Introduction
Microsporidia are unicellular eukaryotic parasites that infect a wide range of animals in terrestrial and aquatic environments (Stentiford et al. Reference Stentiford2013), and they have several genera threatening immunocompromised patients and causing infectious diseases such as diarrhoea and keratoconjunctivitis even in immunocompetent human populations (Chan et al. Reference Chan2003; Didier and Weiss, Reference Didier and Weiss2011). To date, 1300–1500 species of microsporidia have been described (Vávra and Lukeš, Reference Vávra, Lukeš and Rollinson2013), suggesting that this is the largest phylum of intracellular obligate animal parasites, and these are divided into five clades (I–V) according to habitat (Vossbrinck and Debrunner-Vossbrinck, Reference Vossbrinck and Debrunner-Vossbrinck2005).
The higher classification of this phylum has been controversial, but recent molecular analyses suggest that microsporidia are probably a basal branch or sister group of fungi (Capella-Gutiérrez et al. Reference Capella-Gutiérrez, Marcet-Houben and Gabaldón2012; James et al. Reference James2013; Vávra and Lukeš, Reference Vávra, Lukeš and Rollinson2013; Haag et al. Reference Haag2014). The spores of microsporidia have unique features, including a polar tube that functions as an infectious apparatus and coils at the end, a membranous polaroplast that is injected into the target cell to be sporoplasm membrane, and a thick surface composed of an endospore and exospore that surrounds the plasma membrane. These peculiar traits are believed to have evolved as adaptations to their established parasitism of eukaryotic cells and have stimulated scientific interests for a long time (Han and Weiss, Reference Han, Weiss, Heitman, Howlett, Crous, Stukenbrock, James and Gow2017).
In the last decade, Nematocida parisii was isolated from the intestine of the model organism Caenorhabditis elegans and has been intensively studied by Troemel and colleagues (Troemel et al. Reference Troemel2008). They reported the spore exit from intestinal cells (Szumowski et al. Reference Szumowski2014), a ubiquitin-mediated defence mechanism (Bakowski et al. Reference Bakowski2014) and a host strain-dependent clearance of the intracellular pathogens (Balla et al. Reference Balla2015). Besides N. parisii, which infects intestinal cells, Nematocida displodere infects a broad range of tissues such as hypodermis, muscle and neuron in addition to the intestine of C. elegans (Luallen et al. Reference Luallen2016). In addition, Pancytospora epiphaga, which infects hypodermis and muscle of C. elegans, was reported along with other nematode-infecting microsporidia (Zhang et al. Reference Zhang2016).
Soil nematodes appear to be a promising model system to study the intracellular parasites, because of their transparent body and ease in handling. Furthermore, meta-analytic exploration for environmental microsporidian populations in soils and composts suggests that various novel microsporidia infect soil organisms (Ardila-Garcia et al. Reference Ardila-Garcia2013). Thus, we decided to explore soil nematodes in order to discover a novel model to investigate distinct aspects of microsporidian parasites. In this study, we isolated an Oscheius tipulae strain (IMU001) and its novel parasitic microsporidium, the tissue distribution of which is distinct from the known nematode-infective microsporidia. Based on light and electron microscopy and phylogenetic analyses using small subunit rRNA gene (rDNA) sequences, we propose a new microsporidian genus. The following is a description of the new microsporidium we isolated here, which we named Percutemincola moriokae gen. nov., sp. nov.
Materials and methods
Strains
The wildtype strains, Caenorhabditis elegans N2 and O. tipulae CEW1, were provided by the Caenorhabditis Genetics Centre. The C. elegans strain, Is[haf-4::GFP + pRF4(rol-6(su1006))], was established in the previous study (Kawai et al. Reference Kawai2009).
Collection and maintenance of worms
Oscheius tipulae IMU001 was isolated from muddy soil at the base of a plant growing on the riverside in Morioka, Iwate Prefecture, Japan according to the method described in Barrière and Félix (Reference Barrière and Félix2014). About 1 g of soil samples were spread around the bacterial lawn of Escherichia coli OP50 seeded on 9-cm NG agar plates (Brenner, Reference Brenner1974). The samples were then dampened with deionized water. Worms crawling out from the samples were transferred to new 3.5- or 6-cm NG agar plates to separate individual worms. The worms were maintained on NG agar plates seeded with E. coli OP50 at 20 °C.
Light microscopy
Differential interference contrast (DIC) images (Figs 1 and 2) were obtained using a DP70 digital camera on a BX51 microscope (Olympus Corp., Tokyo, Japan). For 4′,6-diamidino-2-phenylindole (DAPI) staining, 1 µg mL−1 DAPI in methanol was prepared using 1 mg mL−1 DAPI solution (Dojindo Laboratory, Kumamoto, Japan) and was used to fix and stain worms. Worms were then washed with M9 buffer (42 mm Na2HPO4, 22 mm KH2PO4, 86 mm NaCl, 1 mm MgSO4, 0.02% (w/v) gelatin) and were mounted on a 2%-agar pad. Fluorescence was observed under a confocal microscope FV1000 (Olympus Corp.). For calcofluor white (CW) staining, adult worms were fixed and stained with a 1:1 mixture of CW M2R (Merk KGaA, Darmstadt, Germany) and 1 M NaOH. The stained worms were mounted on a 2%-agar pad and observed using an epifluorescent microscope through a U-MWU2 filter set (BX51 equipped with BX2N-FL-1; Olympus Corp.). All images were of adult nematodes.

