Introduction
The peach-potato aphid, Myzus persicae (Sulzer) (Hemiptera: Aphididae), is a serious insect pest on a wide range of agricultural and horticultural crops due to its ability to transmit virus diseases and cause substantial direct and cosmetic feeding damage. Control measures still rely heavily on the application of insecticides including significant reliance on pyrethroids, a class of insecticides commonly used in crop protection, animal health and the control of insects endangering human health. Pyrethroids are selectively toxic to invertebrates at low concentrations and account for around 20% of all insecticides used worldwide (McCaffery & Nauen, Reference McCaffery and Nauen2006). However, many species have developed resistance to pyrethroids resulting in insect management problems and increased economic loss to agricultural producers (McCaffery & Nauen, Reference McCaffery and Nauen2006). The primary insecticidal action of pyrethroids results from their disruption of normal sodium channel activation and inactivation kinetics, leading to repetitive neural discharge, convulsive activity, and eventually paralysis and death (Narahashi, Reference Narahashi2000). One type of resistance to pyrethroids is termed knockdown resistance, or kdr, which results from reduced nerve sensitivity to pyrethroids and DDT (Soderlund & Bloomquist, Reference Soderlund, Bloomquist, Roush and Tabashnik1990). Although first reported in 1951 in the house fly Musca domestica Linnaeus (Diptera: Muscidae) (Busvine, Reference Busvine1951), our understanding of the molecular basis of kdr was limited until the recent cloning of sodium channel genes (homologous to the fruit fly para gene) from several species with kdr-type resistance (Miyazaki et al., Reference Miyazaki, Ohyama, Dunlap and Matsumura1996; Williamson et al., Reference Williamson, Martinez-Torres, Hick and Devonshire1996; Dong, Reference Dong1997; Park & Taylor, Reference Park and Taylor1997).
The insect sodium channel, like it's mammalian counterpart, is a large membrane protein comprised mainly of a single 260 kDa polypeptide (the alpha subunit) that contains four repeating and homologous domains (I–IV), with each domain consisting of six hydrophobic transmembrane segments (S1–S6) (see fig. 1a; reviewed in Catterall, Reference Catterall2000). Comparative sequencing studies of sodium channel genes between pyrethroid-resistant and -susceptible insects have identified a small number of point mutations that are associated with pyrethroid nerve insensitivity (reviewed in Soderlund & Knipple, Reference Soderlund and Knipple2003; Davies et al., Reference Davies, Field, Usherwood and Williamson2007). The most common mutation is a leucine to phenylalanine substitution (L1014F) within the domain IIS6 segment of the channel protein that was originally found in kdr housefly strains, M. domestica (Miyazaki et al., Reference Miyazaki, Ohyama, Dunlap and Matsumura1996; Williamson et al., Reference Williamson, Martinez-Torres, Hick and Devonshire1996) and is now known to occur in at least 12 other insect species, ranging from cat fleas to bollworms (see Davies et al., Reference Davies, Field, Usherwood and Williamson2007). The L1014F mutation, and its variants L1014H and L1014S, generally seem to confer moderate levels of resistance to DDT and pyrethroids (10–30 fold). Several other mutations have also been identified in the domain II region of the channel, including M918T and L925I in the IIS4–IIS5 linker, and T929I/V and L932F within the IIS5 transmembrane segment (see Davies et al., Reference Davies, Field, Usherwood and Williamson2007 and references therein). The M918T mutation was the original super-kdr mutation found in the housefly (Williamson et al., Reference Williamson, Martinez-Torres, Hick and Devonshire1996) and all of these additional mutations seem to be associated with enhanced resistance to pyrethroids (Davies et al., Reference Davies, Field, Usherwood and Williamson2007). Finally, a small number of mutations have also been found outside of domain II, usually in domains I or III, although none of these have been observed in more than one species as yet (also reviewed in Soderlund & Knipple, Reference Soderlund and Knipple2003; Davies et al., Reference Davies, Field, Usherwood and Williamson2007).
Here we report on the existence of the methionine-to-threonine mutation (equivalent to M918T) together with L1014F in a field-collected clone of M. persicae, with both mutations being in heterozygous form. Laboratory small-scale and field-simulator bioassay results showed that the presence of both L1014F and M918T in a single clone is able to confer strong levels of resistance to a wide range of Type I and II pyrethroid compounds. The relevance of these results to pyrethroid resistance management in the field is discussed.
