Published online by Cambridge University Press: 09 October 2003
The northern biotype of Echinococcus granulosus occurs in North America and northern Eurasia in life-cycles involving cervids. Previously, cervid isolates of E. granulosus from North America have been characterized using molecular genetic techniques as the G8 genotype. In this study, 5 isolates of E. granulosus were collected from 4 reindeer and 1 moose in north-eastern Finland. DNA sequences within regions of mitochondrial cytochrome c oxidase I (COI) and NADH dehydrogenase I (NDI) genes and the internal transcribed spacer 1 (ITS-1) fragment of the ribosomal DNA were analysed. The mitochondrial nucleotide sequences were identical in all isolates, but high sequence variation was found in the ITS-1 region. Mitochondrial and nuclear sequences of the Finnish cervid E. granulosus and the camel strain (G6) of E. granulosus resembled closely each other. According to phylogenetic analyses, the Finnish isolates have close relationships also with the pig (G7) and cattle (G5) strains. Although some similarities were found with the previously published North American cervid strain (G8), particularly in the NDI sequence and some of the ITS-1 clones, the Finnish E. granulosus form represents a distinct, previously undescribed genotype of E. granulosus. The novel genotype is hereby named as the Fennoscandian cervid strain (G10).
The genus Echinococcus contains 4 generally accepted species: E. granulosus, E. multilocularis, E. vogeli and E. oligarthrus. Particularly one of these species, E. granulosus, is made up of a number of biologically, epidemiologically and genetically distinct entities, whose taxonomical positions are unclear. At present they are referred to as strains (Thompson & McManus, 2001). One of these forms is the cervid strain, or the northern biotype, which is supposed to have a wide geographical distribution, reaching from North America over northern Eurasia as far as Fennoscandia (Rausch, 1995). The life-cycle of the northern form of E. granulosus is primarily maintained by the predator–prey relationship between the wolf (Canis lupus) and ungulates of the family Cervidae. Fourty years ago Sweatman & Williams (1963) proposed, on the basis of morphological evidence, that in fact 2 distinct subspecies of E. granulosus could occur in cervids, namely E. granulosus canadensis in reindeer of Fennoscandian origin and E. granulosus borealis in the North American moose (Alces alces). However, their results became later forgotten, and the northern form is today regarded as a single cervid strain of E. granulosus.
New molecular genetic techniques have been applied successfully for distinguishing the different species of Echinococcus and strains especially within the species E. granulosus (Thompson & McManus, 2001). Genetic variation in Echinococcus has been investigated in either the mitochondrial or the nuclear genome. The generally used genetic markers for the identification of Echinococcus species and strains are the internal transcribed spacer 1 (ITS-1) region of the ribosomal DNA (rDNA) and 2 mitochondrial genome marker genes, the cytochrome c oxidase I (COI) and the NADH dehydrogenase I (NDI). To date, 9 different genotypes (G1–G9) of E. granulosus have been identified. The cervid strain (the genotype G8) was characterized by Bowles, Blair & McManus (1994) using analysis of COI and NDI subsequences and a polymerase chain reaction linked restriction fragment length polymorphism (PCR-RFLP) analysis of the ITS-1 region. Later, also the ITS-1 regions of many Echinococcus species and strains, including the cervid strain, were sequenced to infer phylogenetic relationships within this genus (Bowles, Blair & McManus, 1995; Kedra et al. 1999; van Herwerden, Gasser & Blair, 2000).
The genetically analysed cervid strain material has remained quite limited: ND1 and COI sequences were at first analysed from 4 hydatid cysts (isolates) and all 4 sequenced ITS-1 clones were from 1 of these cysts (Bowles et al. 1994, 1995). All these isolates were collected from a single infected moose in Minnesota, USA. In a very recent study, more mitochondrial genes were analysed from these 4 isolates and 2 new cysts from Alaska, one of which was from a moose and the other from a human patient (McManus et al. 2002). As far as is known, E. granulosus isolates originating from cervids in Eurasia have not earlier been investigated by molecular genetic techniques.
