Introduction
The tea green leafhopper, Empoasca vitis Göthe (Hemiptera: Cicadellidae), is a major pest of the tea plant which is sensitive to light or colors and performs long-distance host orientation using its visual sense, but once within a certain distance of the tea plant is able to detect tea-shoot volatiles using its olfactory sense (Zhao et al., Reference Zhao, Gao, Chen, Cheng and Xu2002; Chen, Reference Chen and Zhou2013).
Currently, studies of the host-finding behavior of E. vitis have mainly focused on observation of how this pest responds to certain colors and host volatiles; however, there has been little research at a morphological or molecular level to understand this tropism (Zhao et al., Reference Zhao, Chen and Cheng2001; Lin et al., Reference Lin, Han, Zhou and Chen2009; Mu et al., Reference Mu, Cui, Ge, Wang, Liu, Yu, Zhang and Han2012; Wang et al., Reference Wang, Li, Wu, Sun, Shi, Ding, Cao and Han2016). The morphological structure of a species generally corresponds to the development of certain functions, such as the development of a compound eye related to precise vision. Similarly, insects with an acute sense of smell have a rich variety of olfactory sensilla (Pitts & Zwiebel, Reference Pitts and Zwiebel2006; Liu et al., Reference Liu, Li and Wang2008; Zhao et al., Reference Zhao, Zheng and Hu2012). Further, both visual and olfactory structures must be able to translate environmental information such as light, colors and odors into neural signals which can control behaviors. In neural signaling pathways, opsins mediate light signal transduction, while olfactory receptors mediate chemical signal transduction (Larsson et al., Reference Larsson, Domingos, Jones, Chiappe, Amrein and Vosshall2004; Terakita, Reference Terakita2005).
In this work, the structure of the compound eye and the sensilla of the antenna and the mouthparts from E. vitis were observed. Three opsin genes (EV_opsin) were described and their expression patterns were investigated. To study the functions of EV_opsin, RNA interference (RNAi) was used to downregulate the expression of EV_opsin in order to assess whether this resulted in the disruption of the host-tropism behavior of adults.
Materials and methods
Scanning electron micrography (SEM) and paraffin section of compound eye
Specimens of E. vitis were collected at the Fujian Agriculture and Forestry University, Fujian, China, and raised in a laboratory. SEM was carried out as described previously by Pitts et al. (Reference Pitts, Fox and Zwiebel2004). Observation of the compound eye by paraffin section and hematoxylin–eosin staining was performed in accordance with the method described in Yan et al. (Reference Yan, Qiao, Zhang, Ma and Cui2011).
RNA isolation and conversion to cDNA
Total RNA from different tissues were extracted using an HP Total RNA Kit (Omega, Savannah, GA, USA) according to the manufacturer's instructions, after grinding with liquid nitrogen. First-strand cDNA was synthesized from 1 µg of total RNA from compound eye in a 20 µl reaction volume using a First Strand cDNA Synthesis Kit (TaKaRa, China).
Cloning and structural analysis of E. vitis opsin genes (EV_opsin)
To clone the long-wavelength (LW) opsin gene, a full length of open reading frame (ORF) was obtained using degenerate primers (LWop-F1, LWop-R1). To clone the ultraviolet-sensitive opsin gene, a short fragment, 3′-ORF region and 5′-ORF region, was amplified using degenerate primers as follows: UVop-F1, UVop-R1; 3′ UVop-F, 3′ UVop-R; 5′ UVop-F, 5′ UVop-R. The PCR reaction volume and conditions were as described previously by Zhang et al. (Reference Zhang, Long, Pengsakul, Wang, Pan, Yu, Fang and Luo2015). To clone the blue-sensitive opsin gene (EV_Bop), a middle fragment was obtained using degenerate primers: Bop-F1 and Bop-R1. The 3′-cDNA region was amplified using nested primers: 3′RACE-outer, 3′RACE-inner; 3′Bop-outer, 3′Bop-inner. Outer PCR assays were performed in a final reaction volume of 20 µl consisting of 0.5 µl of the product of reverse transcription with 3′RACE adaptor primer, 4 µl of 5 × Buffer Fast Pfu (TransGen, China), 0.4 µl Fast Pfu (TransGen, Beijing, China), 2 µl dNTP, 0.2 µl of each out primer (10 µM) and 12.7 µl of double distilled water (ddH2O). The PCR conditions were: 95 °C for 5 min; 35 cycles of 95 °C for 20 s, 58 °C for 30 s and 72 °C for 2 min; and 72 °C for 5 min. Inner PCR assays were performed in the same manner as the outer PCR assays, but the cDNA was replaced with 0.2 µl of the outer PCR products. Amplifications were performed on the same conditions as for the outer PCR. The 5′-cDNA region was amplified using a 5′/3′ RACE Kit, Second Generation (Roche, Switzerland) using the reverse gene-specific primers SP1, SP2 and SP3. The ORF of EV_LWop, EV_UVop and EV_Bop was verified by gene-specific primers, respectively: LWop-F, LWop-R; UVop-F, UVop-R; Bop-F, Bop-R. All the primers used are shown in table 1.