Materials and methods
Aphid clones and rearing
Five M. persicae clones carrying different combinations of L1014F and M918T mutations were investigated (table 1). All clones had similar R3 levels of carboxylesterase resistance (Field et al., Reference Field, Anderson, Denholm, Foster, Harling, Javed, Martinez-Torres, Moores, Williamson and Devonshire1997), to ensure that comparisons of levels of pyrethroid resistance were restricted to the kdr mechanism alone. Each clone was kept in small plastic boxes (Blackman, Reference Blackman, Minks and Harrewijn1988) and reared as before (Anstead et al., Reference Anstead, Williamson, Eleftherianos and Denholm2004).
Table 1. Resistance genotypes and origins of Myzus persicae clones used in leaf-dip, synergism experiments and foliar treatment studies. All clones were used in the leaf-dip bioassays.
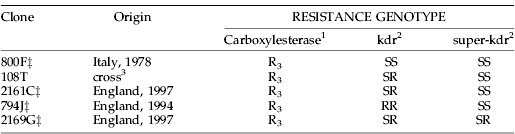
‡ Four clones used in field simulator experiments.
1 Based on an immunoassay (Devonshire et al., Reference Devonshire, Moores and ffrench-Constant1986).
2 Based on direct DNA sequencing of PCR-amplified sodium channel gene fragments from aphid genomic DNA showing variability at the L1014F and M918T sites.
3 Product of a sexual cross in the laboratory between a kdr-SS clone and an -RR clone (Blackman et al., Reference Blackman, Spence, Field, Javed, Devine and Devonshire1996).
Genomic DNA extraction, PCR amplifications and sequencing
Single adult aphids were homogenised in 250 μl of DNA extraction buffer (0.1 M Tris-HCl pH 9, 0.1 M EDTA, 1% SDS) and incubated at 70°C for 30 min. Protein was removed by adding 35 μl KOAc, incubating on ice for 30 min and centrifuging at 12,000 rpm for 15 min. The supernatant was extracted twice with an equal volume of phenol/chloroform and the aqueous phase re-extracted with chloroform. DNA was precipitated by adding half the volume (100 μl) of propan-2-ol and centrifuging at 13, 200 rpm for 5 min. DNA pellets were washed twice with cold 70% ethanol, air-dried and re-suspended in 50 μl TE buffer (10 mM tris[hydroxymethyl] aminomethane chloride (Tris – HCL, pH 8.0), 1 mM EDTA) or sterile distilled water to a final concentration of approximately 100 ng μl−1 and stored at −20°C.
Domain II sodium channel gene fragments (531 bp) were amplified from the genomic DNA templates from each M. persicae clone by two rounds of PCR using primers Aph1, Aph12 and Aph16, as described before (Anstead et al., Reference Anstead, Williamson, Eleftherianos and Denholm2004). PCR fragments were ethanol precipitated and DNA pellets reconstituted in TE buffer. DNA sample concentration was evaluated on 1.5% agarose gels, and sequencing was performed using the ABI Prism® BigDyeTM terminator cycle sequencing ready reaction kit and primers Aph5 and Aph15 as previously described (Anstead et al., Reference Anstead, Williamson, Eleftherianos and Denholm2004). Sequence data were aligned and analysed using Vector NTI (Informax Inc.).
Insecticides
Formulated pyrethroids used in leaf-dip bioassays and foliar spray assays included the Type II compounds: deltamethrin (Decis®, 25 g l−1 EC; Agrevo, UK); cypermethrin (Toppel®, 100 g l−1 EC; United Phosphorus, UK); lambda-cyhalothrin (Hallmark®, 50 g l−1 EC; Syngenta, UK); tau-fluvalinate (Maverik®, 240 g l−1 EC; Syngenta, UK), and the Type I compounds: bifenthrin (Capture®, 250 g l−1 EC; FMC, UK); permethrin (Permasect®, 230 g l−1 EC; Mitchell Cotts Chemicals, UK) and tefluthrin (Pestanal®, 240 g l−1 EC; Fluka, UK). For leaf-dipping, all formulations were diluted to the required concentration in distilled water containing 0.01% ‘Agral’ (Syngenta, UK), a non-ionic surfactant added to improve leaf-wetting and to compensate for the loss of formulant at low insecticide concentrations.