In Finland, E. granulosus occurs in the north-eastern part of the country as a rare parasite of reindeer and wild animals (Maijala et al. 2002; Hirvelä-Koski et al. 2003). Our aim in this study was to genotype the isolates of E. granulosus collected from Finland by determining the gene marker sequences mentioned above and to resolve the phylogenetic position of the Finnish E. granulosus type among the genus Echinococcus. The genetic diversity of E. granulosus reflects differences in infectivity for distinct hosts. Thereby, the strain identification of the Finnish E. granulosus isolates is of importance, when the risk posed to the human population by this parasite is evaluated.
Five isolates of E. granulosus from north-eastern Finland were examined. For the purposes of this study, an E. granulosus isolate refers to the protoscoleces obtained from a single hydatid cyst. Four cysts were collected from 4 reindeer (Rangifer tarandus tarandus) during routine meat inspection in slaughterhouses. The infected reindeer were from 3 separate grazing areas (Sodankylä, Salla and Kuusamo). The fifth cyst was found from a moose, which was shot by hunters in Kuusamo. The cysts were stored frozen at −20 °C. Protoscoleces were aspirated from the melted cysts and rinsed several times with saline. Then 25–30 mg portions of protoscoleces from each isolate were used for DNA extraction. DNA was prepared from individual isolates using the DNeasy™ Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions.
From each isolate, the mitochondrial NDI and COI gene fragments and the ITS-1 region of the nuclear rDNA were amplified by the polymerase chain reaction (PCR) using previously published oligonucleotide primers (Bowles & McManus, 1993 a, b; Bowles, Blair & McManus, 1992). The PCR was performed in a final 100 μl volume containing a template (5 μl), 125 μM of each dNTP (Finnzymes, Espoo, Finland), 1 mM MgCl2, 10 pmol of each primers and 2 units of Taq DNA polymerase in reaction buffer (Amersham Pharmacia Biotech, Piscataway, NJ, USA). The PCR cycle program consisted of an initial denaturation step at 94 °C for 10 min followed by 35 cycles of 60 sec at 94 °C, 54 °C, and 72 °C, and a final extension step at 72 °C for 10 min. The sizes of the amplification products were assessed by electrophoresis in 1·5% (w/v) Tris–borate/EDTA (TBE) agarose gels and ethidium bromide staining.
Before sequencing, the mitochondrial PCR fragments were purified by the QIAquick® PCR purification Kit (Qiagen). The ITS-1 amplification products were excised from the agarose gels, purified (QIAquick® Gel Extraction Kit, Qiagen) and subsequently cloned using the Topo® TA Expression Kit (Invitrogen, Groningen, Netherlands). The plasmid DNA was purified (Wizard® Plus Minipreps DNA Purification System, Promega, Madison, WI, USA) and precipitated with ethanol and sodium acetate using standard methods (Sambrook & Russell, 2001). The sequences of the PCR fragments and plasmid inserts were obtained automatically by using the ABI Prism Dye Terminator Sequencing Kit (Applied Biosystems, Foster City, WI, USA) and the ABI Prism 3100 Genetic Analyzer. The sequencing primers for the mitochondrial sequences were the same as used for the PCR. The ITS-1 sequences were obtained using the plasmid-specific sequencing primer T7 (Proligo, Boulder, CO, USA) and primers described by Bowles & McManus (1993 b) and Bowles et al. (1995).
Previously published sequences of Echinococcus isolates were used as the reference material (Table 1). The sequences were aligned using ClustalW implemented at the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw) (Thompson, Higgins & Gibson, 1994). Prior to the phylogenetic analysis, GeneDoc sequence analysis and editor program (http://www.psc.edu/biomed/genedoc/) was used to delete all sites with insertions, deletions or ambiguous bases (Nicholas, Nicholas & Deerfield, 1997).

The quartet-based maximum-likelihood programme TREE-PUZZLE was used to infer quartet puzzling trees (Schmidt et al. 2002). The HKY substitution model (Hasegawa, Kishino & Yano, 1985) was in effect and parameter estimation was done using the neighbour-joining tree. For each dataset, the gamma distribution parameter alpha, nucleotide frequencies and the transition/transversion ratio were estimated. Support for internal nodes was assessed using 1000 puzzling steps. The TreeView program was used to illustrate the phylogenetic trees (Page, 1996).