Fig. 1. Percutemincola moriokae infection of O. tipulae strain and its host specificity. (A–F) The field-collected nematode (O. tipulae IMU001) was observed at the adult stage under a DIC microscope. White arrows in each panel indicate a distinct region devoid of intestinal granules (A), emerging spores (B), coexistence of the Pe. moriokae and intestinal granules (C), intestinal cytosol packed with rod-shaped spores (D), hypodermal infection (E) and spores in the intestinal lumen (F). (G) Laboratory-bred O. tipulae CEW1 infected by Pe. moriokae spores harboured spores in intestinal and hypodermal cells. White and black arrowheads indicate intestinal and hypodermal Pe. moriokae spores, respectively. (H) No intracellular infection was observed in laboratory-bred C. elegans, albeit a number of spores can be observed in the intestinal lumen (black arrows) after mixed culture. Bars = 20 µm.

Fig. 2. Type specimen of Pe. moriokae. (A–D) Semi-ultrathin sections of Plain Resin-embedded O. tipulae IMU001 harbouring Pe. moriokae were stained with toluidine blue for the type specimen. White arrowheads in (A) and (B) and an arrow in (B) indicate intestinal cells and the intestinal lumen that contain Pe. moriokae spores, respectively. Black arrowheads in (C) and (D) indicate hypodermal spores. Bar = 20 µm.
Polymerase chain reaction (PCR) amplification of partial rDNA sequences
In order to identify the nematode species that we collected, we performed genomic PCR based on the C. elegans standard protocol (Fay, Reference Fay2013). Worms were picked from an NG agar plate and individually transferred into PCR tubes containing 5 µL TE buffer. An equal volume of lysis buffer (0.2 mg mL−1 proteinase K, 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 2.5 mm MgCl2, 0.45% NP-40, 0.45% Tween 20) was added to each tube and incubated successively for 1 hr at 65 °C then 15 min at 95 °C to obtain genomic templates. For the amplification of worm 18S rDNA, nematode-specific primer pairs (SSU18A: 5′-AAAGATTAAGCCATGCATG-3′ and SSU26R: 5′-CATTCTTGGCAAATGCTTTCG-3′) were used (Barrière and Félix, Reference Barrière and Félix2014).
We also conducted sequence analyses of the novel microsporidium. First, we obtained a partial sequence of the microsporidian rDNA through semi-nested PCR using primer Micro33R (5′-TAGAGACCGTTGTAGTTCCG-3′) as a forward primer (Ardila-Garcia and Fast, Reference Ardila-Garcia and Fast2012) and universal primers 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Eden et al. Reference Eden1991) and 1391R (5′-GACGGGCGGTGWGTRCA-3′) (Sahl et al. Reference Sahl2008) as reverse primers. To further amplify sequences neighbouring the rDNA fragment, we generated primers Micrs922F (5′-ACACCACAAGGGGTGGATTG-3′) and Micrs1326R (5′-TCTCCGCGCGACCGCTCTGT-3′), and paired them with 1492R and microsporidia universal primer V1 (5′-CACCAGGTTGATTCTGCCTGAC-3′) (Zhu et al. Reference Zhu1993) for PCR, respectively.
Sequence analyses
The PCR products were treated with exonuclease I (Wako Pure Chemical Industries, Ltd., Osaka, Japan) and shrimp alkaline phosphatase (Promega Corp., Fitchburg, WI) and subjected to sequence analyses unless multiple bands were observed by agarose gel electrophoresis. For the samples with multiple bands, the band of the predicted size was excised and purified using Wizard® SV Gel and PCR Clean-Up System (Promega Corp.). The sequences of the DNA fragments were analysed using BigDye® Terminator v3.1 (Thermo Fisher Scientific, Waltham, MA) and Applied Biosystems 3130xl Genetic Analyzer (Thermo Fisher Scientific).
Nematode species identification
Using the small subunit (SSU) rDNA sequence obtained via the host genomic PCR, we performed a nucleotide BLAST (BLASTN) search against ‘others (nr etc)’ in the NCBI Nucleotide collection (nr/nt) database (http://www.ncbi.nlm.nih.gov). Results from the BLAST query revealed 100% alignment (847/847 bp) between our entire amplified sequence and a portion of the SSU rDNA sequence from Oscheius tipulae CEW1 (accession No. KP756939). Morphology of the nematode IMU001 was subsequently compared with known morphologic traits of O. tipulae. Based on the analyses we concluded that the nematode is a strain of O. tipulae.
Preparation of spore isolates
Fully-grown infected worms were collected from two 9-cm NG agar plates using S-basal solution (100 mm NaCl in 50 mm potassium phosphate buffer (pH 6.0)). The worms were centrifuged at 2000 × g for 1 min and washed twice with 5 mL of S-basal. The worms were separated from free spores and bacteria in the suspension by filtration through an Isopore™ Membrane with 10 µm pore size (Merk KGaA). The residual worms on the membrane were washed with S-basal and resuspended in 2 mL of S-basal. The worms were homogenated using a dounce-type tissue grinder (Kimble Chase, Vineland, NJ), and then sequentially filtered through 10-, 3-, and 1.2-μm pore size Isopore™ Membranes. The residues on the 1.2-μm membrane were retrieved in 1 mL of S-basal. For freezing stocks, glycerol was added to a 15% total volume. The spore isolates were aliquoted and frozen at −80 °C until use.
Mixed culture test
Caenorhabditis elegans was grown with infected O. tipulae IMU001, which were continuously releasing infectious spores. We used a C. elegans strain expressing GFP-fused HAF-4 (Is[haf-4::GFP + rol6(su1006)]), to enable quick discrimination of the C. elegans from the O. tipulae, based on the strain's intestinal expression of GFP and roller phenotype.
