineages. We identify alternate plant hosts that could be used by WCM, particularly in the absence of wheat. The potential implications of these results for the spread of WSMV are discussed.
Materials and methods
Mite collections
Eriophyoid mites were collected from 44 locations across major wheat growing regions of mainland Australia (southern Western Australia, Victoria, New South Wales Australia Capital Territory and southern Queensland) and from Tasmania. Mites were collected directly from the crease of wheat grains sampled at late dough stage (late spring–early summer in 2005 and 2006). In addition, 22 potential alternate plant hosts were screened for the presence of WCM. Upon collection, live mites were placed in 0.5 ml microcentrifuge tubes and stored at −80°C.
DNA extraction and polymerase chain reactions
Single mites were extracted using a Chelex method following Carew et al. (Reference Carew, Goodisman and Hoffmann2004). Briefly, microcentrifuge tubes containing mites were centrifuged at 14,000 rpm for 5 min to ensure mites were at or near the base of the tube. Next, 3 μl of Proteinase K (Roche) was added to each tube and mites were crushed using a plastic pestle moulded from a pipette tip. Then, 100 μl of 5% Chelex (Bio Rad) solution was added, tubes were mixed, and then incubated, firstly for 1.0–1.5 h at 55°C, followed by 8 mins at 90°C. Mite extractions were then cooled on ice and stored at −20°C.
Polymerase chain reactions (PCRs) were performed to amplify ~450 bp of the nuclear ribosomal internal transcribed spacer 1 (ITS1), ~400 bp of the mitochondrial 16S rRNA gene, and ~340 bp of the nuclear ANT gene. We also investigated the mitochondrial cytochrome oxidase I (COI) gene for WCM species identification; but found, in single individuals, we were able to amplify two alternate copies of COI, based on the primers selected. However, the target of this study was to identify WCM species, not an in-depth study of COI or possible pseudogenes; so we did not pursue COI further. All PCRs were performed in 25 μl volumes containing 1×immolase buffer (Bioline), 200 mM of each dNTPs, 2 mM MgCl2, 1 μg μl−1 Bovine Serum Albium (BSA), 0.5 μM forward and reverse primers (table 1), 0.75 units of immolase DNA polymerase (Bioline), and 6 μ l of Chelex DNA extraction supernatant (taken from just above the resin after centrifugation at 13,000 rpm for two minutes). The ITS1 fragment was amplified using a PCR profile consisting of an initial denaturation at 95°C for 7 min followed by ten cycles of a touch down consisting of 95°C for 20 s, 55–45°C (decreasing the annealing step by 1°C each cycle) for 30 s, 72°C for 30 s followed by 30 cycles of 95°C for 20 s, 45°C for 45 s and 72°C for 30 s (increasing the elongation step by two seconds each cycle). The mitochondrial 16S rRNA fragment was amplified using a PCR profile consisting of an initial denaturation at 95°C for 7 min, then 40 cycles of 95°C for 20 s, 53°C for 45 s and 72°C for 30 s (increasing the elongation step by two seconds each cycle). The ANT fragment was amplified using a PCR profile consisting of an initial denaturation at 95°C for 7 min, then 40 cycles of 95°C for 20 s, 50°C for 45 s and 72°C for 30 s (increasing the elongation step by two seconds each cycle). An aliquot of 5 μl of the PCR amplification product was run on a 1.5% agarose gel, stained with ethidium bromide and visualized under UV light. PCR product sizes were estimated using Hyper Ladder II (Bioline).
Table 1. PCR primers used to identify sequence variation in WCM.

a Primers developed for this study.
Sequence analysis
The ITS1 and 16S rRNA PCR products were sequenced in forward and reverse directions using the same PCR primers used in amplification and then aligned and edited in Sequencher version 3.2.1 (Genecodes). Sequencing of ANT products was carried out using only the forward primer, due to consistently poor sequence quality from the reverse primer. All products were first cleaned using the QIAquick PCR purification kit (QIAGEN) and sequenced on an ABI3100 capillary sequencer (Applied Biosystems). All sequences used in phylogenetic analyses were submitted to Genbank (Accession numbers EU734723–U734741). Consensus sequences and related Aceria species sequences from Genbank were analysed with MEGA 3.1 (available from www.megasoftware.net), using a distance-based method, neighbour-joining (NJ), and a character-based method, maximum parsimony. Neighbour-joining trees were constructed using the distance calculated from the Kimura-2-parameter (K2P) model. Maximum parsimony analysis was conducted with max-mini branch and bound searches. For both methods, all gaps were deleted and all codon positions were considered. Bootstrap resampling was applied using 1000 replicates. Pairwise sequence divergences were calculated under a K2P model. We also performed Bayesian reconstruction and found this did not alter the tree topology or improve the resolution of relationships between species.