To assess the effect of the tree construction method to tree topologies, also neighbour-joining and maximum parsimony methods were applied using the version 2.1 of the MEGA program (Kumar et al. 2001). Pairwise distances for the neighbour-joining trees were calculated using the Kimura 2-parameter model and a transition/transversion ratio of two. For maximum parsimony analysis all nucleotide changes were weighed equally and the branch-&-bound algorithm was used for the tree searching. One thousand replicates of bootstrapping were used for neighbour-joining and maximum parsimony analysis to get relative support for internal nodes.
The COI and NDI fragments were identical in all analysed Finnish cervid isolates. The sequences have been submitted to the GenBank database with the accession numbers AF525457 and AF525297. The percentages of nucleotide sequence identities determined for both genes between the Finnish isolates and the other Echinococcus species and strains are shown in Table 2. Both mitochondrial sequences of the Finnish isolates were unique and differed from those of all the previously published Echinococcus species and strains.
Table 2. Pairwise nucleotide sequence identities of partial CO1 (366 bp) and ND1 (443 bp) genes between the Finnish Echinococcus granulosus form and the other strains and species of Echinococcus (See Table 1 for abbreviations.)

The COI sequence (366 bp) of the analysed isolates is most similar to that of the camel (G6) and pig (G7) strains, differing only in 1 and 2% of positions in the nucleotide sequence alignment, respectively. Ambiguous nucleotides typical for the COI sequence of the G8 genotype (Bowles et al. 1994) were not detected. These ambiguous sites complicate the sequence comparison, but still the sequence of the G8 genotype seems to differ considerably from that of the Finnish isolates. When the COI sequences are compared, the genotypes most different from the Finnish isolates are G1–G4, which differ in 8–9% of positions in the nucleotide sequence alignment. This is almost as much as the difference to the 3 other Echinococcus species (E. multilocularis, E. vogeli and E. oligarthrus, 10% each). Nucleotide identity percentage relationships are rather similar in the NDI sequence (443 bp) comparison, except that the G8 genotype resembles very much the Finnish isolates differing in 3% of positions (13 different nucleotides, 9 silent). On the other hand, there are even more nucleotide differences between the NDI sequence of the Finnish strain and the G1–G4 strains of E. granulosus (12–15%) or the other species of Echinococcus (14–17%). The G6 and G7 genotypes are still very similar to the Finnish isolates differing in 3 and 4% of nucleotides, respectively.
Between the Finnish isolates and the G6–G7 genotypes, the COI amino acid sequences differ in 1% and the NDI amino acid sequences in 3% of positions. The NDI amino acid sequence of the G8 genotype differs also 3% from that of the Finnish isolates. The amino acid differences are highest between the Finnish isolates and the G1–G3 genotypes; 6–7% in the COI sequences and 15–16% in the NDI sequences. The amino acid sequences of the other Echinococcus species differ less from those of the Finnish isolates, in 4–6% of sites in the COI sequence and in 11–13% in the NDI sequence.
For each isolate examined, an ITS-1 PCR fragment was visualized on an ethidium bromide gel as a single band of approximately 1 kb. The PCR products contained several distinct ITS-1 variants, and a total of 15 different clones were obtained. The longest obtained sequence was 1071 bp and the shortest 943 bp. Seven clones were chosen for further analyses (accession numbers are shown in Table 1), and 8 were ignored because they differed only slightly from 3 of the 7 analysed clones, namely c1, e1 and e3. Percentages of nucleotide sequence identities determined for ITS-1 fragments between the Finnish clones and other Echinococcus species and strains are presented in Table 3. To simplify the data set, only one or two representative examples per strain or species from a number of published ITS-1 variants were chosen for the analyses. As an exception to this, all known G8 variants were taken into the comparison, because this genotype was presumed to be the closest relative of the Finnish cervid E. granulosus isolates.