Electron microscopy
Uninfected and infected worms were collected separately in M9 buffer containing 50 mm NaN3. The worms were fixed and stained according to a previously described C. elegans procedure (Kawai et al. Reference Kawai2009). Fixation was performed successively with fixation solution (2.5% (w/v) glutaraldehyde and 1% (w/v) paraformaldehyde in 0.05 M sodium cacodylate-HCl (pH 7.3), 0.1 M sucrose, 0.1 mm MgCl2) and postfixation solution (1% (w/v) osmium tetroxide in 0.1 M cacodylate buffer). After ethanol dehydration and Plain Resin (Nisshin EM, Tokyo, Japan) embedding, the ultrathin sections were prepared. The sections were stained with 1% (w/v) uranyl acetate and Reynold's lead citrate. A transmission electron microscope H-7650 (Hitachi, Ltd., Ibaraki, Japan) was used for visualization.
Preparation of type specimen
Semi-ultrathin sections of infected worms embedded in Plain Resin were dried and then stained with 1% toluidine blue on a heated plate (60–70 °C). The slides were washed with water and air dried.
Phylogenetic analysis
A contig of 1203 bp flanked by the universal V1 and 1492R primer sequences of the novel microsporidium was obtained by DNA sequencing. The rDNA sequence was submitted to DNA Data Bank of Japan under accession no. LC136798. We performed BLASTN searches using the obtained small subunit rDNA sequence as a query against the nr/nt nucleotide collection database to roughly identify the obtained sequence.
For phylogenetic analysis of the microsporidian sequence, we first data-mined representatives of each microsporidian clade (five clades in total (Troemel et al. Reference Troemel2008; Sapir et al. Reference Sapir2014)) from GenBank, obtaining a total in-group operational taxonomic unit (OTU) of 64 species, including the sequences of the novel microsporidium (Pe. moriokae) and two outgroups (Conidiobolus coronatus and Basidiobolus ranarum).
We aligned the sequences using the online version of MAFFT (ver. 7.221; Computational Biology Research Consortium [http://mafft.cbrc.jp/alignment/server/index.html]) with the Q-INS-i setting to allow for consideration of secondary structures in rDNA during alignment. Other parameters were set at default settings (BLOSUM62; 200PAM/K = 2; Gap opening penalty = 1.53). The alignment result was edited using the online version of Gblocks (ver. 0.91b; Institut de Biologia Evolutiva (CSIC-UPF) [http://molevol.cmima.csic.es/castresana/Gblocks_server.html]) using the least stringent settings. GBlocks retained 1020 aligned sites, which were then used for phylogenetic inference. Maximum Likelihood (ML) phylogenetic analysis was conducted using the GUI version of RAxML (Stamatakis, Reference Stamatakis2006; Silvestro and Michalak, Reference Silvestro and Michalak2012), with 1000 bootstrap (BS) replications, using the GTRGAMMA model.
To confirm the stability of the topology, we also conducted other phylogenetic analyses using different methods (Maximum Parsimony = MP; Neighbour Joining = NJ; and Minimum Evolution = ME), all performed in MEGA 7 (Kumar et al. Reference Kumar, Stecher and Tamura2016). In all three analyses: (1) all positions with <30% site coverage were eliminated, and (2) in order to assess topology robustness, bootstrap searches with 1000 replications were conducted. MP analysis was done using the Subtree–Pruning–Regrafting (SPR) algorithm. For NJ analysis, a distance matrix was computed using the Maximum Composite Likelihood method. Similar to NJ, the distance matrix for ME analysis was computed using the Maximum Composite Likelihood method, and the subsequent tree search was conducted under the Close-Neighbour-Interchange (CNI) algorithm. Only the ML tree, with bootstrap replication numbers of all three analyses written on each node, is shown in Fig. 6. MP, ME and NJ individual trees are provided as supplementary data.
Lifespan assay and time course of the tissue distribution of spores
Uninfected worms reared on one to three 9-cm NG agar plates were collected and resuspended in 5 mL of S-basal and bleached by adding 500 µL of 5% NaClO and 200 µL of 5 M NaOH. The remaining eggs were washed with S-basal three times and incubated at 20 °C overnight to acquire synchronized L1 larvae. The spore isolates (containing approximately 150 000 spores) and controls (15% glycerol in S-basal) were poured on NG agar plates seeded with OP50. The synchronized larvae were then spread on the plates. The plates were incubated at 20 °C. For the lifespan assay, worms were reared on the same media for 5 days and then transferred to spore-free NG agar plates. Dead worms were counted every 2 or 3 days. All the worms tested died within the experimental period. To analyse the time course for the tissue distribution of spores, 10 or 11 worms were sampled every day from a 3- to 7-day culture for each experiment. The worms were stained with CW to detect mature spores surrounded by cell walls. Worms that contain any mature spores in hypodermis and in intestine were rated. Statistical tests for the worm lifespan was performed through the log-rank test.
Results
Occurrence and light microscopy
Wild nematodes collected from soil samples were observed under a differential interference contrast (DIC) microscope. A novel nematode infective microsporidium strain (IMUm001) was discovered inside the body of an IMU001-strain nematode, which was isolated from a soil sample in Morioka, Iwate Prefecture, Japan (Figs 1 and 2). In unstained living worms, a locally cleared region devoid of intestinal granules was observed before sporulation of the parasite (Fig. 1A and B). Spores were found in hypodermal and intestinal cells and also in the intestinal lumen (Fig. 1C–F). Spores located in intestinal cells were slightly more slender (averaging 1.0 µm wide and 3.8 µm long) than those in hypodermal cells (averaging 1.3 µm wide and 3.2 µm long) (Figs 1A–F and 2). Under routine culture conditions, about half of IMU001 were positive for the microsporidian spores, suggesting active infection (Table 1). Stains for chromosomes by DAPI and for β-glucan-containing cell walls by CW revealed the monokaryotic nucleus in the middle and the cell wall at the periphery of spores, respectively (Fig. 3).