Table 1. Primers used for PCR, RACE, qRT-PCR and dsRNA synthesis.

All the PCR products were checked on 1.0–2.0% (w/v) agarose gels and visualized using ethidium bromide and UV illumination to confirm the product size. The PCR products were purified using a Gel Extraction Kit (Omega, USA). The purified DNA was sub-cloned into a pMD19-T vector (TaKaRa, China), and then transformed into competent Escherichia coli DH5α. The products were bidirectionally sequenced (BGI, Guangzhou, China) and blasted on the GenBank Database. The TMHMM Server v. 2.0 was used to predict the transmembrane domain. The functional sites of the opsin amino acid sequences were analyzed with PROSITE Scan. The neighbor-joining tree was constructed using MEGA 6.0 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013).
Tissue expression pattern of EV_opsin
First-strand cDNA was synthesized from 1 µg total RNA of different tissues in a 20 µl reaction volume using PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, China). Quantitative real-time PCR (qRT-PCR) assays were performed on the Bio-Rad CFX96 to detect the tissue expression pattern of EV_opsin in a final reaction volume of 10 µl consisting of 1 µl cDNA diluted at 1:10, 2 × SYBR Premix EX Taq II (TaKaRa, China), 5 µl, the three specific primers (10 µM as per table 1) consisting of 0.3 µl EV_LWop, 0.3 µl EV_UVop, 0.35 µl (EV_Bop) and 0.45 µl (β-actin) with ddH2O added to make up the volume to 10 µl. Reactions without the cDNA template served as a negative control and the reactions were run in triplicate for each sample. The qRT-PCR program was conducted using a two-step method as follows: 95 °C for 30 s at 40 cycles: 95 °C for 5 s, and 60 °C for 30 s, with fluorescence acquisition at 80 °C for 10 s. The expression of the EV_opsin gene in specific tissue was presented as 2−ΔΔCt (Schmittgen et al., Reference Schmittgen, Zakrajsek, Mills, Gorn, Singer and Reed2000; Livak & Schmittgen, Reference Livak and Schmittgen2001; Schmittgen, Reference Schmittgen2001).
Antibody production and immunolocalization
The peptides (EAQSTSDGASVGSG from EV_LWop, NEQEPKHDNVSTTT from EV_UVop and ATEKTQTSDNAPSA from EV_Bop) were synthesized, coupled to keyhole limpet hemocyanin, and used to generate sequence-specific, peptide-affinity purified polyclonal antibodies in mice (Kunming strain) as described by Yu et al. (Reference Yu, Wang, Zhang, Fang and Luo2014). The immunized mice were maintained in the Laboratory Animal Center, Xiamen University, and the studies were conducted adhering to the guidelines for animal husbandry and also approved by the Ethics Committee of Xiamen University according to the Regulations for the Administration of Affairs Concerning Experimental Animals (approved by the State Council of the People's Republic of China; Permit Number: XUMLAC2012-0122). The procedures for immunolocation were carried out as described in Zhang et al. (Reference Zhang, Long, Pengsakul, Wang, Pan, Yu, Fang and Luo2015), except that skim milk was replaced by bovine serum albumin.