Table 3. Pairwise nucleotide sequence identities of the ITS-1 region between the clones of the Finnish isolates (EgFin) and the clones from the other strains and species of Echinococcus (The boundaries for the ITS-1 region were adapted from van Herwerden et al. (2000). See Table 1 for abbreviations.)

The ITS-1 sequence was found to be very divergent between and within the isolates of this and previous studies. Some clones differ from each other in about the same amount of nucleotide positions, as do the most dissimilar strains of E. granulosus or the different species of Echinococcus. The ITS-1 clones of the Finnish isolates can be divided into 2 groups, of which the sequences of the first group have the highest identity percentages with the ITS-1 sequences of the G5 genotype and the large fragment of G6. The second group resembles mainly the G7 genotype, the small fragment of the G6 and some clones of the G8 genotype.
The quartet puzzling trees based on an alignment of the COI and NDI sequences are presented in Fig. 1. A similar topology was found using the maximum parsimony and neighbour-joining methods (data not shown). The only difference between the COI and NDI trees in Fig. 1 is the location of the horse strain (G4) of E. granulosus. In both trees, the Finnish strain clusters with the pig (G7), camel (G6) and bovine (G5) strains in a well-supported clade. The cervid (G8) strain is very closely related to the Finnish isolates as well as to the pig and camel strains in the tree produced from the NDI data set. The COI sequence of G8 was ignored in the analysis, because it contains many ambiguous nucleotides and differs actually from the COI sequence of G1 only by these ambiguities (Bowles et al. 1994).

Fig. 1. Uprooted quartet puzzling trees of Echinococcus species and strains based on an alignment of partial mitochondrial COI and NDI genes (COI, log likelihood=−1054·0; NDI, log likelihood=−1639·0). The figures represent the relative support of the nodes using 1000 puzzling steps. Nodes with less than 50% support are collapsed. The distance scale is proportional to the number of mutations per site. ‘Eg’, E. granulosus, ‘EgFin’, the Finnish E. granulosus isolates, ‘Em’, E. multilocularis, ‘Eo’, E. oligarthrus, ‘Ev’, E. vogeli. The identity codes for the strains of E. granulosus and E. multilocularis are the same as in Table 1.
The quartet puzzling tree produced from the analysis of the ITS-1 multiple sequence alignment (649 bp) is presented as a phylogram in Fig. 2. Two distinct monophyletic groups containing clones from the Finnish isolates can be found: some clones (c1, e1 and e3) group with the cattle (G5) strain, large fragment of the camel (G6) strain, and the other clones (a1, b2 and a2) with the small fragment of the camel strain and one cervid (G8) strain clone. One Finnish clone (d1) is an outlier of these groups in the quartet puzzling phylogram, but in the maximum parsimony and neighbour-joining trees it clusters with the first mentioned group. Otherwise, these two methods yielded trees that were mostly similar to the tree presented in Fig. 2. The phylogeny of most of the cervid (G8) strain ITS-1 variants is rather distinct from that of the Finnish isolates: 2 cervid strain clones group with the sheep (G1) and pig (G7) strains and 1 of the cervid strain clones seems to form its own evolutionary lineage.

Fig. 2. Uprooted quartet puzzling tree of Echinococcus species and strains based on an alignment of nuclear ITS-1 rDNA sequences (log likelihood=−2225·6). The figures represent the relative support of the nodes using 1000 puzzling steps. Nodes with less than 50% support are collapsed. The distance scale is proportional to the number of mutations per site. ‘Eg’, E. granulosus, ‘EgFin’, the Finnish E. granulosus isolates, ‘Em’, E. multilocularis, ‘Eo’, E. oligarthrus, ‘Ev’, E. vogeli. The identity codes for the strains and the ITS-1 clones are shown in Table 1.