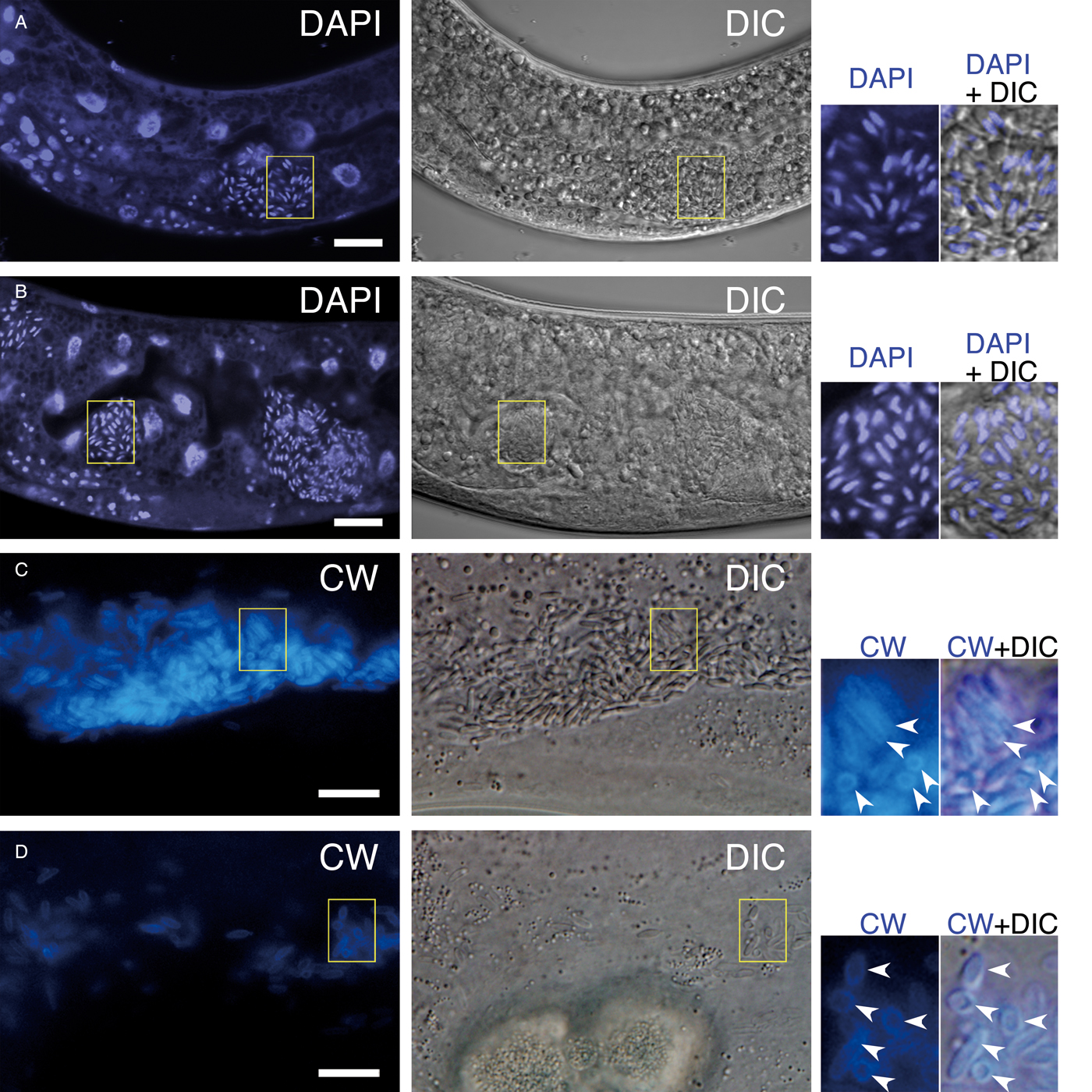
Fig. 3. DAPI-positive nuclei and CW-positive cell walls of Pe. moriokae. (A, B) DAPI staining of infected IMU001 worms. The rod-shaped spores showed intracellular fluorescence. (C, D) The intestinal (C) and hypodermal (D) images of the infected worms stained with CW. Yellow boxes denote regions magnified in the right-side panels. Arrowheads in (C) and (D) indicated CW fluorescence surrounding each spore. Bars = 10 µm.
Table 1. Infection rate and the growth temperature

Host identification and host specificity
We identified the IMU001 host species through sequence analysis of a segment of its SSU rDNA, which we amplified from the nematode's extracted genomic DNA using nematode-specific primers (SSU18A and SSU26R (Barrière and Félix, Reference Barrière and Félix2014)). BLAST searches indicated that the entire 847 bp amplicon sequence was a perfect match for the corresponding O. tipulae rDNA sequence (Sequence identity = 100.0%, e-value = 0.0, accession No. KP756939). Oscheius tipulae is a common species in the family Rhabditidae, the same family as C. elegans. The nematode IMU001 shares characteristics with the reference O. tipulae strain CEW1 (Félix, Reference Félix2006), i.e. the mode of reproduction (self-fertile hermaphrodites and males), growth rate (5–6 days to be fertile from the embryonic stage at 20 °C) and morphology (smaller body and longer rectum than C. elegans). Furthermore, O. tipulae is distributed worldwide including Japan and known to be abundant in soil (Baïlle et al. Reference Baïlle, Barrière and Félix2008). Based on our results, we thus concluded that IMU001 is a strain of O. tipulae.
