Introduction
Cryptosporidium is an opportunistic zoonotic parasite that can infect many animals more than 150 species of animals, including humans (Kotloff, Reference Kotloff2017; Khan et al., Reference Khan, Shaik and Grigg2018). Cryptosporidium is common in ruminants and can cause watery diarrhoea in juvenile ruminants in particular (Santin, Reference Santin2013). It is well known that small ruminants, such as sheep and goats, play a significant role in some zoonotic infections (Casemore, Reference Casemore1989). Cryptosporidiosis causes huge economic losses to farmers, and the lack of effective prevention and/or treatment in livestock is an unresolved issue (Ryan et al., Reference Ryan, Fayer and Xiao2014). Cryptosporidiosis is also a major cause of foodborne and waterborne disease outbreaks in high-income countries and is also the main cause of diarrhoea in young children in low- and middle-income countries (Zahedi and Ryan, Reference Zahedi and Ryan2020; Yang et al., Reference Yang, Guo, Xiao and Feng2021). In South Asia and sub-Saharan Africa, Cryptosporidium infection causes diarrhoea in close to 2.9–4.7 million children under 2 years of age each year (Kotloff et al., Reference Kotloff, Nataro, Blackwelder, Nasrin, Farag, Panchalingam and Levine2013; Sow et al., Reference Sow, Muhsen, Nasrin, Blackwelder, Wu, Farag and Levine2016). Currently, no effective vaccine has been developed for the treatment of cryptosporidiosis (Ryan et al., Reference Ryan, Zahedi and Paparini2016).
A few years ago, Cryptosporidium was designated as one of the 24 most destructive foodborne parasites by the expert committees of the Food and Agriculture Organization of the United Nations (FAO) and World Health Organization (WHO) (Bouwknegt et al., Reference Bouwknegt, Devleesschauwer, Graham, Robertson and van der Giessen2018). The clinical diagnosis of cryptosporidiosis is done using detection methods such as microscopy of acid-fast-stained preparations, immunofluorescent antibody staining, nested polymerase chain reaction, enzyme-linked immunosorbent assay and immunochromatography (Chalmers and Katzer, Reference Chalmers and Katzer2013; Checkley et al., Reference Checkley, White, Jaganath, Arrowood, Chalmers, Chen and Houpt2015; Xiao and Feng, Reference Xiao and Feng2017; Adeyemo et al., Reference Adeyemo, Singh, Reddy and Stenstrom2018). Currently, 44 Cryptosporidium species and more than 120 genotypes have been recognized (Feng et al., Reference Feng, Ryan and Xiao2018; Ryan et al., Reference Ryan, Feng, Fayer and Xiao2021). So far, many Cryptosporidium species and genotypes have been discovered in sheep using molecular tools. Cryptosporidium xiaoi, Cryptosporidium parvum and Cryptosporidium ubiquitum are the 3 dominant Cryptosporidium species in sheep (Guo et al., Reference Guo, Li, Ryan, Feng and Xiao2021). The 2 major Cryptosporidium species in sheep responsible for zoonotic infections in humans are C. parvum and C. ubiquitum (Xiao and Feng, Reference Xiao and Feng2008).
Cryptosporidiosis causes losses in the sheep breeding industry, and threatens human health by spreading through water sources. A systematic review and meta-analysis were conducted to evaluate cryptosporidiosis infection in sheep worldwide. The potential risk factors including region, age, the presence or absence of diarrhoea and Cryptosporidium species/genotypes were also analysed. The results describe the regional distribution characteristics of Cryptosporidium species and subtypes and may contribute to the prevention and control of Cryptosporidium infection.