Restriction fragment length polymorphism analysis
An RFLP test was designed based on ITS1 sequences to identify the two major WCM lineages. Restriction digests were preformed using Hha I (Fermentas) and Acc I (New England BioLabs). Digests were carried out in 20 μl volumes with 10 μl of PCR product, 1×the recommended buffer, 5 μg Bovine Serum Albium (BSA) and three units of restriction endonuclease. Restriction digests were incubated at 37°C overnight. Digest products were separated via electrophoresis for two hours at 100 V on 3.5% agarose gels, stained with ethidium bromide and visualized under UV light. The size of digest fragments was estimated using a 50 bp ladder (Promega).
Results
Evidence for two lineages
Partial sequences for ITS1, 16S rRNA and ANT from WCM were obtained from major wheat growing states and territories in Australia (Queensland, New South Wales, Victoria, Western Australia, Australian Capital Territory and Tasmania). A total of 80 WCM were sequenced for ITS1, 54 for 16S rRNA and 88 for ANT. To enable comparison of sequences in individual mites, we also sequenced 30 mites for all three regions.
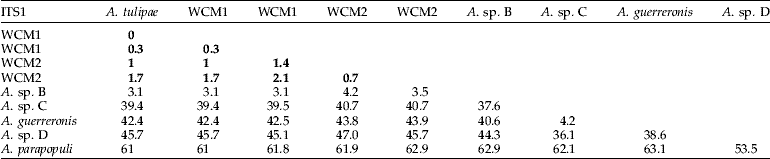
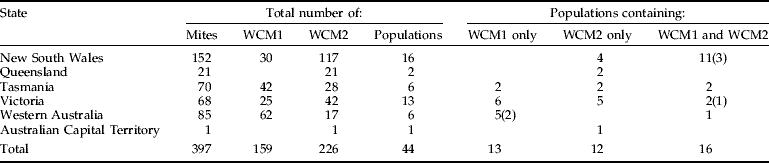
Sequence variation indicated that WCM formed two distinct lineages separated by an average sequence divergence of 1.4% (and 3–4 indels) for ITS1 and 4.4% for the 16S rRNA gene (fig. 1, table 2). Both WCM lineages (herein referred to as WCM1 and WCM2) were widespread throughout Australia, occurring in all states, with the exception of Queensland where only WCM2 was found. This was also confirmed by RFLP analysis of the ITS1 region of 397 WCM sampled from wheat (table 3). WCM1 and WCM2 were characterized by four unique sequences for ITS1, two sequences for WCM1 separated by <0.3% and two sequences for WCM2 separated by <0.7%. An exact match between ITS1 sequence from WCM1 in Australia and A. tulipae in Canada indicates that these are the same species of mite. This suggests that the Canadian specimen may have been misnamed as A. tulipae. However, ITS sequences from species confirmed to be A. tulipae would be needed to verify this. A single sequence was found to represent WCM2 for the partial 16S rRNA gene, while there were two sequences for WCM1 separated by <1.1%. An average sequence divergence of 4.4% separated WCM1 and WCM2 for the 16S rRNA. Trees for ITS1 and 16S rRNA were congruent (fig. 2). In 100% of cases, individuals identified as WCM1 (or WCM2) based on ITS1 were identical to WCM1 (or WCM2) identified from 16srRNA sequences; there was no mixing of lineages. Moreover, for WCM1, there are two ITS1 and two 16S rRNA sequences. We compared all WCM1 individuals (total of 16) containing both ITS1 and 16SrRNA sequences and found that, in all but one case, the groupings were identical.