RNAi of EV_opsin
Primers (table 1) with a T7 promoter sequence were designed according to the ORF of EV_opsin and enhanced green fluorescent protein (EGFP) plasmid for the templates of double-strand (ds) RNA synthesis. Then the dsRNA of EV_LWop (559 bp), EV_UVop (500 bp), EV_Bop (574 bp) and EGFP (574) (as a control) were synthesized using the T7 RiboMAX™ Express RNAi System (Promega, USA). The dsRNA obtained was suspended in RNase-free water and checked by electrophoresis on a 1.0% agarose gel.
Two control groups (ddH2O and dsEGFP) and three treatment groups (dsEV_LWop, dsEV_UVop and dsEV_Bop) were adopted for EV_opsin RNAi. The dsRNA was adjusted to a final concentration of 300 ng µl−1 with an artificial diet (Sasaki et al., Reference Sasaki, Hayashi and Ishikawa1991) which was continuously fed to groups of 30 E. vitis, and the test repeated three times, with the diet being changed every 2 days. The survival rate was recorded on the second, fourth and sixth day and the adult females were taken out for transcription level testing according to the qRT-PCR method described above, every 2 days at 14:00 p.m. in the treatment groups and on the sixth day in the control groups.
Determination of tropism behavior and its response to host color after EV_opsin RNAi
In order to determinate the tropism behavior of E. vitis toward its host (tender tea leaves) color and odors, square-chamber and cylindrical-chamber tests were conducted using facilities in which internal chambers were able to be created by dividing the sidewalls and lids with glass panels or screens. The tests were conducted in a white room under a 65 lumens light. The testing methods were in accordance with those described by Zhao et al. (Reference Zhao, Gao, Chen, Cheng and Xu2002). To verify whether the E. vitis were attracted by the host's color, white artificial tea leaves were used as a control and were placed on one side of a glass panel on one side of a square chamber (fig. 1a, chamber 3), while green tea leaves were placed on the other side (chamber 2). The insects were put into the center (chamber 1) and the numbers of insects in chambers 2 and 3 containing the green and white leaves were recorded every 15 min after adaption for 10 min until all the insects had left chamber 1 (fig. 1a).

Fig. 1. Behavioral measuring device. (a) Square-chamber testing facility; (1) releasing room, (2) and (3) recording room. (b) Cylindrical-chamber testing facility; (1) releasing room, (2) and (4) selecting room, (3) and (5) recording room. The green tea leaves were collected in the field, while the white is an artificial host as the same size as the natural host. ‘Wind’ represents the condition of ventilation; the box around the green tea leaves is transparent plastic wrap which was able to prevent the influence of host volatiles.
To detect whether host odors attracted E. vitis, tea leaves covered with plastic wrap instead of the artificial host were placed in one side chamber of the screen covered square chamber and ventilation allowed (fig. 1a). The experiment was run over a period of 60 min. To detect whether distance affected the ability of E. vitis to find its host using color and odor, a cylindrical-chamber test was carried out as described above but with two intervening chambers added to increase the distance between the insects and the host/control (fig. 1b) with the number of insects being recorded in the outermost chambers (chambers 3 and 5). The tests were conducted after the E. vitis specimens had been ingesting the artificial diet with EV_opsin dsRNA for 6 days. The treatment and control groups all consisted of 30 insects and the test was repeated three times.
Statistical analysis
The tropism rate (TR) was calculated based on the number of insects attracted out of the total number of insects, expressed as a percentage and was used to evaluate the preference of E. vitis for host odors and color. The declined TR was defined as (TRT−50%)/(TRC−50%), where TRT and TRC represent the TR in the treatment and control groups, respectively; a figure of 50% is assumed to indicate that the E. vitis specimens have no preference for their host's color or odor. All values were expressed as means ± standard deviation (SD). Significant differences between means from the data were tested using one-way analysis of variance followed by Duncan's multiple comparison test using SPSS 13.0 (SPSS, Inc., Chicago, Illinois, USA). A P value <0.05 was considered to be significant.
Results
Structure of compound eye, sensilla of antenna and mouthparts
SEM observation showed that the E. vitis specimens had a pair of large compound eyes which occupied two-thirds of their heads and were each composed of 500–600 hexagonal ommatidia (fig. 2a–d). The rhabdom was located immediately under the crystalline cone showing that the structure of the compound eye in E. vitis constituted a photopic eye (fig. 3a, b). There were only three sensilla trichodea (ST) on one antenna (fig. 2e, f), while there were many ST and sensilla basiconica on the mouthparts (fig. 2g–i). In addition, two important olfactory organs, the maxillary palpus and the labial palpus were absent in E. vitis.