Materials and methods
Search strategy and selection criteria
Five literature databases were used to search for studies on the global prevalence of Cryptosporidium in sheep, namely, PubMed, Web of Science, the China National Knowledge Infrastructure (CNKI), VIP Chinese Journals Database and Wanfang Data. All published studies so far on Cryptosporidium in sheep from 15 September 2021 onwards were retrieved. In PubMed, the search formula used was ‘[((((sheep) OR (ovine)) OR (ram)) OR (ewe)) OR (small ruminant)) AND ((Cryptosporidium) OR (cryptosporidiosis)]’. In Web of Science, the search formula used was ‘[((((TS = (sheep)) OR TS = (ovine)) OR TS = (ram)) OR TS = (ewe)) OR TS = (small ruminant)) AND ((TS = (Cryptosporidium)) OR TS = (cryptosporidiosis)]’. In the 3 Chinese databases, ‘sheep’ (Chinese) and ‘Cryptosporidium’ (Chinese) were used as keywords. Selected eligible studies were designed and analysed according to the preferred reporting items of the systematic reviews and meta-analysis (PRISMA) statement (Table S1).
The following clauses were used as the criteria for article exclusion: (1) the purpose of the study was not the prevalence of Cryptosporidium in sheep; (2) the total number of sheep tested and the number of sheep that tested positive were not provided; (3) there was no clear testing method; (4) the sample was a mixture of specimens from multiple sheep; (5) the study sample size was less than 20; (6) the study was a review or case report. For articles whose full text was not available, the first author was not contacted for more research information and/or statistics.
Quality assessment
Established methods were used to evaluate the quality of studies (Guyatt et al., Reference Guyatt, Oxman, Vist, Kunz, Falck-Ytter and Alonso-Coello2008). Studies were awarded points for aspects that were suitable to assess the quality studies. A study received 1 point for each of the following items: a clear research goal, a clear detection method, a clear research period, analysis involving 3 or more influencing factors and a sample size of more than 200. Studies with scores of 4 or 5 points were considered to be of high quality; studies with scores of 2 or 3 points were considered to be of medium quality; studies with scores of 0 or 1 point were classified as low quality.
Data extraction
All titles, abstracts and full texts were separately screened by 2 authors (YCC and HKQ) and the data were independently extracted. Disagreements were resolved by discussion with JYH. The following information was extracted by YCC and HKQ: study ID (the first author and publication date), country, sampling time, detection method, total samples, positive sample, prevalence, study quality and Cryptosporidium species/genotypes (Table S2).
Statistical analysis
All data were analysed using Stata version 14.0. Because of high heterogeneity (I 2 > 50%, P < 0.1), a random-effects model was used for meta-analysis. Sensitivity, subgroup and meta-regression analyses were conducted to investigate potential sources of heterogeneity [code: gen ser = sqrt (rate × (1 − rate)/n); metan rate ser, random label (namevar = study); metaninf rate ser, label (namevar = study) random; metan rate ser, label (namevar = study) by (region) random xlabel (0, 0.1, 0.2, 0.3); metareg logr region, wsse (selogr) bsest (reml)]. To test the stability of the data, a sensitivity analysis of the data was done. The effect of selected studies on the pooled prevalence by excluding single studies sequentially was evaluated (Wang et al., Reference Wang, Liu, Liu, Li, Zhang, Zhao and Zhu2018). The overall study was evaluated using forest plots, and the publication bias of the study was evaluated using a funnel plot and Egger's tests (Egger et al., Reference Egger, Davey Smith, Schneider and Minder1997). The following potential sources of heterogeneity were examined: region (Asia compared with other regions), age (<3 months compared with the other age group), the presence or absence of diarrhoea (diarrhoea compared with no diarrhoea) and Cryptosporidium species/genotypes (C. parvum compared with the other species/genotypes). If multiple detection methods were involved in the research, microscopy results were the first choice (Robinson and Chalmers, Reference Robinson and Chalmers2020). Microscopic detection is a simple, fast and cheap method for the detection of Cryptosporidium oocysts (Robinson and Chalmers, Reference Robinson and Chalmers2020). Immunofluorescence microscopy is generally considered the gold standard in laboratories in Europe and the United States.