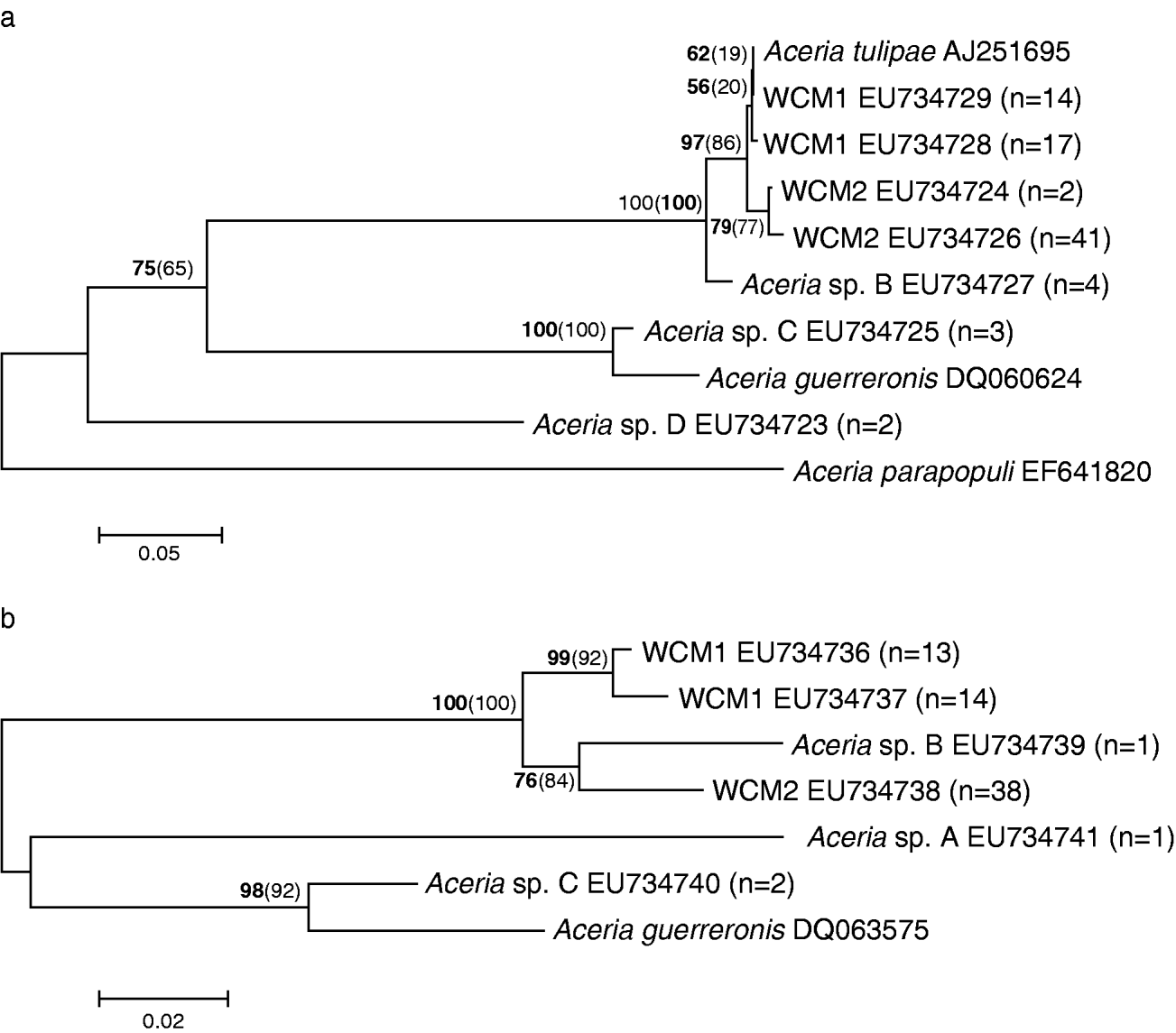
Fig. 1. Consensus neighbour joining trees for WCM and related Aceria species for (a) 350 bp of ITS1 and (b) 352 bp of 16S rRNA. Bootstrap values for NJ (bolded) and parsimony (in parenthesis) are based on 1000 replicates. Sample size (n) indicates the number of individuals sequenced for each consensus sequence. Genbank accession numbers are given for each sequence.