Leaf-dip bioassays
These dose-response tests were conducted at a range of insecticide concentrations increasing logarithmically from 0.01 to 1000 ppm, following a previously established method (Foster et al., Reference Foster, Denholm and Thompson2002). All bioassays were scored after 48 h exposure. Each bioassay was done twice with results pooled and subjected to probit analysis using the POLO computer programme (LeOra Software, Berkeley, California). EC50 values were calculated in ppm of the active ingredient. Statistical significance of EC50 values was based on non-overlap of 95% confidence intervals (Robertson & Preisler, Reference Robertson and Preisler1992). Resistance factors were calculated by dividing EC50 values computed for each aphid clone by the corresponding EC50 for the laboratory reference non-kdr clone (800F).
Synergism bioassays
The same protocol was followed as previously described, with the exception that aphids were topically treated with piperonyl butoxide (PBO) in acetone (0.25 μl per aphid) prior to exposure to pyrethroids. The synergist concentration chosen (100 ppm) was the maximum concentration that gave no mortality of insects when applied alone and served as a control. Groups of synergist-treated aphids were kept at room temperature for 2 h prior to placement on pyrethroid-treated leaves, after which they were held at 20°C under ambient daylight conditions. Synergism experiments were performed at six pyrethroid concentrations (0.01, 0.1, 10, 100 and 1000 ppm) with two replicates of ten aphids in each, and each bioassay was repeated twice. Mortality was assessed 48 h after treatment. The replicated results were pooled and analysed by probit analysis as previously described. To assess the degree of synergism in each clone, synergism ratios were calculated by dividing the EC50 values of the pyrethroid alone by the corresponding EC50 values for pyrethroid plus PBO.
Field simulator experiments
Two separate experiments were done on Chinese cabbage (var chinensis cv. ‘Tip-Top’) grown in field simulators, as previously described (Foster et al., Reference Foster, Denholm and Thompson2002). Each experiment used four M. persicae clones carrying different kdr and super-kdr genotypes (table 1). Lambda-cyhalothrin was applied as an aerosol either at the recommended field application rate for brassicas (50 g active ingredient l−1 or 2×, 3×, 5× and 10× field rate, or no treatment (control). All treatments were applied at the volume of 400 l ha−1. Post-treatment counts of live aphids (both adults and nymphs) on each plant were made three days later. Mean survival ratios relative to the pre-treatment number were calculated per clone per plant replicate. The resulting experimental data were analysed using generalized linear models adjusting for any potential effects of plant position and simulator.
Results
Analysis of M. persicae sodium channel domain II sequences
Sodium channel gene sequences from the kdr-RR clone (794J) and the kdr-SS clone (800F) have been previously determined by sequencing cDNA templates of para genes using mRNA of pooled aphid individuals (Martinez-Torres et al., Reference Martinez-Torres, Devonshire and Williamson1997, Reference Martinez-Torres, Foster, Field, Devonshire and Williamson1999). To facilitate the detection of sodium-channel resistance mutations in single aphids, genomic DNA rather than RNA was isolated from individual M. persicae and used as the template for PCR. Since this region of the gene contains two introns, a short 63 bp intron after threonine952 and a larger 976 bp intron after valine1015 (see fig. 1b; Genbank accession AM711603), we used aphid-specific primers upstream of the known super-kdr sites (Aph1; fig. 1b) and downstream of the kdr L1014F mutation site (Aph12 and Aph16, both within intron 2) in a two-step, nested PCR to generate single fragments of 530 bp that contained all the known domain II mutation sites.

Fig. 1. (a) Diagram of the Myzus persicae voltage-gated sodium channel α-subunit showing the four main repeating homologous domains (I–IV), the proposed membrane folding of the trans-membrane segments (S1–S6) within each domain and the locations of the two pyrethroid resistance-associated mutations, (*) kdr and (▲) super-kdr, identified in clones 794J and 2169G. (b) Nucleotide and amino acid sequences of the IIS4–S6 region of the Myzus persicae sodium channel gene, showing the positions of the L1014F (kdr) and M918T (super-kdr) mutations (as in (a) with codons boxed) and sequences of the two introns that occur within this region (lowercase). The primers used for PCR amplifications and DNA sequencing are marked by arrows. The sequence shown is that for clone 800F; the sequences of the other clones used in this study are identical except at the kdr and super-kdr codons (see text for details).
The sequences of these 530 bp gene fragments were compared for five different M. persicae clones: 800F, 108T, 2161C, 794J and 2169G (see table 1 for clone origins and genotypes). As expected, the non-kdr clone (800F), which had previously been analysed (Martinez-Torres et al., Reference Martinez-Torres, Foster, Field, Devonshire and Williamson1999), did not contain the leucine-to-phenylalanine (L1014F) mutation. The L1014F mutation was, however, present in the other four clones; 794J was homozygous for this mutation (TTC/TTC), as found before (Martinez-Torres et al., Reference Martinez-Torres, Foster, Field, Devonshire and Williamson1999), while clones 108T, 2161C and 2169G were heterozygous for the wild-type and kdr alleles (CTC/TTC).