Results
Characteristics of studies
A total of 1434 records were initially identified, and 190 potentially relevant studies were selected for full-text search after screening of the title and abstract. Of these, 11 were review studies, 4 were case reports, 13 had incomplete information or only provided prevalence, 5 did not examine sheep, 11 had sample sizes less than 20 and 20 did not have full text available. In total, 126 articles of sufficient quality were considered for meta-analysis (Fig. 1).

Fig. 1. Flow diagram of the selection of eligible studies.
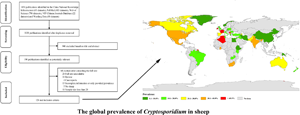
The selected studies came from 41 countries (Fig. 2, Table 1). Fifty-three datasets originated from Asia [Azerbaijan (n = 1), China (n = 28), Cyprus (n = 1), India (n = 3), Iran (n = 6), Iraq (n = 1), Jordan (n = 1), Kuwait (n = 2), Pakistan (n = 3), Turkey (n = 7)]. Thirty-three datasets were from countries in Europe [Belgium (n = 1), Czech Republic (n = 1), France (n = 2), Greece (n = 3), Ireland (n = 1), Italy (n = 2), Macedonia (n = 1), Norway (n = 1), Poland (n = 2), Romania (n = 1), Serbia (n = 1), Spain (n = 9) and the United Kingdom (n = 8)]. Twelve datasets were from countries in Africa [Algeria (n = 4), Egypt (n = 1), Ethiopia (n = 1), Ghana (n = 1), Kenya (n = 1), South Africa (n = 1), Tunisia (n = 1), Zambia (n = 1)]. Eleven datasets were from countries in North America [Canada (n = 1), Greenland (n = 1), Grenada (n = 1), Mexico (n = 4), Trinidad and Tobago (n = 1), United States (n = 3)]. Six datasets were from South America [all 6 datasets were from Brazil (n = 6)]. Eleven datasets were from countries in Oceania [Australia (n = 9), New Zealand (n = 1), Papua New Guinea (n = 1)] (Tables 1 and 2). The age of sheep was <3 months in 52 datasets, 3–12 months in 12 datasets and >12 months in 31 datasets. Most datasets did not contain any information on health status. Diarrhoea in sheep was reported in 17 datasets, and no diarrhoea was reported in sheep in 8 datasets (Table 2).

Fig. 2. Map of Cryptosporidium infection in sheep across the world. Prevalence ranges are shown in different colours. [The figure was designed by Arcgis 10.2, and the original vector diagram imported in Arcgis was adapted from Natural Earth (http://www.naturalearthdata.com).]
Table 1. Prevalence calculated from pooled published data of Cryptosporidium infection in sheep

Table 2. Pooled prevalence of Cryptosporidium infection in sheep across the world

Note: aOther including C. bovis, C. scrofarum, C. andersoni, C. hominis, C. canis, C. ryanae, C. suis, C. fayeri, C. meleagridis, C. muris, Cryptosporidium spp., sheep genotype I.
Cryptosporidium infection in sheep by region
Overall, the estimated Cryptosporidium prevalence in sheep ranged from 14.8% [95% confidence interval (CI) 12.9–16.7%] to 29.8% (95% CI 20.5–39.2%) with substantial heterogeneity (I 2 = 98.5%, P < 0.001). On the global scale, pooled estimated prevalence of Cryptosporidium infection in sheep was 18.9% (95% CI 17.2–20.6%, 7836/47 585) (Table 2). On 6 continents (Table 2, Figs 3–8), the infection rates of Cryptosporidium in sheep were 14.8% (Asia), 20.2% (Europe), 21.7% (Africa), 29.8% (North America), 20.3% (South America) and 19.1% (Oceania). The highest number of studies on Cryptosporidium infections in sheep originated from Asia (n = 53), while the infection rate in Asia was the lowest among the 6 continents, with values below 10.0% in China and Iran. Europe had the second most positive samples, with a total of 13 countries reporting Cryptosporidium infection in sheep. The highest prevalence rate was reported for Cyprus [76.9% (95% CI 63.1–90.8%)], and the lowest prevalence rate was in Ethiopia [1.2% (95% CI 0.4–2.1%)] (Table 1).