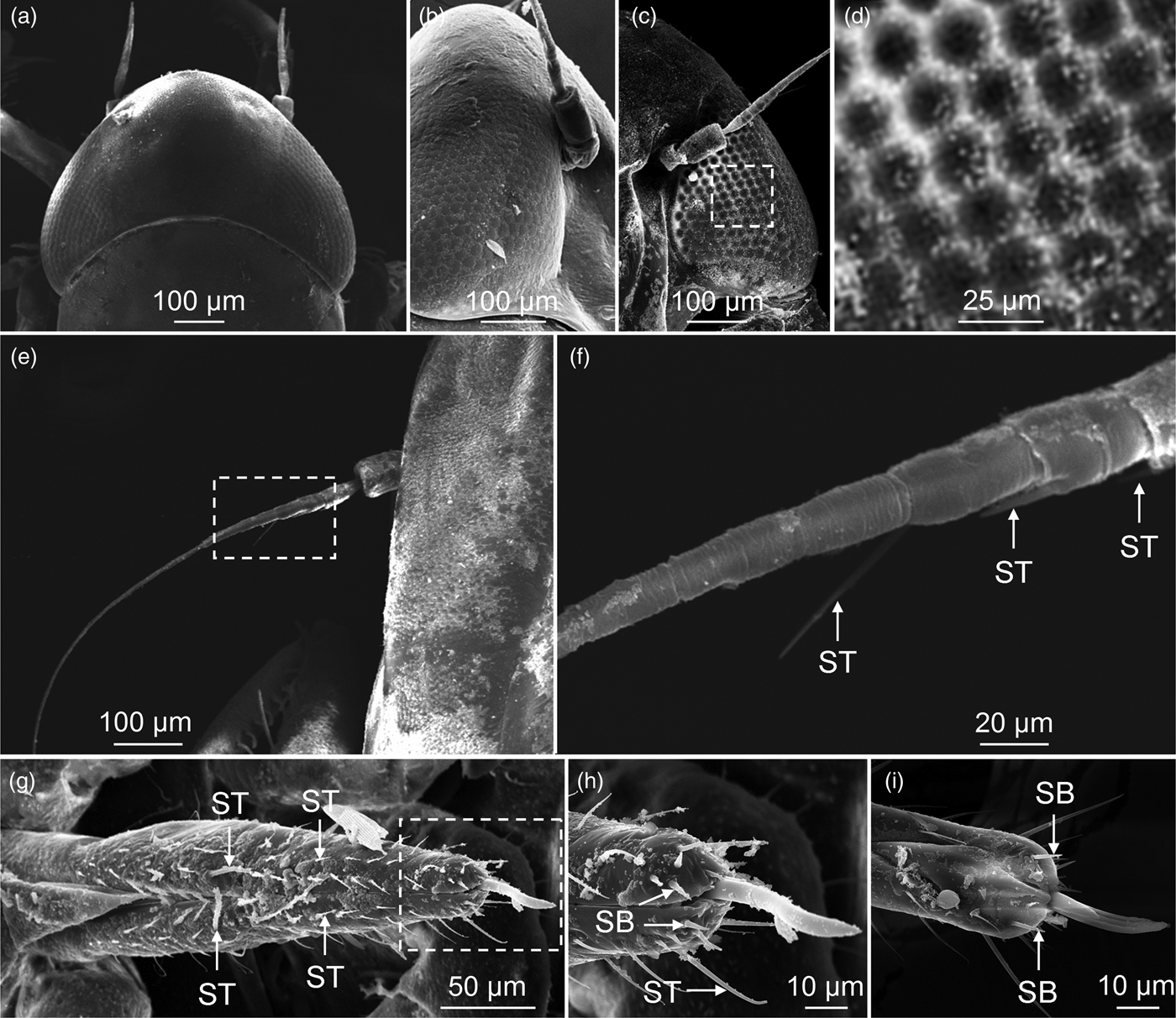
Fig. 2. SEM of sensory organs of E mpoasca vitis. (a) Dorsal view of compound eye. (b) Side view of compound eye. (c) Ventral view of compound eye. (d) Enlarged figure of dotted box in (c) shows hexagonal ommatidia. (e) Overview of antenna. (f) Enlarged figure of dotted box in (e) shows three sensilla trichodea (ST). (g) Dorsal view of mouthparts and ST distribution on surface of the labium. (h) Enlarged figure of dotted box in (g). Apical of the mouthparts shows ST and sensilla basiconica (SB), and SB in the dorsal side (i). ST and SB are considered important chemical sensilla of the insect.