Further sequencing through the IIS4–S6 region fragment of clone 2169G also revealed a T to C substitution in the ATG codon of amino acid 918 (numbering according to the M. domestica sequence, Genbank X96668). This additional mutation was also heterozygous in 2169G individuals and generates the methionine to threonine (M918T) mutation, termed super-kdr, originally found in housefly (Williamson et al., Reference Williamson, Martinez-Torres, Hick and Devonshire1996). This mutation was not present in any of the other clones investigated, and no other base substitutions were identified in either the exon or intron sequences of any clone.
Resistance to pyrethroids in leaf-dip bioassays
The slopes of probit lines from leaf-dip bioassays were relatively shallow (table 2). Since all aphids within each clone were genetically identical, this must be attributed to the bioassay method rather than genetic heterogeneity in response. The kdr-RR/skdr-SS clone (794J) was resistant to all pyrethroids tested, showing 23- to 55-fold resistance against the Type II pyrethroids (deltamethrin, cypermethrin, lambda-cyhalothrin and tau-fluvalinate) and 19- to 73-fold resistance against the Type I pyrethroids (bifenthrin, permethrin and tefluthrin) relative to the 800F clone. Notably, there was significant variation in response to each compound between the three clones carrying kdr-SR genotypes. EC50 values for the field clone with a heterozygous kdr and super-kdr genotype (2169G) were significantly and consistently greater (RFs up to 251 to Type II and 455 to Type I pyrethroids) than those of the two kdr-SR/skdr-SS clones that had either been generated by a sexual cross in the laboratory (108T) or collected from the field (2161C). These two clones did not differ significantly in response, though the latter showed consistently larger RFs than the former (table 2). EC50 values for the double heterozygote (2169G) were significantly higher than those shown by the kdr-RR clone (794J) for all the pyrethroids tested.
Table 2. Summary of leaf-dip bioassay results for Myzus persicae clones treated with pyrethroid insecticides.

AI, active ingredient; CI, confidence interval; RF, resistance factor. Values followed by the same letter (a–d) do not differ significantly (P<0.05).
1 Effective concentration to give 50% dead or with irreversible symptoms of poisoning.
2 Resistance factor=EC50 for clone; EC50 for 800F.
Bioassays including PBO
Piperonyl butoxide (PBO) was used to obtain in vivo evidence for possible metabolic resistance to pyrethroids in the clones investigated. Pre-treatment with PBO had no synergistic effect on the toxicity of lambda-cyhalothrin or permethrin in the kdr-SS standard clone (800F) with synergism ratios (SRs) of 1.1 and 0.8, respectively (table 3). The synergism of both pyrethroids with PBO was also very slight in the two kdr-SR/skdr-SS clones (2161C and 108T). However, in the double heterozygote clone (2169G) and the kdr-RR/skdr-SS clone (794J), SRs were higher (up to 5-fold).
Table 3. Summary of synergism bioassay results for Myzus persicae clones treated with PBO and lambda-cyhalothrin or permethrin.

AI, active ingredient; CI, confidence interval; RF, resistance factor; SR, synergist ratio. Values followed by the same letter (a–d) do not differ significantly (P<0.05).
1 Effective concentration to give 50% dead or with irreversible symptoms of poisoning.
2 Resistance factor=EC50 for clone; EC50 for 800F.
3 Synergist ratio=EC50 for clone treated with insecticide alone; EC50 for clone treated with insecticide plus PBO.
Resistance to lambda-cyhalothrin applied as foliar sprays
The predicted survival ratios (PSRs) of aphids for each clone, relative to pre-treatment numbers, following foliar applications of lambda-cyhalothrin at different concentrations (up to 10× recommended field rate) are shown in fig. 2. The kdr-SS/skdr-SS, standard clone (800F) was fully controlled by all treatments. The kdr-SR/skdr-SS (2161C) and kdr-RR/skdr-SS (794J) clones showed significant inverse slopes for PSR vs. dose rate (2161C: b=0.21±0.017, P<0.0001; 794J: b=0.07±0.017, P=0.0472). However, greater than four times the field rate was required before the PSR of the kdr-SR/skdr-SS clone fell below 1. One of the PSRs fell below 1 in either the kdr-RR/skdr-RR clone or the kdr-SR/skdr-SR clone (2169G). The non-significant slope for PSR vs. dose rate for clone 2169G (b=0.03±0.017, P=0.4318) showed that this genotype was effectively immune to all the insecticide treatments.