Fig. 3. Forest plot of the prevalence estimates of Cryptosporidium infection in sheep in Asia.

Fig. 4. Forest plot of the prevalence estimates of Cryptosporidium infection in sheep in Europe.

Fig. 5. Forest plot of the prevalence estimates of Cryptosporidium infection in sheep in Africa.

Fig. 6. Forest plot of the prevalence estimates of Cryptosporidium infection in sheep in North America.

Fig. 7. Forest plot of the prevalence estimates of Cryptosporidium infection in sheep in South America.

Fig. 8. Forest plot of the prevalence estimates of Cryptosporidium infection in sheep in Oceania.
Prevalence according to age, presence or absence of diarrhoea and Cryptosporidium species/genotypes
Subgroup analysis according to age showed that the Cryptosporidium infection rate in sheep <3 months was 27.8% (95% CI 23.3–32.4%, 3284/11 938). This was significantly higher than in sheep 3–12 months [17.2%; 95% CI 11.7–22.7%, 614/4054, odds ratio (OR) 2.13, P < 0.05] and in sheep >12 months of age (10.2%; 95% CI 7.8–12.6%, 895/8392, OR 3.18, P < 0.05) (Table 2). The infection rate for sheep with diarrhoea was 35.4% (95% CI 26.8–44.0%, 844/1915), while the infection rate for sheep without diarrhoea was 11.3% (95% CI 7.1–15.5%, 176/1691) (Table 2). Fourteen Cryptosporidium species/genotypes (C. parvum, C. ubiquitum, C. xiaoi, Cryptosporidium bovis, Cryptosporidium scrofarum, Cryptosporidium andersoni, Cryptosporidium hominis, Cryptosporidium canis, Cryptosporidium ryanae, Cryptosporidium suis, Cryptosporidium fayeri, Cryptosporidium meleagridis, Cryptosporidium muris and sheep genotype I) were detected in sheep globally. The prevalence rate of C. parvum was 4.8% (95% CI 4.0–5.6%, 722/16 354), of C. xiaoi 6.6% (95% CI 5.3–8.0%, 1301/15 667) and of C. ubiquitum 3.2% (95% CI 2.6–3.9%, 720/19 655) (Table 2). The distribution of the Cryptosporidium species/genotypes found in sheep across the 6 continents was as follows: C. parvum was most common in Europe while C. xiaoi was most common in Oceania, Asia and Africa and C. ubiquitum was most common in North America and South America (Fig. 9). Proportions of subgenotype families IIa and IId of C. parvum were 35.1% (141/402) and 32.1% (129/402). The most common subgenotypes IIdA20G1 and IIaA15G2R1 of C. parvum were reported in 15.4% (62/402) and 19.7% (79/402) of positive samples. Subgenotype family IId was most common in Oceania, Asia and Europe, whereas IIa was obviously distributed evenly in all evaluated regions (Table 3). Proportions of subgenotype families XIIa and XIId of C. ubiquitum were 90.0% (216/240) and 3.8% (9/240). XIIa was most common in Oceania, Europe, Asia and Africa, whereas XIId was found in Oceania (Table 4).

Fig. 9. Diversity of Cryptosporidium species/genotypes in sheep across the world. The horizontal axis shows the 6 continents, and the vertical axis shows the number of species/genotypes.
Table 3. Cryptosporidium parvum subtypes in sheep

Table 4. Cryptosporidium ubiquitum subtypes in sheep

Sensitivity analysis and publication bias
Sensitivity analysis showed that the analysis was reliable (Figs S1–S6). A funnel plot was used to measure the publication bias of the selected studies. An asymmetrical funnel plot, with some points falling outside the funnel, indicates publication bias. The funnel plot showed obvious asymmetry (Fig. 10), and P < 0.001 was obtained using Egger's test (Table S3), indicating that obvious publication bias was detected.