Fig. 3. Structure of compound eye of E mpoasca vitis. (a) and (b) Vertical and transverse section show internal structure of compound eye. (Co) Cornea, (CC) crystalline cone, (RC) retinal cell, (Rh) rhabdom.
Molecular cloning and sequence analysis of three E. vitis opsin genes (EV_opsin)
A full length of 1146 bp ORF of E. vitis LW opsin (EV_LWop; GenBank accession number KF938691) was obtained (fig. 4a, lane 1), as well as 1155 bp ORF of ultraviolet-sensitive opsin (EV_UVop; GenBank accession number KJ123697; fig. 4a, lane 2) and 1547 bp cDNA including 324 bp 3′-untranslated region (UTR) and 110 bp 5′-UTR of blue-sensitive opsin (EV_Bop; GenBank accession number KJ813817; fig. 4a, lane 3). Their putative product proteins consist of 381, 384 and 379 amino acid residues, respectively.

Fig. 4. Empoasca vitis opsin gene (EV_opsin) cloning and the predicted transmembrane structure. (a) Amplification products of ORF from E. vitis opsin gene. (M) DNA ladder mixture marker, lane (1), (2) and (3) the PCR product of EV_LWop, EV_UVop and EV_Bop ORF, respectively. (b), (c) and (d) Predicted seven transmembrane structure of EV_LWop, EV_UVop and EV_Bop, respectively.
The deduced proteins contained seven transmembrane domains (fig. 4b–d), a retinal binding site and G-protein binding sites that were typical of opsin genes. The results of phylogenetic analysis of the amino acid sequence of opsins from other known insect species revealed that EV_opsin cluster in the main visual opsin clades of insects as would be expected: EV_LWop in the LW, EV_UVop in the short-wavelength and EV_Bop in the middle-wavelength (fig. 5). They showed a close relationship with Nephotettix cincticeps Uhler and Sogatella furcifera Horvath (69–88% identities) rather than other insect orthologues.

Fig. 5. Neighbor-joining tree of EV_opsin orthologs from different insects. Six orders of insect including Hemiptera, Lepidoptera, Hymenoptera, Coleoptera, Diptera and Orthoptera were used here. Species and accession number are shown in the figure. Numerical values indicate bootstrap support for each node. Bootstrap support values were based on 1000 replicates. NJ tree shows EV_opsin cluster in the main visual opsin clades of insects as expected: EV_LWop in the long-wavelength (LW) opsin, EV_UVop in the short-wavelength (SW) opsin and EV_Bop in the middle-wavelength (MW) opsin.
Tissue distribution and immunolocalization of EV_opsin
qRT-PCR analysis indicated that EV_opsin were abundantly expressed in the compound eye rather than in other tissues (fig. 6a–c). The expression of EV_opsin in adult E. vitis was detected by immunofluorescence antibody staining using anti-EV_opsin antibody. EV_LWop was mainly distributed in the proximal end of the rhabdom at all areas and in the middle of the rhabdom in the central area of the compound eye (fig. 7a, b); similarly, EV_UVop showed the same pattern as EV_LWop and was also expressed in the distal end of some rhabdom (fig. 7c). However, EV_Bop was concentrated on the proximal end and middle of the rhabdom in the central area of the compound eye (fig. 7d). Based on the expression features of EV_opsin, it is speculated that two or three opsins are co-expressed in each photoreceptor cell.