Fig. 2. Survival ratios of Myzus persicae clones to lambda-cyhalothrin applied as aerosol foliar sprays at the recommended field rate for brassicas and two, three, five and ten times field rate (kdr-SS/super-SS, ...○...; kdr-SR/super-SS, - - -○- - -; kdr-RR/super-SS, – –○– –; kdr-SR/super-SR, —○—).
Discussion
Point mutations in the insect para sodium channel gene that reduce the sensitivity to pyrethroids have been identified in several insect species (Soderlund & Knipple, Reference Soderlund and Knipple2003; Davies et al., Reference Davies, Field, Usherwood and Williamson2007 and references therein). It has been previously shown that the L1014F change represents the primary mutation conferring a base level of resistance to pyrethroids and DDT in certain M. persicae clones (Martinez-Torres et al., Reference Martinez-Torres, Foster, Field, Devonshire and Williamson1999). However, our leaf-dip bioassay results showed that the kdr/super-kdr double heterozygote clone (2169G), not previously tested with pyrethroid insecticides, had significantly greater resistance to both Type I and Type II pyrethroid compounds compared to a clone homozygous for L1014F but lacking M198T. Furthermore, experiments applying foliar sprays to aphids on whole plants showed that 2169G was effectively immune to lambda-cyhalothrin (and probably other pyrethroids). Thus, the present data imply that L1014F and/or M918T exhibit substantial dominance in M. persicae or that pyrethroid resistance can be enhanced further by other mechanisms or additional mutations in the sodium channel gene. Our DNA sequencing results support a hypothesis that M918T, now known to be present in M. persicae, enhances the phenotypic expression of kdr resistance, as has been demonstrated in other insect species (Williamson et al., Reference Williamson, Martinez-Torres, Hick and Devonshire1996; Guerrero et al., Reference Guerrero, Jamroz, Kammlah and Kunz1997).
Pyrethroid-resistant insects that possess super-kdr-type mutations within the sodium channel protein are known to exhibit greater resistance to the more potent Type II pyrethroids (Farnham & Khambay, Reference Farnham and Khambay1995a,Reference Farnham and Khambayb) than insects containing the kdr mutation alone. In the current study, Type II or α-cyano compounds (deltamethrin, cypermethrin and lambda-cyhalothrin) appeared to be more strongly resisted by the kdr/super-kdr heterozygote than the Type I pyrethroids (bifenthrin, permethrin and tefluthrin), a structure-activity relationship that follows the pattern previously described for super-kdr house flies (Farnham et al., Reference Farnham, Murray, Sawicki, Denholm and White1987) and diamondback moths (Schuler et al., Reference Schuler, Martinez-Torres, Thompson, Denholm, Devonshire, Duce and Williamson1998).
The observation that the kdr/super-kdr heterozygote clone 2169G was considerably more resistant to pyrethroids than clone 794J, which is homozygous for L1014F but lacks M918T, is of particular interest. These two mutations are homologous to those in super-kdr strains of house flies (Williamson et al., Reference Williamson, Martinez-Torres, Hick and Devonshire1996), and the elevated levels of resistance in doubly heterozygous aphids approximated the enhanced resistance in super-kdr vs. kdr house fly strains. A similar situation has been reported in Drosophila melanogaster Meigen (Diptera: Drosophilidae) (Pittendrigh et al., Reference Pittendrigh, Reenan, ffrench-Constant and Ganetzky1997). Unexpectedly, however, the enhancement of resistance over that conferred by L1014F in homozygous form was observed in a clone heterozygous for both mutations. In other species, kdr and super-kdr alleles have tended to be recessive or incompletely recessive in expression (Farnham et al., Reference Farnham, Murray, Sawicki, Denholm and White1987; Roush & McKenzie, Reference Roush and McKenzie1987), an observation that is inconsistent with the high levels of resistance in clone 2169G. It was inferred that the large increase in resistance to deltamethrin could arise not only in super-kdr double house fly mutants, but also when comparable mutations were expressed on different sodium channel polypeptides in heterozygotes. However, in super-kdr house fly mutants and aphid double heterozygotes, the two lesions reside in the same polypeptide. In contrast, in para heterozygotes, the lesions reside in different polypeptides (Pittendrigh et al., Reference Pittendrigh, Reenan, ffrench-Constant and Ganetzky1997). The increase in resistance in super-kdr house fly strains is presumed to reflect a combined deficit in pyrethroid binding by the mutated channels. This explanation seems inadequate to account for the phenotype of M. persicae kdr/super-kdr heterozygotes. However, this phenotype could be reconciled with the alternative view that the sodium channel mutations characterized in the current study confer resistance to pyrethroids by a mechanism other than direct alteration of the binding site, with resistance being mediated via functional alterations of the encoded sodium channel polypeptide. Direct electrophysiological studies of the properties of susceptible and resistant sodium channels would help to resolve these various possibilities.