Fig. 10. Funnel plot for examination of publication bias of the prevalence estimates of Cryptosporidium in sheep across the world.
Sources of heterogeneity by meta-regression analysis
The result indicated substantial heterogeneity (I 2 = 98.5%, P < 0.001). Univariate meta-regression analysis was conducted to further identify the source of heterogeneity. The results showed that age (P < 0.001) and presence or absence of diarrhoea (P = 0.001) were factors that fostered heterogeneity. Region (P = 0.069) and species/genotypes (P = 0.128) were not related to heterogeneity (Table 2).
Discussion
A reliable estimate of the prevalence of Cryptosporidium in sheep was produced through a meta-analysis based on datasets comprising a large population of sheep (n = 47 585) across 41 countries on 6 continents. In Europe, the highest infection rate was 49.0% (95% CI 39.3–58.8%) in Ireland (Mirhashemi et al., Reference Mirhashemi, Zintl, Grant, Lucy, Mulcahy and De Waal2016), while the lowest rate was 2.3% (95% CI 0.0–7.0%) in the Czech Republic (Kotkova et al., Reference Kotkova, Nemejc, Sak, Hanzal, Kvetonova, Hlaskova and Kvac2016). Cryptosporidium infection in sheep differs not only between countries but also in different regions of the same country. In China an infection rate of only 0.9% (3/318) was reported for sheep in Xinjiang (Qi et al., Reference Qi, Zhang, Zhao, Jing, Guan, Luo and Zhang2019), while a much higher infection rate of 13.1% (49/375) was found in sheep in Inner Mongolia (Ye et al., Reference Ye, Xiao, Wang, Wang, Amer, Roellig and Feng2013).
In the age subgroup analysis, the Cryptosporidium infection rate in sheep under 3 months of age was significantly higher than in sheep 3–12 months of age and in sheep older than 12 months. This is supported by studies in China (Wang et al., Reference Wang, Feng, Cui, Jian, Ning, Wang and Xiao2010; Zhang et al., Reference Zhang, Chen, Zou, Hou, Sun, Li and Zhu2020), though other studies documented slightly divergent findings. Ye et al. (Reference Ye, Xiao, Wang, Wang, Amer, Roellig and Feng2013) reported that the Cryptosporidium infection rate in sheep aged 3–12 months was higher (26.7%; 20/70) than in sheep less than 3 months of age (18.4%; 16/87) or in sheep older than 12 months (6.1%; 13/213). Mi et al. (Reference Mi, Wang, Huang, Mu, Zhang, Jia and Chen2018) investigated Cryptosporidium infection in sheep in 10 provinces in China and confirmed that the highest infection rate occurred between 3 and 12 months of age (34.6%; 116/335), followed by sheep less than 3 months of age (28.2%; 108/383) and older than 12 months of age (22.4%; 71/317). In general, Cryptosporidium infection in sheep under 3 months of age is paid more attention. However, high rates of Cryptosporidium infection in other age groups suggest that different management measures in different geographical areas may play a role. The global prevalence of Cryptosporidium infection in sheep suffering diarrhoea was approximately 3 times higher than in sheep without diarrhoea (P < 0.05). However, most of the publications considered in our analysis did not mention whether feces samples represented sheep with diarrhoea and inadequate data collection may also affect the stability of the results. Due to the limited amount of suitable data, the relationship between infection and diarrhoea should be interpreted with caution. The results showed that feces of healthy sheep may also contain Cryptosporidium oocysts. Therefore, prevention of Cryptosporidium transmission in healthy sheep should not be neglected. In a 2020 study from Algeria, a Cryptosporidium infection rate of 100% (280/280) in neonatal lambs with diarrhoea was reported (Dahmani et al., Reference Dahmani, Ouchene, Dahmani, Ouchene-Khelifi and Oumouna2020). Therefore, timely control of sheep diarrhoea may also be an effective way to prevent the spread of Cryptosporidium.