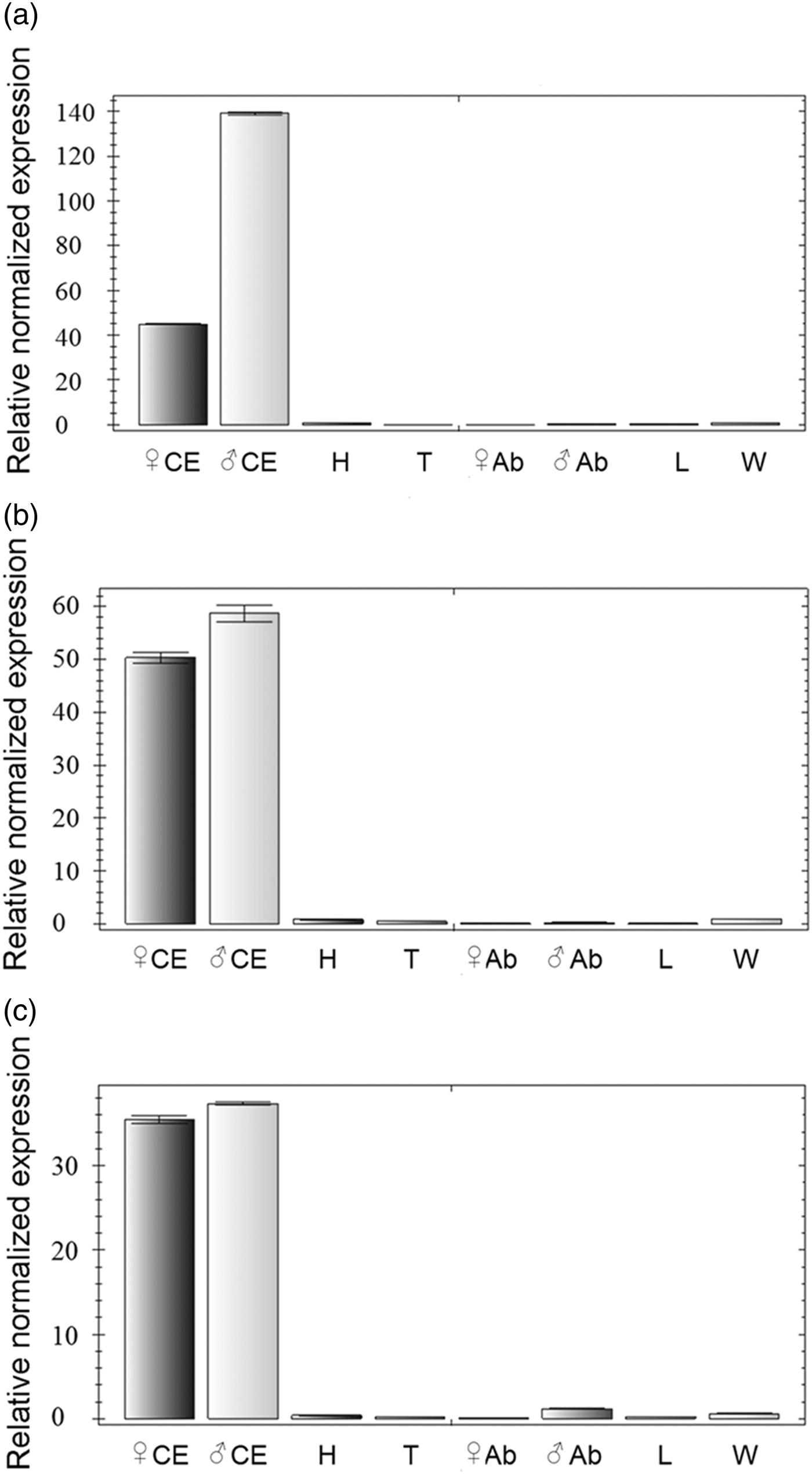
Fig. 6. Tissue specificity of EV_opsin. Tissue distributions of EV_LWop (a), EV_UVop (b) and EV_Bop (c) using qRT-PCR. The abbreviations in the figures are compound eyes (CE), head without CE (H), thorax (T), abdomen (Ab), leg (L) and wing (W).

Fig. 7. Immunolocalization of EV_opsin in compound eye of E mpoasca vitis. (a) and (b) Immunolocalization of EV_LWop. (c) and (d) Immunolocalization of EV_UVop and EV_Bop, respectively. Red fluorescent was anti-EV_opsin marked with Cy3-labled secondary antibody, blue fluorescence was nuclear marked with Hoechst. (PRh), (MRh) and (DRh) Proximal end, middle part and distal end of rhabdom, respectively.