Fig. 2. The number of base pair differences observed between sequences of WCM1, WCM2 and Aceria sp. B over 269 bp of the ANT sequence. Circles are proportional to the number of identical sequences (n). The number of mites with each ANT sequence, Genbank accession numbers and lineage type as assessed with the ITS1 RFLP test are given.
Table 2. Kimura-2-parameter (K2P) sequence divergence (%) between WCM and related species for (a) ITS1 and (b) 16S rRNA. Bolded values indicate pairwise sequence divergence between wheat curl mite sequences.
(a)
(b)

Table 3. The distribution of WCM identified through PCR-RFLP of ITS1 on wheat from states around Australia. The total number of mites that were tested with the ITS1 RFLP test, the number WCM1 and WCM2 found, and the populations examined are given. The number of populations that only had WCM1, WCM2 and both lineages are presented. The number of populations that contained individuals the showed both WCM1 and WCM2 profiles are in parenthesis.
Limited sequence variation was observed among WCM for ANT with all nucleotide substitutions silent. Due to this low variability, a haplotype network was constructed based on the number of nucleotide differences separating each genotype (fig. 2). A maximum of five nucleotide differences separated different WCM ANT sequences. One sequence represented WCM2, while four sequences were detected for WCM1. Overall, while more sequence variation was found in WCM1, there was no structure between sequences within lineages. The only sequence variation found in WCM2 was for two individuals from Western Australia, a population separated by >1000 km from other WCM populations analyzed. We were able to confirm one of the lineages (WCM1), collected from the La Trobe University (Melbourne) glasshouses, as being A. tosichella, based on morphological criteria (R.B. Halliday, personal communication).
The ITS1 RFLP test was successful as a means of rapidly identifying WCM1 and WCM2. Two composite RFLP profiles were observed using Acc I and Hha I, fragment sizes for WCM1 were 343 bp/152 bp for Acc I, and 318 bp/177 bp for Hha I. For WCM2, fragment sizes were 493 bp/90 bp/61 bp for Acc I and 493 bp for Hha I. However, a small number of individuals <4% (12 out of 396) appeared to show both WCM1 and WCM2 ITS1 RFLP profiles. Sequence data for 16S rRNA indicated these extractions contained single individuals of either WCM1 or WCM2 rather than two mites. While ANT sequence data largely agreed with 16S rRNA, on two occasions it differed. In one case, there was a heterozygote for WCM1 and WCM2 and, in the other, showed the opposite species assignment to 16S rRNA. The occasional mixing of ITS1 profiles combined with the two contrasting ANT sequences might indicate limited genetic exchange between the lineages that results in mixing of nuclear DNA. The RFLP tests also showed WCM1 and WCM2 occurred both allopatrically and sympatrically (table 3). Wheat curl mite that showed both WCM1 and WCM2 profiles for the ITS1 RFLP test were from both allopatric and sympatric populations. Furthermore, most of these individuals fell within the WCM1 group most closely related to WCM2 for ANT (fig. 2).
Alternate hosts
Several varieties of wheat were screened for the presence of WCM, including ‘Marombi’, ‘Whistler’, ‘Wegdetail’, ‘Diamondbird’, ‘Whyla’, ‘H45’, ‘Ventura’, ‘McKellar’ and ‘Tennant’. On all wheat varieties, we only found WCM with no other eriophyoid mites detected. There appeared to be no varietal preferences between WCM1 and WCM2 (data not shown). Furthermore, both WCM1 and WCM2 were found on a variety of alternate plant hosts (table 4). WCM1 was detected on 11 alternate host plant species and WCM2 on 14 alternate host plant species. For the first time, we record the presence of WCM on the following alternate plant hosts: Avena fatua; Bothriochloa macra; Bromus diandrus; Bromus hordeaceus; Bromus catharticus; Dactylis glomerate; Digitaria sanguinalis; Hordeum leporinum; Lolium rigidum; Panicum effusum; Setaria jubiflora; Setaria verticillate; Urochloa panicoides; Vulpia bromoides; and also triticale (a wheat/rye hybrid). These plant species are predominantly summer grasses, which are likely to provide a ‘green bridge’ for WCM between wheat-growing seasons.