We prepared spore isolates from a homogenate of microsporidium-infected IMU001 worms and spread the isolates on E. coli OP50-seeded NG agar plates to examine their infectivity. We then cultured uninfected O. tipulae IMU001 and CEW1, and wild type C. elegans (N2) on the plates to observe spore formation. Spore formation was detected in IMU001 and CEW1 worms, but not in C. elegans (Fig. 1G and H). In order to expose the C. elegans to a larger amount of spores, we examined a mixed culture, consisting of C. elegans and infected IMU001 worms, which constantly shed infectious spores. Neither intestinal nor hypodermal cells of the C. elegans harboured spores regardless of their existence within the intestinal lumen (Fig. 1H). These data demonstrate that the microbe horizontally infects O. tipulae and exhibits host specificity.
Structural characteristics of spores
Under ultramicroscopic observation, rod-shaped spores could be clearly identified in intestinal and hypodermal cells (Fig. 4A). The intestinal cells of uninfected worms were filled with granular organelles of variable electron-density and morphology (Fig. 4B), similar to those seen in C. elegans (Kawai et al. Reference Kawai2009). While the electron-lucent granules remained, most of the electron-dense granules disappeared in intestinal cells that harboured the spores.
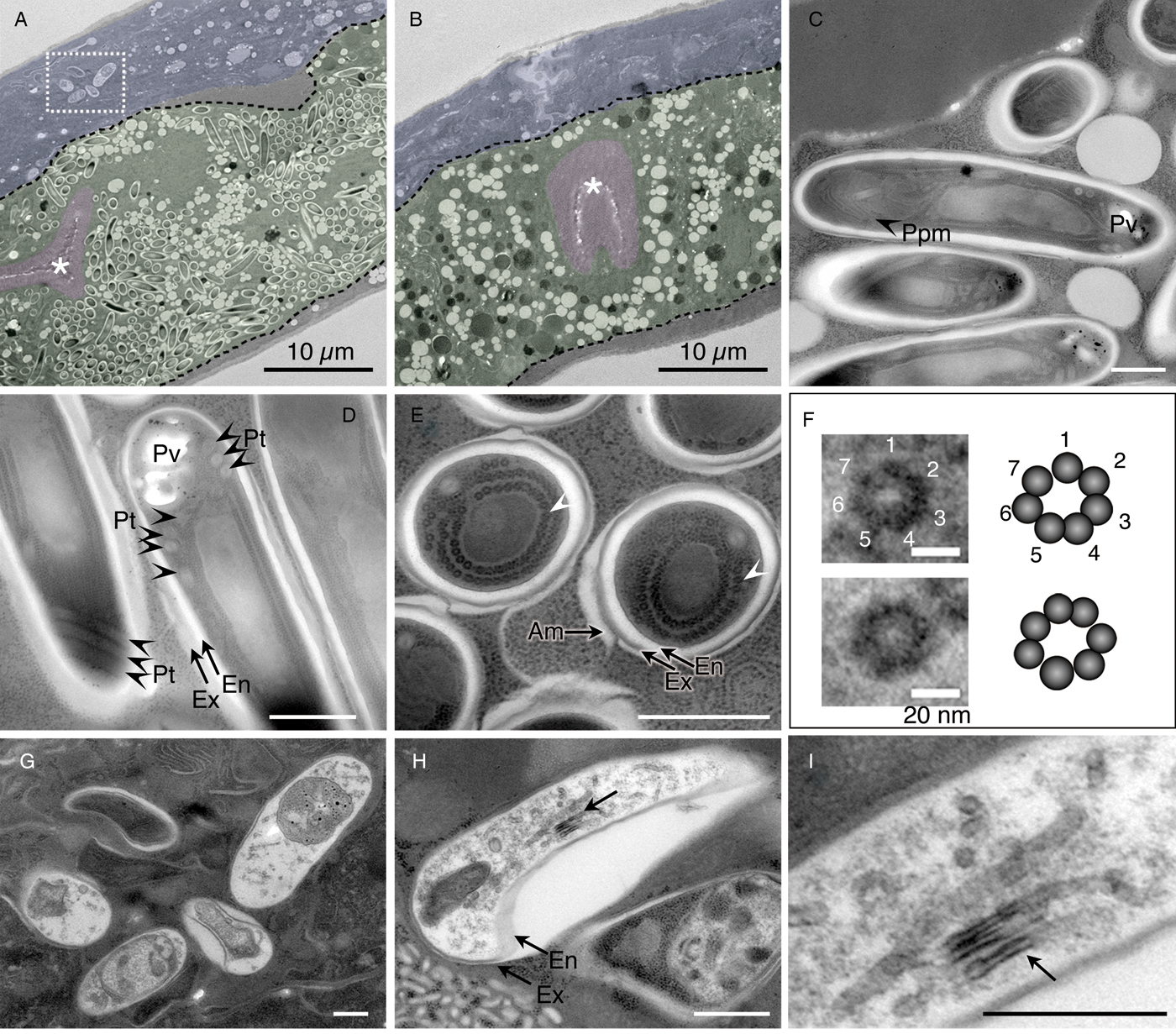
Fig. 4. Percutemincola moriokae-infection induced an alteration in the host intestine and morphological variations between intestinal and hypodermal Pe. moriokae spores. (A–I) Fine structures of Pe. moriokae infection in O. tipulae IMU001. (A and B) Longitudinal sections of the infected and uninfected worms. Tissues are labelled by false colour overlay (green for intestinal cells, magenta for the intestinal lumen and microvilli, and blue for the hypodermis). The rod-shaped spores were observed in the intestinal and hypodermal cells of the infected worm (A) but not in the uninfected worm (B). Broken black lines outline the basal membrane of the intestinal cells. The box in (A) indicates hypodermal spores. Asterisks show the intestinal lumen. (C–E) Longitudinal sections (C, D) and a cross section (E) of the intestinal spores. A subset of spores was surrounded by additional (potentially host-derived) membranes (E). Arrowheads indicate the small electron-dense dots in round arrangement. (F) Magnified images of the dots in (E) and their schematic images. (G–I) Morphological variations of spores in the hypodermal cells. Some spores contain tubular structures (arrows) and a large cavity between the endospore and exospore (H). The magnified image of (H) is shown in (I). Am, additional membrane. En, endospore. Ex, exospore. Pt, coiled polar tube. Ppm, polaroplast membrane. Pv, posterior vacuole. Bars in A, B = 10 µm, in C, D, E, G, H, I = 500 nm, in F = 20 nm.