Evaluation of the RNAi effect against EV_opsin
The identification of EV_opsin dsRNA and EGFP dsRNA was performed by 1% agarose gel electrophoresis and produced high-quality results with the dsRNA being of the size expected (fig. 8). The control and treatment groups showed high mortality (20.0–44.4%) on the second day and the survival rate of dsEV_LWop, dsEV_UVop and dsEV_Bop groups were 58.9, 46.7 and 61.1%, respectively, after continuous feeding of EV_opsin dsRNA to E. vitis for 6 days (fig. 9). To demonstrate whether or not the opsin genes were downregulated, qRT-PCR was performed on mRNA purified from both the control and treatment groups. It was found that the transcription level of EV_opsin mRNA was significantly reduced with progressively longer periods of feeding. On the fourth and sixth days, the dsEV_LWop and dsEV_Bop groups were reduced by 56.2, 77.3, 65.3 and 70.0%, respectively (P = 0.016, 0.004, 0.007, 0.007) (fig. 10a, c). The dsEV_UVop group was reduced by 40.0% on the sixth day compared with control groups (P = 0.031) (fig. 10b).

Fig. 8. Identification of EV_opsin dsRNA products by agarose gel electrophoresis. (M) and (M1) different DNA ladder mixture markers. Lane (1), (2) and (3) (4) the synthetic dsRNA of EV_LWop (559 bp), EV_UVop (500 bp), EV_Bop (574 bp) and EGPF (574 bp) show similar sizes as expected.

Fig. 9. Survival rate of Empoasca vitis after feeding with dsRNA. *P < 0.05.

Fig. 10. Transcription level detection of EV_opsin after RNA interference. (a), (b) and (c) Variations of EV_opsin transcription levels after RNAi. The control groups (1) and (2) are insects continuously fed the artificial diet containing ddH2O and dsEGFP, respectively, for 6 days; (3), (4) and (5) are insects which ingested the EV_opsin dsRNA on the second, fourth and sixth days. *P < 0.05.
Tropism behavior and its response after EV_opsin RNAi
The tropism behavioral determination demonstrated that E. vitis was strongly attracted by tea leaves compared with the artificial models over the course of the 1 h experiment (69.23%) in the square-chamber testing facility without any influence by the host odors (fig. 11a), suggesting that this tropism response was caused by the host color. The TR of E. vitis to tea leaves (60.9%) was higher than that which resulted when the leaves were covered with plastic wrap (39.1%) under the ventilation condition (fig. 11b) (P = 0.002), which suggests that the host odors were able to attract the pest over a short distance (~20 cm). However, cylindrical-chamber test showed that the TR showed no statistical difference between the natural condition and the covered host plant under ventilation, although the insects gathered on the host-plant side (60.9%) more than on the artificial host side without any influence from host odors (fig. 11b) (P = 0.002). These results indicate that host color is attractive to the pest over a long distance (~50 cm) but the odor is not. After E. vitis continuously ingested the EV_opsin dsRNA for 6 days, the TR to host color was effectively impaired by 67.6 and 29.5% in the dsEV_LWop and dsEV_Bop treatment groups (P = 0.001 and 0.049), but there were no statistically significant changes in the dsEV_UVop group (P = 0.883) (fig. 11c).

Fig. 11. Determination of tropism behavior. (a) Validation of whether the host color attracts Empoasca vitis and the estimated reaction time (testing in square chamber). (b) How odors, distance and color affect host-finding behavior in E. vitis (testing in square chamber and cylindrical chamber, respectively). (c) Behavioral response to the host color after continuous feeding of E. vitis with dsRNA over 6 days. *P < 0.05.