Table 4. Numbers of mites found on alternate plant hosts. The number of sites that each host was collected from; the number of WCM1, WCM2 and other Aceria species found on each host; and the total number of mite collected from each host. Mites were typed using a PCR-RFLP test based on ITS1. The number of mites examined using PCR-RFLP includes those that failed to amplify for ITS1. Aceria sp. C and Aceria sp. D yielded similar PCR-RFLP profiles and could not be confidently distinguished and are shown in the same column.

a Mites from these plant species were identified as A. tosichella based on morphology (R.B. Halliday, personal communication).
Comparison with other Aceria
Four putative Aceria species (Aceria sp. A, Aceria sp. B, Aceria sp. C and Aceria sp. D) were discovered on alternate grass hosts, in some instances together with WCM; but none of these species were found on wheat. These additional Aceria species were sequenced for comparison to WCM. We also included sequences from Genbank for A. tulipae (Kumar et al., Reference Kumar, Fenton, Duncan, Jones, Sreenivasulu and Reddy2001), A. parapopull and the coconut mite, A. guerreronis (Navia et al., Reference Navia, de Moraes, Roderick and Navajas2005) in the analyses. However, only Aceria sp. B could be amplified for all three gene regions. Aceria sp. A only amplified for the 16S rRNA gene, Aceria sp. C for ITS1 and 16S rRNA, and Aceria sp. D only for ITS1. Wheat curl mite (including A. tulipae from Canada) formed a single clade for ITS1 and were separated by <3.1% sequence divergence from other Aceria species (table 2a, fig. 1a). Similarly, the 16S rRNA sequences showed that WCM were separated by >5% sequence divergence from other Aceria species, although Aceria sp. B clustered within the WCM clade (table 2b, fig. 1b). Aceria sp. B also separated by at least four nucleotide substitutions from WCM for the partial ANT gene sequences (fig. 2).
Discussion
The presence of two WCM lineages with limited genetic exchange indicates that in Australia A. tosichella may represent a complex of two closely related species. No evidence of genetic exchange between lineages was seen when comparing sequences for 16S rRNA and ITS1, but a small number of individuals displayed both WCM1 and WCM2 ITS1 RFLP profiles, suggesting genetic exchange is possible. There was ample opportunity for genetic mixture between WCM1 and WCM2, as they were often found in sympatry; some populations even had both lineages co-existing on the same grain of wheat. Despite close contact, the WCM lineages were almost always distinct, suggesting if WCM reproduce sexually that hybridisation is uncommon. Sequence divergence separating WCM1 and WCM2 was low for ITS1 although insertion/deletions also characterised this separation. The sequence divergence separating the WCM1 and WCM2 lineages for 16S rRNA was more indicative of WCM1 and WCM2 representing two species. The average divergence of 4.4% was only marginally lower than the sequence divergence separating Aceria sp. B from the WCM lineages, which ranged from 5.0 to 6.5%.
Our DNA evidence for two putative WCM species is supported by morphological studies. In Europe and Canada, two types of very similar mites that differed in body length, width and height dimensions have been found on wheat (Shevchenko et al., Reference Shevtchenko, De-Millo, Razvyazkina and Kapkova1970). Intraspecific morphological variation of A. tosichella observed overseas (Sukhareva, Reference Sukhareva1981; Skoracka & Kuczyński, Reference Skoracka, Kuczyński, Naukowa and Ignatowicz2006), has also been seen in Australia (Frost, Reference Frost1995; Halliday & Knihinicki, Reference Halliday and Knihinicki2004). In particular, Frost (Reference Frost1995) suspected that two morphs of WCM observed on wheat could correspond to two different species. Our attempts to link morphological variation with the two molecular lineages have been unsuccessful, and this was complicated by the small size of the mites and the inability to gain both DNA and morphological information from the same individual. Therefore, it remains unclear if the morphological variation identified in earlier work is evidence of intraspecific morphological plasticity or due to the presence of two different species.
The presence of two distinct WCM lineages has implications for the control of WCM and raises questions about their ability to transmit WSMV. Both WCM lineages are widely distributed within Australia; however, it is uncertain if one or both lineages are vectors of WSMV. This could be determined by directly testing for the presence of WSMV in the mites with an RNA-based assay and through controlled transmission experiments. However, even if both lineages are vectors of WSMV, there could be differences in their transmission efficiency (e.g. species differences in the duration mites remain viruliferous after acquiring the virus). A thorough understanding of the associations between each WCM lineage and WSMV under field conditions will be important for the development of effective management options, which will likely require quick and easy-to-use tools to correctly distinguish mite lineages.