The spores in the intestine showed characteristics of microsporidian spores, including coiled polar tubes, a thick surface composed of endo- and exospores, and lamellar polaroplast membranes, which are accordion-like membranous stacks that serve as sporoplasm membranes after the invasion (Fig. 4C and D). The polar tube of the novel microsporidium had 3–4 coils (Fig. 4D), which is relatively low-coiled (Weber et al. Reference Weber, Deplazes, Mathis, Versalovic, Carroll, Funke, Jorgensen, Landry and Warnock2011). Although a subset of spores showed an exospore that directly interacted with the host cytosol (Fig. 4C and D), another subset had an additional outermost membrane (Fig. 4E), implying derivation from the host membranous system.
We observed small electron-dense dots in circular arrangements within the sporoplasm of examined cross sections (Fig. 4E and F). These dots seemed to correspond to ribosomes, based on an ordinal schematic diagram depicting generalized microsporidian spores (Cali, Reference Cali1991). However, in higher magnification, it was revealed that each dot consisted of seven small particles in a circular arrangement (Fig. 4F). Moreover, our images indicate that the dots were each approximately 10 nm in size, too small to be identified as eukaryotic ribosomes, which are typically 25–30 nm.
The spores in the hypodermis were generally electron lucent and displayed no detectable structure, except for obvious electron-dense monokaryotic nuclear and tubular bodies (Fig. 4G–I). The spores exhibited diverse morphology from bending rod to planular shapes. Despite the morphological differences between spores in the hypodermis and those in the intestine, they were unapparent in DIC images (Fig. 1), although we observed slight differences in fluorescent intensity following staining with CW; i.e. fluorescence in the hypodermis was slightly weaker than in the intestine (Fig. 3C and D).
Structural characteristics of pre-spore stages
Monokaryotic nucleus with developed endoplasmic reticulum and irregular cell shape were found in merogonial stages (Fig. 5A). The dense surface coat was visible in sporonts which contain a lot of free ribosomes in the cytoplasm (Fig. 5B–D). In sporulation, tubular structures appear in the proximity of outside of a dense surface coat (Fig. 5E–G). A developing polar tube is elongated along with fibrous materials that circumvent a temporally formed anterior vacuole (Fig. 5H).

Fig. 5. A–H. Fine structures of pre-spore stages of Pe. moriokae. (A) Monokaryotic nuclei indicated by (N) were observed at merogonial stage. (B–D) The dense surface coat was visible at sporogonial stage. White arrow indicates the dense surface coat. (E–G) Uninucleate sporoblasts were associated with tubular structures (white arrowheads) in proximity of outside of the plasma membrane. (H) Immature spores temporarily form anterior vacuole. Developing polar tube (arrow) elongates along with fibrous materials and circumvents the anterior vacuole (asterisk). The boxes in (B) and (E) are magnified in (C) and (F), respectively. Bars in A = 2.0 µm, in B, C, E, F, G, H = 500 nm, in D = 100 nm.
Molecular phylogeny
We obtained partial SSU rDNA sequence spanning from the sequence for a microsporidian universal forward primer V1 (Zhu et al. Reference Zhu1993) to a universal reverse primer 1492R (Eden et al. Reference Eden1991; Harris et al. Reference Harris, Kellye and Pace2004). Phylogenetic analyses placed the novel microsporidium in microsporidian clade IV, which also includes human pathogens such as the Enterocytozoon and Vittaforma genera (Fig. 6, Supplementary Fig. S1–S3). The sequence formed a strongly supported monophyletic group (BS value for ML, MP, NJ, ME = 100%) with unidentified environmental microsporidia (KC111784 and FJ755987, Ardila-Garcia et al. Reference Ardila-Garcia2013) and Pancytospora epiphaga (KX424959, Zhang et al. Reference Zhang2016). BLASTN search also supported the phylogeny: IMUm001 is suggested to be related to KC111784 (e-value = 0.0, sequence identity = 96%), FJ755987 (e-value = 0.0, sequence identity = 95%), and KX424959 (e-value = 0.0, sequence identity = 95%). Our analyses also indicate that this monophyletic clade forms a larger clade with the recently identified nematode pathogens genus Enteropsectra and Pancytospora philotis and insect pathogens Orthosomella and Liebermannia (BS values for ML, NJ, ME and MP are 86, 100, 100 and 72%, respectively).