As mentioned in a previous review, C. parvum was the dominant species in sheep in Europe, while C. xiaoi was the dominant species in Australia and C. ubiquitum appeared to dominate in the Americas and Asia (Ryan et al., Reference Ryan, Fayer and Xiao2014). Compared with previous study the prevalence of C. xiaoi in Asian sheep appeared to increase in recent years. Altogether C. parvum, C. xiaoi and C. ubiquitum are still the dominant species in sheep. Other species/genotypes that occasionally infected sheep such as C. hominis (Ryan et al., Reference Ryan, Bath, Robertson, Read, Elliot, McInnes and Besier2005; Pritchard et al., Reference Pritchard, Marshall, Giles, Mueller-Doblies, Sayers, Marshall and Chalmers2008; Kaupke et al., Reference Kaupke, Michalski and Rzeżutka2017), C. canis (Zhang et al., Reference Zhang, Jian, Li, Ma, Karanis, Qigang and Karanis2018), C. fayeri (Ryan et al., Reference Ryan, Bath, Robertson, Read, Elliot, McInnes and Besier2005), C. meleagridis (Zucatto et al., Reference Zucatto, Aquino, Inacio, Figueiredo, Pierucci, Perri and Bresciani2015), C. muris (Mahdi and Ali, Reference Mahdi and Ali2002; Kotkova et al., Reference Kotkova, Nemejc, Sak, Hanzal, Kvetonova, Hlaskova and Kvac2016; Zhang et al., Reference Zhang, Chen, Zou, Hou, Sun, Li and Zhu2020), C. suis (Ryan et al., Reference Ryan, Bath, Robertson, Read, Elliot, McInnes and Besier2005; Goma et al., Reference Goma, Geurden, Siwila, Phiri, Gabriel, Claerebout and Vercruysse2007), C. andersoni (Ryan et al., Reference Ryan, Bath, Robertson, Read, Elliot, McInnes and Besier2005; Wang et al., Reference Wang, Feng, Cui, Jian, Ning, Wang and Xiao2010; Sweeny et al., Reference Sweeny, Ryan, Robertson, Yang, Bell and Jacobson2011; Koinari et al., Reference Koinari, Lymbery and Ryan2014; Yang et al., Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014; Hijjawi et al., Reference Hijjawi, Mukbel, Yang and Ryan2016) and C. bovis (Ryan et al., Reference Ryan, Bath, Robertson, Read, Elliot, McInnes and Besier2005; Pritchard et al., Reference Pritchard, Marshall, Giles, Mueller-Doblies, Sayers, Marshall and Chalmers2008; Yang et al., Reference Yang, Jacobson, Gordon and Ryan2009; Smith et al., Reference Smith, Chalmers, Mueller-Doblies, Clifton-Hadley, Elwin, Watkins and Giles2010; Mirhashemi et al., Reference Mirhashemi, Zintl, Grant, Lucy, Mulcahy and De Waal2016; Kaupke et al., Reference Kaupke, Michalski and Rzeżutka2017; Squire et al., Reference Squire, Yang, Robertson, Ayi and Ryan2017) may play a role in zoonotic transmission. Cryptosporidium ryanae (Mirhashemi et al., Reference Mirhashemi, Zintl, Grant, Lucy, Mulcahy and De Waal2016), C. scrofarum (Ryan et al., Reference Ryan, Bath, Robertson, Read, Elliot, McInnes and Besier2005; Koinari et al., Reference Koinari, Lymbery and Ryan2014; Yang et al., Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014) and sheep genotype I (Sweeny et al., Reference Sweeny, Ryan, Robertson, Yang, Bell and Jacobson2011; Yang et al., Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014) were also detected in sheep, but there was currently no evidence of human infection by these species/genotypes. The XIIa subgenotype family of C. ubiquitum was found in sheep in Iran (Firoozi et al., Reference Firoozi, Sazmand, Zahedi, Astani, Fattahi-Bafghi, Kiani-Salmi and Akrami-Mohajeri2019), Italy (Dessi et al., Reference Dessi, Tamponi, Varcasia, Sanna, Pipia, Carta and Scala2020), Spain (Díaz et al., Reference