Discussion
The size of the compound eyes and the number and structure of ommatidium are the results of long adaption to the ecological environment. For instance, the photopic eye in the cricket (Gryllus bimaculatus De Geer) is suitable for diurnal activities while the scotopic eye in the lacewing (Chrysopa pallens Rambur) is appropriate for activity in the daytime and in the evening. However, the degraded compound eyes found in burrowing insect are a result of long-term living in darkness (Wang, Reference Wang and Li2004; Zhang et al., Reference Zhang, Zhu, Fan and Wei2007; Henze et al., Reference Henze, Dannenhauer, Kohler, Labhart and Gesemann2012). The photopic eye was observed in E. vitis by paraffin serial sections suggesting that this pest is a diurnal insect. Some previous studies have confirmed that different agricultural pests have diverse taxis spectra when exposed to light. For instance, blue was more attractive to thrips (Frankliniella occidentalis Pergande) (Wu et al., Reference Wu, Xu, Zhang, Zhang and Zhu2007), yellow was the preferred color for Bemisia tabaci Gennadius against other colors (Blackmer & Byrne, Reference Blackmer and Byrne1993), while E. vitis preferred amber yellow, lake water blue and gem green rather than snow white and reddish orange (Zhao et al., Reference Zhao, Chen and Cheng2001). In the present study, E. vitis demonstrated a significant preference for tea leaves compared with artificial white host models, suggesting that the color green is an important factor in host orientation.
In this study, three visual related genes (EV_LWop, EV_UVop and EV_Bop) were isolated and found to play an important role in the control of the E. vitis tropism to colors. From the results of transmembrane prediction, functional site testing, phylogenetic analysis, and the high expression of proteins in the rhabdom of the compound eye, it was deduced that the encoded proteins are insect opsins and have a vital function in the detection of green, UV and blue.
RNAi by direct injection is the usual method applied in functional research on opsins in larger insects (Leboulle et al., Reference Leboulle, Niggebrügge, Roessler, Briscoe, Menzel and Ibarra2013; Tamaki et al., Reference Tamaki, Takemoto, Uryu, Kamae and Tomioka2013). In this study, EV_opsin dsRNA was mixed into an artificial diet and then continuously fed to the E. vitis specimens. The results showed that the transcripts of EV_opsin were successfully downregulated. Thus, we consider that this method is suitable for opsin functional analysis in small piercing-sucking insects. After the ingestion of EV_LWop dsRNA for 6 days, the tropism to host color was significantly impaired, while the tropism in the dsEV_UVop group remained unchanged. All of these results suggest that LW opsin dominated the use of color vision in host-finding activity and that EV_LWop may mediate the green light signal transduction pathway. The range of the spectral absorption peak of LW opsin in other related diurnal insects such as winged pea aphids (Acyrthosiphon pisum Harris), crickets (G. bimaculatus) and honeybees (Apis mellifera carnica Pollmann) are 510–540 nm (Döring et al., Reference Döring, Kirchner, Skorupski and Hardie2011; Henze et al., Reference Henze, Dannenhauer, Kohler, Labhart and Gesemann2012; Leboulle et al., Reference Leboulle, Niggebrügge, Roessler, Briscoe, Menzel and Ibarra2013). Moreover, the tea leaves used in this behavioral assessment are close to pure green (515–535 nm), further supporting the hypothesis that EV_LWop has a decisive role in host-color communication.
In regard to olfaction, some previous studies have demonstrate that olfactory organs which have developed diverse sensilla in insects such as mosquitoes are able to effectively detect host odors over long distances (Pitts & Zwiebel, Reference Pitts and Zwiebel2006; Syed & Leal, Reference Syed and Leal2007; Zhang et al., Reference Zhang, Long, Pengsakul, Wang, Pan, Yu, Fang and Luo2015). However, low concentrations of odor could not be perceived by E. vitis on account of the small number of olfactory sensilla on their antennae and the absence of a maxillary palpus and a labial palpus. In common with other leafhoppers (Zhao et al., Reference Zhao, Dai, Zhang and Zhang2010), abundant gustatory sensilla were observed in E. vitis mouthparts showing that this pest may have efficient gustation. Taken together, the morphological, molecular and behavioral studies lead to a conclusion that the developed compound eye in E. vitis plays a leading role in host location, and the LW opsin significantly affects the tropism to host color; the lack of olfactory sensilla results in long-distance odors not being able to be detected until the insect is close to the host plant, when it is able to finally judge the quality and fitness of the plant food by gustation in order to decide whether to stay or leave.
Acknowledgements
This project was supported by the National Sharing Service Platform for Parasite Resources (TDRC-22).