At this stage, one of the most practical methods to reduce the build-up of WCM numbers and the risk of WSMV is to control green bridges that allow mites to persist between seasons when wheat is unavailable. Wheat curl mite are reliant on their host plant not just for food but also for the high humidity provided by protected parts of the plant or within the rolled leaves WCM create. In cooler conditions, mites can survive for up to two weeks away from hosts but die within a few hours if exposed to low humidity and moderate to high temperatures (Oldfield & Proeseler, Reference Oldfield, Proeseler, Lindquist, Sabelis and Bruin1996). Spraying roadside verges and grassy weeds within paddocks over the summer months is recommended in areas at risk of WSMV. However, the alternate plant hosts that may harbour the different lineages need to be determined for effective control.
In the literature, there are reports of over 60 alternate plant hosts that can be colonised by A. tosichella. We record the presence of WCM on an additional 15 alternate plant hosts. However, the presence of WCM does not necessarily mean WCM can colonise a particular alternate host (Skoracka & Kuczyński, Reference Skoracka, Kuczyński, Naukowa and Ignatowicz2006). Some of the alternate plant hosts have previously been tested for the colonisation ability of WCM, although no assumptions can be made due to the confusion arising from the existence of a WCM species complex. Mites have previously been shown to be incapable of colonising Avena fatua (Slykhuis, Reference Slykhuis1955; Somsen & Sill Jr., Reference Somsen and Sill1970) or Dactylis glomerate (Connin, Reference Connin1956; Somsen & Sill Jr., Reference Somsen and Sill1970; Harvey et al., Reference Harvey, Seifers and Martin2001). Conjecture also exists over the ability of WCM to colonise Digitaria sanguinalis (Slykhuis, Reference Slykhuis1955; Connin, Reference Connin1956; Harvey et al., Reference Harvey, Seifers and Martin2001) while WCM has a poor ability to colonise Setaria verticillate (Slykhuis). Our record of WCM on Eragrostis cilianensis is supported by Somsen & Sill Jr. (Reference Somsen and Sill1970) who reported that mite infestations were common on this grass throughout Kansas, particularly in the summer period.
Some of the alternate host plants identified in this study as harbouring WCM have previously been tested for their susceptibility to WSMV. Dactylis glomerata was found to be immune to WSMV (Somsen & Sill Jr., Reference Somsen and Sill1970), whereas Avena fatua, Cynodon dactylon, Digitaria sanguinalis, Eragrostis cilianensis, Setaria verticillate and triticale have all been reported as being susceptible to WSMV (Slykhuis, Reference Slykhuis1955; Connin, Reference Connin1956; Somsen & Sill Jr., Reference Somsen and Sill1970; Sanchez-Sanchez et al., Reference Sanchez-Sanchez, Henry, Cardenas-Soriano and Alvizo-Villasana2001). Unfortunately, until the alternate WCM host plants identified in this study are tested further for the colonisation ability of both WCM1 and WCM2, they must all be considered as potential refuges for the mites and virus.
The development of satisfactory control techniques relies on understanding the relationship between WCM, their host plants and WSMV. This study has revealed the existence of two lineages of WCM distributed throughout Australia, and at least one of the lineages also exists in other parts of the world. Further research is needed to understand the implications of finding two putative species for the control of WSMV. In other countries, WCM is also associated with the transmission of other viruses, including the High Plains Virus (Slykhuis, Reference Slykhuis1956). Associations between the lineages and all viruses transmissible by WCM need to be considered. We now have a simple RFLP-based test for distinguishing the lineages, and the next step is a comprehensive assessment to test transmission, map distributions and increase our understanding of the biology of these lineages.
Acknowledgements
The authors wish to thank Paul Parker, Peter Mangano, Bruce Halliday and Danuta Knihinicki for contributions to the project. This research was supported by the Grains Research and Development Corporation, with infrastructure provided through the Australian Research Council via their Special Research Centre Scheme. Funding was also provided by the Tasmanian Department of Primary Industries and Water.