Díaz, Navarro, Prieto, Perez-Creo, Vina, Diaz-Cao and Morrondo2018), Ghana (Squire et al., Reference Squire, Yang, Robertson, Ayi and Ryan2017), Kuwait (Majeed et al., Reference Majeed, El-Azazy, Abdou, Al-Aal, El-Kabbany, Tahrani and Xiao2018), China (Li et al., Reference Li, Cai, Cai, Wu, Li, Lei and Xiao2016; Mi et al., Reference Mi, Wang, Huang, Mu, Zhang, Jia and Chen2018) and Australia (Yang et al., Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014), while XIId was only detected in Australian sheep (Yang et al., Reference Yang, Gardner, Ryan and Jacobson2015). It was reported from Canada, Turkey, the United States and the United Kingdom that C. ubiquitum XIIa subgenotypes were found to infect humans, and XIId subgenotypes were also found to infect humans in the United States (Li et al., Reference Li, Xiao, Alderisio, Elwin, Cebelinski, Chalmers and Feng2014). IIaA15G2R1 was the most widely distributed subtype found in sheep across the world (Geurden et al., Reference Geurden, Thomas, Casaert, Vercruysse and Claerebout2008; Díaz et al., Reference Díaz, Quilez, Chalmers, Panadero, Lopez, Sanchez-Acedo and Diez-Banos2010, Reference Díaz, Quílez, Prieto, Navarro, Pérez-Creo, Fernández and Morrondo2015, Reference Díaz, Navarro, Prieto, Perez-Creo, Vina, Diaz-Cao and Morrondo2018; Smith et al., Reference Smith, Chalmers, Mueller-Doblies, Clifton-Hadley, Elwin, Watkins and Giles2010; Ye et al., Reference Ye, Xiao, Wang, Wang, Amer, Roellig and Feng2013; Koinari et al., Reference Koinari, Lymbery and Ryan2014; Paz e Silva et al., Reference Paz e Silva, Lopes, Bresciani, Amarante and Araujo2014; Yang et al., Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014; Majeed et al., Reference Majeed, El-Azazy, Abdou, Al-Aal, El-Kabbany, Tahrani and Xiao2018; Mammeri et al., Reference Mammeri, Cartou, Chevillot, Thomas, Julien, Vallée, Polack, Follet and Adjou2019) and was the dominant C. parvum subgenotype infecting sheep. In recent years, there was an increasing occurrence of C. parvum IIa in humans, especially in Colombia and Mexico (Urrea-Quezada et al., Reference Urrea-Quezada, Gonzalez-Diaz, Villegas-Gomez, Durazo, Hernandez, Xiao and Valenzuela2018; Higuera et al., Reference Higuera, Villamizar, Herrera, Giraldo, Vasquez, Urbano and Ramirez2020). Feng et al. (Reference Feng, Ryan and Xiao2018) reported IId subtypes mainly in lambs and goat kids in some European and Middle Eastern countries. In our analysis, other IId subtypes, including IIdA15G1 (Papanikolopoulou et al., Reference Papanikolopoulou, Baroudi, Guo, Wang, Papadopoulos, Lafi and Xiao2018; Qi et al., Reference Qi, Zhang, Zhao, Jing, Guan, Luo and Zhang2019; Bordes et al., Reference Bordes, Houert, Costa, Favennec, Vial-Novella, Fidelle and Razakandrainibe2020), IIdA16G1 (Papanikolopoulou et al., Reference Papanikolopoulou, Baroudi, Guo, Wang, Papadopoulos, Lafi and Xiao2018; Sahraoui et al., Reference Sahraoui, Thomas, Chevillot, Mammeri, Polack, Vallee and Adjou2019), IIdA18G1 (Yang et al., Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014, Reference Yang, Gardner, Ryan and Jacobson2015; Mi et al., Reference Mi, Wang, Huang, Mu, Zhang, Jia and Chen2018), IIdA19G1 (Yang et al., Reference Yang, Jacobson, Gardner, Carmichael, Campbell, Ng-Hublin and Ryan2014; Mi et al., Reference Mi, Wang, Huang, Mu, Zhang, Jia and Chen2018), IIdA24G1 (Imre et al., Reference Imre, Luca, Costache, Sala, Morar, Morariu and Darabus2013), and other subgenotypes were also detected in sheep. Cryptosporidium xiaoi