The northern biotype of E. granulosus seems to be an ecologically rather uniform group. There are only a few, if any, differences in the life-cycle and hosts of the parasite in North America and northern Fennoscandia. In North America the most important intermediate hosts for the hydatid cyst stage of the cestode are the moose and the caribou (Rangifer tarandus sspp.) (Miller, 1953; Barrett & Dau, 1981). The definitive hosts for the adult tapeworm are the wolf and the coyote (Canis latrans) but also dogs can become infected. In Fennoscandia the cycle of the parasite was earlier essentially synantropic involving dogs and semi-domesticated reindeer (e.g. Söderhjelm, 1946; Ronéus, 1974). However, according to recent evidence, at least in Finland, the wolf acts today as the principal definitive host (Hirvelä-Koski et al. 2003). Only some hydatid cysts have been found from the moose in Finland, and the importance of the moose in the life-cycle of the parasite is still unclear (Maijala et al. 2002; Oksanen, unpublished data).
Despite these similar features in the life-cycles, the E. granulosus of cervid origin does not necessarily constitute such a homogeneous group as has been believed. E. granulosus isolated from reindeer in Canada was described as E. granulosus subspecies canadensis by Webster & Cameron (1961). Reindeer were earlier introduced into north-western Canada from the Laplandic Norway. Later Sweatman & Williams (1963) made a comprehensive biological and morphological comparison between E. granulosus forms occurring in the sheep-dog cycle in New Zealand, the reindeer-dog cycle of Laplandic origin in north-western Canada and the moose-wolf cycle in North America. They suggested, that these forms represent distinct subspecies, E. granulosus granulosus, E. g. canadensis and E. g. borealis, respectively. The most distinctive morphological characters were found from E. g. canadensis.
According to the current study there are obvious genetic differences between the North American and the Fennoscandian E. granulosus forms of cervid origin. The Finnish cervid isolates examined in this study share some molecular similarity with the previously characterized cervid (G8) genotype in the mitochondrial NDI gene and in some variants of nuclear ITS-1 sequence. However, differences in these sequences and particularly in the mitochondrial COI sequence and in some of the ITS-1 clones are so obvious that the Finnish E. granulosus variant apparently represents a distinct genotypic group, which we hereby wish to designate as the genotype G10, the Fennoscandian cervid strain of E. granulosus. This strain is probably related to the earlier described canadensis form, and further studies have been initiated to collect material for detailed morphological analyses. Furthermore, more cervid isolates should be analysed genetically to determine the geographical distribution of the G8 and G10 genotypes.
The Fennoscandian cervid strain resembles closely the camel strain (G6) in both mitochondrial and nuclear sequences. Phylogenetic analysis of the mitochondrial sequences shows that the Fennoscandian cervid and the camel strains comprise an evolutionarily divergent monophyletic group with the pig strain (G7) and the cattle strain (G5) (Fig. 1). NDI sequence of G8 is also related to this group, but the COI fragment of G8 resembles more the sheep strain (G1) (Bowles et al. 1994). However, this COI subsequence of G8 is ambiguous in number of positions, which complicates the comparison. Very recently, a non-ambiguous COI fragment of G8 was published but, unfortunately, it is from a different part of this gene (McManus et al. 2002). In the same publication, other new mitochondrial sequences of the G8 genotype and some other strains were also compared. The alignment contained altogether 1674 nucleotide sites. The picture of the phylogeny of the genus Echinococcus is thus becoming clearer. Further analyses of the mitochondrial genome will provide more data on the phylogenetic relationships of the Fennoscandian cervid strain.
In phylogenetic analyses the ITS-1 variants of the Fennoscandian cervid strain seem to become divided mainly into 2 distinct evolutionary lineages, which are closely related to the camel strain ITS-1 variants. One of these lineages is related also to the cattle strain ITS-1 sequence, and the other to one of the ITS-1 variants of G8. The validity of the use of the ITS-1 fragment for strain identification or phylogenetic analyses of Echinococcus species and strains was questioned by Kedra et al. (1999) on the basis of high sequence divergence found within both G7 and G8 genotypes (only 1 example of the 21 published ITS-1 variants of G7 is shown in the phylogram in Fig. 2). This conclusion is supported by our results – polymorphism of ITS-1 rDNA sequence copies was found also in the Fennoscandian cervid strain. Moreover, the ITS-1 sequence data in the databanks is insufficient for comprehensive comparison of the Echinococcus species, as some ITS-1 variants of the known isolates, namely sheep strain large, E. oligarthrus large and E. multilocularis small PCR fragments (Bowles & McManus, 1993 b), have never been sequenced.