Fig. 6. Phylogenetic placement of the novel microsporidium, Pe. moriokae. Maximum likelihood phylogenetic tree based on the partial SSU rDNA sequences indicate that Pe. moriokae is most likely a member of clade IV and closely related to unidentified microsporidia and JUm1396. Percutemincola moriokae is shown in bold letters (IMUm001). BS values higher than 50% by four different methods (maximum likelihood, neighbour-joining, minimum evolution and maximum parsimony) are shown at each corresponding node. BS value of 100% for all examined methods are indicated by closed squares. Letters in parentheses are GenBank accession numbers. IMUm002 was obtained from a nematode strain (IMU002) isolated from the same soil sample as of IMU001 and IMUm001.
Influence of the microsporidian (IMUm001) infection on host lifespan and infection time course
To investigate the effect of the microsporidia infection on the host, we analysed the longevity of the IMU001 worms cultured with or without the IMUm001 spore isolate. The median lifespan of the worms cultured with spores was 9–11 days, which was 6 days shorter than that of the worms without spores (Fig. 7A).

Fig. 7. Comparison of lifespan of O. tipulae IMU001 in the presence and absence of Pe. moriokae infection and time course of the tissue distribution of spores. (A) Oscheius tipulae survival rate was examined after synchronized L1 larvae were reared in the presence (●) or absence (□) of Pe. moriokae spores until day 5. Dead worms were counted (n = 82 with, n = 67 without spore fraction). All examined worms were indicated as 100%. A significant difference in worm lifespan was observed (P < 0.001 by the log-rank test). (B) The time course of spore distribution of Pe. moriokae in O. tipulae IMU001. The worms were stained with CW to detect the mature spores in the cells. The plots indicate the mean values of duplicate experiments and their differences from the measured values are displayed as error bars. Expt.1 and 2 indicate independent experiments.
To elucidate the infection process, we analysed the spore formation time course. After bleaching, newly hatched L1 larvae (day 0) were grown with the spore isolate on NG agar plates seeded with E. coli OP50. The spore-forming rate was examined daily from days 3 through 7 via CW staining (Fig. 7B). An increase in hypodermal spores was first observed on day 4, while an increase in intestinal spores was first seen on day 5, suggesting tissue-dependent differences in the infection process.
Description
Phylum Microsporidia Balbiani, 1882
Percutemincola, a new genus
Diagnosis: The closest relatives are Pancytospora and Enteropsectra and form a group of nematode-infective microsporidia, based on SSU rDNA phylogeny. Monokaryotic arrangement of nuclei exists throughout the life-cycle. Spores contain a low coiled polar tube and posterior vacuoles. Infectious spores appear first in hypodermis and disseminate to intestinal cells. The genus name ‘Percutemincola’ is a combination of the terms for ‘through’, ‘skin’, and ‘invader’ in Latin (Per-cutem-incola).
Type species: Pe. moriokae
Percutemincola moriokae, new species
Diagnosis: Like the genus. Anterior vacuoles are only visible during polar filament development in sporont. Tubular structures are formed in the proximity of sporont. Small electron-dense dots located around nuclei of spores. The sporonts are not internalized in the host endoplasmic reticulum, unlike Liebermannia (Sokolova et al. Reference Sokolova, Lange and Fuxa2007; Reference Sokolova2009). Spore size is 2.9–4.6 µm × 0.8–1.2 µm in intestinal cells and 2.5–5.0 µm × 1.0–2.0 µm in hypodermal cells.
Host species: Oscheius tipulae (Lam & Webster, 1971)
Type locality: The type species was isolated from wet soil on the riverside of the Shizukuishi River, Morioka, Iwate Prefecture, Japan [141°8′23″ East; 39°41′45″ North] in September 2010.
Infection site: hypodermal cells and intestinal cells.
Transmission: Horizontal transmission. Vertical transmission is unknown.
Pre-spore stages: Monokaryotic meront shows irregular cell shape. Sporont contains a lot of free ribosomes. In sporulation, tubular structures appear in the proximity of outside of a dense surface coat and developing polar tube elongates along with fibrous materials and circumvents a temporally formed anterior vacuole.
Spore: The intestinal spore contains a polar tube with 3–4 coils, a polaroplast, posterior vacuoles but lacks anterior vacuoles. Thick exospore and endospore layers coat each spore. Some of these spores have an additional membrane as their outermost layer. The exospore and endospore layers sometimes form a cavity between them. Sporogony takes place in the hypodermis and intestine, as CW-staining was detected in both tissues.
Type specimen: Semi-ultrathin sections of Pe. moriokae strain IMUm001-infected O. tipulae IMU001 were deposited at the National Museum of Nature and Science, Tokyo (voucher No. NSMT-Pr 367 and NSMT-Pr 368).
Etymology: The specific name ‘moriokae’ is derived from the type locality.
Discussion
We isolated a nematode-infective microsporidium from a wild O. tipulae strain IMU001. The novel microsporidium infects the hypodermal and intestinal cells of its host. Phylogenetic analysis using SSU rDNA sequences indicates that the novel microsporidium along with Pa. epiphaga and unidentified microsporidia KC111784 and FJ755987 form a sister relationship with other nematode-infective microsporidia in clade IV. Since most nematode-infective microsporidia infection initiated in the intestines, we propose a new genus Percutemincola and new species Pe. moriokae, to classify the new species, of which spores first appeared in the hypodermis of the host nematode.
Comparison of Pe. moriokae with other microsporidia
One of the most studied nematode-infective microsporidium is N. parisii, which causes a deleterious effect on host C. elegans, and is classified to clade II (Troemel et al. Reference Troemel2008). Percutemincola moriokae infects both hypodermal and intestinal cells and form locally cleared region devoid of intestinal granules. In spite of light microscopic resemblance of those microsporidia, electron microscopy revealed differences such as the number of turns in the coils of the polar tube and existence of membrane encompassing spore cluster, which is a character of N. parisii spore.