generally did not infect humans (Guo et al., Reference Guo, Li, Ryan, Feng and Xiao2021). Although C. xiaoi was detected in HIV/AIDS patients (Adamu et al., Reference Adamu, Petros, Zhang, Kassa, Amer, Ye and Xiao2014), the authors did not describe C. xiaoi as zoonotic. In earlier studies, Robertson (Reference Robertson2009) found little evidence that human cryptosporidiosis was contracted from sheep and the transmission of cryptosporidiosis between sheep and animal breeders was only occasionally suspected (Mahdi and Ali, Reference Mahdi and Ali2002). In spite of this, many Cryptosporidium species/genotypes infecting sheep were found to be zoonotic. Overall, the Cryptosporidium infection rate in sheep slightly increased from 13.5% (1837/13 631) during 2011–2015 to 19.0% (2746/14 458) during 2016–2021 (Table S2). Therefore, measures should be considered to reduce and/or mitigate risks associated with contact between breeders and sheep to prevent the transmission of Cryptosporidium from sheep to humans.
Five databases were selected for comprehensive literature retrieval, which covers a large time span and a large total sample size. However, there are also some limitations associated. First, there were only few reports on Cryptosporidium infection in sheep from some countries, with some countries only having 1 report published within the last 30 years. As a result, Cryptosporidium infections in sheep in these countries are not yet fully understood. Second, all available data were not fully included; there were some data associated with unpublished literature, conference abstracts and other data sources that were not considered suitable for this analysis. Moreover, some articles could not be downloaded in full, and these publications were thus excluded. Third, when it came to explaining the source of heterogeneity, only certain factors were analysed. Other aspects such as sheep breed and feeding may also have contributed to data heterogeneity. However, even with these constraints, the results of the current study are close to the true global prevalence of Cryptosporidium in sheep.
Conclusions
This analysis shows that Cryptosporidium infections in sheep are widespread (18.9%) globally and can lead to disease and, consequently, huge economic losses in the sheep breeding industry. Risk factors related to cryptosporidiosis in sheep, such as age, should be accounted for so that farmers can apply effective management plans according to local conditions that may differ between geographical regions and environments, and prevent zoonotic transmission. In conclusion, the result provides a theoretical basis for the prevention and control of Cryptosporidium infection.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182022001196.
Data availability
All data generated or used during the study appear in the submitted article.
Acknowledgements
We thank Accdon-LetPub Editor for editing the English text of a draft of this manuscript.
Author's contributions
L. X. Z. conceived and designed the study; Y. C. C., J. Y. H. and J. Q. L. conducted the study; H. K. Q., Y. C. C. and J. Y. H. collected and analysed the data; Y. C. C. and L. X. Z. wrote the manuscript. All the authors have read and approved the final version of the manuscript.
Financial support
This research was funded by NSFC-Henan Joint Fund Key Project (U1904203) and Leading Talents of the Central Plains Thousand Talents Program (19CZ0122).
Conflict of interest
None.
Ethical standards
Not applicable.