The strain variation of E. granulosus influences the pathogenicity of the parasite to humans. It may also affect the prognosis in patients with echinococcosis. In North America the cervid strain of E. granulosus causes the so called sylvatic hydatid disease in humans. According to many reports from Alaska and Canada this form of cystic echinococcosis has a more benign nature than the pastoral hydatid disease and, as a distinction from the pastoral form, the sylvatic form more commonly affects lungs than liver (Meltzer et al. 1956; Wilson, Diddams & Rausch, 1968; Pinch & Wilson, 1973; Moore et al. 1994). Severe complications are very rare but, quite recently, a lethal anaphylactic reaction in a patient with a liver cyst and a case with peritoneally disseminated hydatidosis were reported in Alaska (Castrodale et al. 2002). The causative agent of the last-mentioned case was confirmed as the G8 genotype (McManus et al. 2002). Only few epidemiological studies or even case reports of human echinococcosis have been reported from the Fennoscandian countries. On the basis of some of these reports also the Fennoscandian type of hydatid disease may have the same kind of organ involvement and mild nature as the North American sylvatic type (Söderhjelm, 1945; Rein, 1957). However, also some serious cases with gigantic cysts or atypically located cysts outside the lungs and the liver have been reported in reindeer-breeding Laplanders (Cederberg, 1946; af Schultén, 1890). Two different cervid forms of E. granulosus may differ from each other in their virulence and pathogenetic properties in humans. Thus, the results of the large epidemiological studies from North America can not be assumed to be directly comparable when evaluating the pathogenicity of the hydatid parasite in Fennoscandia.
The authors would like to thank Antti Oksanen (National Veterinary and Food Research Institute of Finland, Oulu Regional Unit) for providing parasite material for analysis and for commenting on the manuscript. Acknowledgement of financial support from the Academy of Finland, the Sigrid Jusélius Foundation and the Helsinki University Central Hospital Funds (EVO) is also made.

Table 1. Hosts and geographical origins of the analysed Finnish Echinococcus granulosus isolates and the Echinococcus species and strains used as reference material

Table 2. Pairwise nucleotide sequence identities of partial CO1 (366 bp) and ND1 (443 bp) genes between the Finnish Echinococcus granulosus form and the other strains and species of Echinococcus

Table 3. Pairwise nucleotide sequence identities of the ITS-1 region between the clones of the Finnish isolates (EgFin) and the clones from the other strains and species of Echinococcus
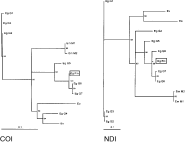
Fig. 1. Uprooted quartet puzzling trees of Echinococcus species and strains based on an alignment of partial mitochondrial COI and NDI genes (COI, log likelihood=−1054·0; NDI, log likelihood=−1639·0). The figures represent the relative support of the nodes using 1000 puzzling steps. Nodes with less than 50% support are collapsed. The distance scale is proportional to the number of mutations per site. ‘Eg’, E. granulosus, ‘EgFin’, the Finnish E. granulosus isolates, ‘Em’, E. multilocularis, ‘Eo’, E. oligarthrus, ‘Ev’, E. vogeli. The identity codes for the strains of E. granulosus and E. multilocularis are the same as in Table 1.
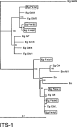
Fig. 2. Uprooted quartet puzzling tree of Echinococcus species and strains based on an alignment of nuclear ITS-1 rDNA sequences (log likelihood=−2225·6). The figures represent the relative support of the nodes using 1000 puzzling steps. Nodes with less than 50% support are collapsed. The distance scale is proportional to the number of mutations per site. ‘Eg’, E. granulosus, ‘EgFin’, the Finnish E. granulosus isolates, ‘Em’, E. multilocularis, ‘Eo’, E. oligarthrus, ‘Ev’, E. vogeli. The identity codes for the strains and the ITS-1 clones are shown in Table 1.