Except for marine nematodes, most nematode-infective microsporidia are intestinal cell specialists (Zhang et al. Reference Zhang2016), which are considered to infect intestine through fecal-oral transmission, while Pe. moriokae infects hypodermis too. One Nematocida species N. displodere also infects multiple tissues of C. elegans (Luallen et al. Reference Luallen2016). Nematocida displodere are thought to access the tissues from intestinal lumen and the spores are released when the host bursts. The time course study of Pe. moriokae infection revealed that mature spores appear earlier in hypodermal cells than in intestinal cells, suggesting a possibility that the infection route involves the free spores first infecting the hypodermis and then spreading to the intestine. Another possibility is also conceivable that infection occurs independently in both tissues but sporulation is delayed in intestinal cells.
Pancytospora epiphaga and Pancytospora philotis, which are the close relatives of Pe. moriokae, show different host specificity and tissue tropism. Pa. epiphaga infects hypodermis and muscle (but not intestine) of C. elegans and C. brenneri, while Pa. philotis infects intestine of O. tipulae (Zhang et al. Reference Zhang2016), implying that they acquired the distinct infection mechanisms and host specificity. Because Pa. epiphaga and Pe. moriokae may adopt the relevant infection mechanism to infect hypodermis of nematodes, according to future studies, a taxonomic transfer of Pa. epiphaga to Percutemincola should probably be considered.
Many microsporidian species are reported to have polymorphic spores, some of which seem to be target-cell specific (Vávra et al. Reference Vávra1998; Reference Vávra2006; Sokolova and Fuxa, Reference Sokolova and Fuxa2008). Percutemincola moriokae showed dimorphism between spores in hypodermal and intestinal cells, implying that the morphology of the hypodermal spores is relevant to their target cells or to the discharged structure.
Percutemincola moriokae-O. tipulae as a new model to study microsporidia–host relationship
The relationship between Pe. moriokae and O. tipulae can be used to examine various interesting traits of microsporidia as follows. First, Pe. moriokae is a member of clade IV along with most human-infective microsporidia, such as Encephalitozoon cuniculi, Encephalitozoon hellem, and Enterocytozoon bieneusi. Therefore, Pe. moriokae may serve as a potential model not only for invertebrate but also mammalian pathogens. For example, the infection mechanism of Pe. moriokae to ectodermal epithelium is of interest since it might be useful as a model to study microsporidial keratoconjunctivitis. Studying the difference in the infection mechanisms among nematode-infective microsporidia could also be useful to elucidate the core mechanism of such infections.
Second, although the intestinal cells of O. tipulae and C. elegans show similar morphological characters (e.g. large polarized cells, apical microvilli, and granular organelles) (Fig. 4, Kawai et al. Reference Kawai2009; McGhee, Reference McGhee2007), Pe. moriokae was not able to infect C. elegans. This specificity may be attributed to a C. elegans defence mechanism against Pe. moriokae or an incompatibility of the infection mechanism. A wild C. elegans strain from Hawaii eliminates N. parisii when inoculated at a young larval stage, suggesting the importance of parasite to host-strain compatibility (Balla et al. Reference Balla2015). It would be interesting to investigate if Pe. moriokae infection of C. elegans is prohibited before or after the invasion.
Third, obvious host responses are provoked by the infection. In C. elegans, the intestinal granules consist of various granular organelles, each of which has a distinct function, e.g. lipid storage and lysosome-related functions (Tanji et al. Reference Tanji2016). The variations in electron density of the intestinal granules in uninfected O. tipulae are reminiscent of C. elegans intestine. These granules reduced in number with Pe. moriokae infection, while many electron-lucent granules (potentially lipid droplets) remained, suggesting that the infection affects cell physiology, decreasing the electron-dense granular organelles in the intestine. Perturbation of the intestinal granules will provide a good indicator of the host responses. In regards to host responses, it is interesting that a subset of spores shows an additional outermost membrane. Sporogony of microsporidia can occur in two ways, depending on species: sporogony within the host-derived membranous surroundings (like N. parisii), or sporogony in direct contact with the host cytosol (Franzen and Müller, Reference Franzen and Müller1999; Szumowski et al. Reference Szumowski2014). Percutemincola moriokae exhibits the latter type because many spores exist directly within the cytoplasm. Therefore, when considered in the context of a host defence mechanism, the formation of additional membranes is probably to sequester parasites.
Finally, recent advances in sequence-based forward genetics (Sarin et al. Reference Sarin2008; Chu et al. Reference Chu2014) and CRISPR/Cas9-based genome editing mediated by ribonucleoprotein injection (Cho et al. Reference Cho2013) have facilitated research on non-model organisms, such as O. tipulae. However, such type of experiment prefers organisms that can be easily handled in a laboratory-environment and readily observed in each experiment. Percutemincola moriokae and its host, O. tipulae, fulfill such requirements for a model system. Therefore, altogether, our model system is useful for further studies to understand the various aspects of microsporidian infection.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182018000628
Acknowledgements
We thank Ms. Reiko Aoyama, Ms. Ayumi Hosoda and Ms. Kikuko Kawano for their support on the experiments. Ms. Satomi Nishikori supported sample collection in the field. We thank all of the staff in the Technical Support Center for Life Science Research in Iwate Medical University, for their support in preparing ultrathin sections and the type specimen. Laboratory-bred nematode strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflicts of interest
None
Ethical standards
Not applicable