An additional explanation for the enhanced pyrethroid resistance in 2169G aphids could be the presence of other, undisclosed mutations in the sodium channel protein and/or other independent resistance mechanisms. For example, mutations have been found outside of domain II that seem to have a direct role in resistance, such as the valine-to-methionine (V421M) mutation in IS6 of Heliothis virescens (Fabricius) (Lepidoptera: Noctuidae) and the phenylalanine-to-isoleucine (F1538I) mutation in IIIS6 of the cattle tick, Boophilus microplus (Zhao et al., Reference Zhao, Park and Adams2000; Tan et al., Reference Tan, Liu, Wang, Huang, Chen, Gurevitz and Dong2005). It has been demonstrated that mutations in positions outside of the main transmembrane domains can also act as ‘modifiers’ of resistance and are capable of enhancing the effects of some of the primary mutations. For example, two cockroach mutations, glutamic acid-to-lysine (E434K) and cysteine-to-arginine (C764R), found in the first intracellular linker connecting domains I and II (Liu et al., Reference Liu, Valles and Dong2000), do not alter sodium channel sensitivity to pyrethroids themselves but, instead, are able to dramatically enhance the insensitivity conferred by the primary kdr mutation (L993F, cockroach equivalent to L1014F) when all three mutations were expressed together in recombinant channels in Xenopus oocytes (Tan et al., Reference Tan, Liu, Tsai, Valles, Goldin and Dong2002). Likewise, the E434K and C764R mutations were also shown to enhance the effect of the H. virescens V421M mutation when co-expressed in oocytes (Liu et al., Reference Liu, Tan, Valles and Dong2002). Similar kdr-modifier mutations may also be present in M. persicae. Given the limited number of M. persicae field populations that have been analysed so far, it seems unlikely that the full range of primary and secondary mutations capable of conferring pyrethroid target-site resistance have been identified. Because the current sequencing analysis of the sodium channel gene has been limited to the domain IIS4–IIS6 region of the channel where the most common mutations are located, a more extensive sequencing survey in pyrethroid-resistant field-collected clones may reveal alternative mutations in other regions of the M. persicae gene.
Over-expression of metabolic detoxification enzymes may also be involved in resistance to pyrethroids. Pre-treatment with PBO to suppress certain detoxification enzymes did not substantially reduce RFs for lambda-cyhalothrin or permethrin in the 2169G clone. However, it has been suggested that PBO does not suppress all forms of metabolic detoxification (Scott, Reference Scott, Roush and Tabashnik1990). Nonetheless, it is now clear that M918T is present in M. persicae sampled at a range of localities (Anstead et al., Reference Anstead, Williamson and Denholm2005; Fenton et al., Reference Fenton, Malloch, Woodford, Foster, Anstead, Denholm, King and Pickup2005), though always in conjunction with L1014F, suggesting that the mutations act in a complementary manner to enhance the pyrethroid resistance phenotype. A PCR-based assay for discriminating between alleles at both mutation loci has also been developed to assist with the genotypic characterisation of aphids and to monitor the incidence and dynamics of target-site resistance to pyrethroids in M. persicae (Anstead et al., Reference Anstead, Williamson, Eleftherianos and Denholm2004). It would also be valuable to determine whether M918T confers substantial pyrethroid resistance on its own, by identifying field populations that contain this mutation only, generating such clones in laboratory crossing experiments or through site-directed mutagenesis, coupled with functional or binding assays and in vitro expression of the modified gene.
Acknowledgements
We thank Diana Cox and Barbara Hackett for assistance with aphid rearing and biochemical characterization of the aphid clones and Alan Devonshire for useful discussion and advice. Ioannis Eleftherianos was supported by a grant from IKY (Greek State Scholarships Foundation). Steve Foster was supported by a SAPPIO-Link project (Sustainable Arable Protection through Precision Input Optimisation). Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the United Kingdom.







