Introduction
Chironomids (Diptera) are often the most abundant group of insects in freshwater environments worldwide (Pinder, Reference Pinder, Mittler, Radovsky and Rash1986). The family Chironomidae includes over 10,000 species, which are distributed from the tropics to the Arctic in lakes, streams and puddles (Coffman & Ferrington, Reference Coffman, Ferrington, Merritt and Cummings1984; Cranston, Reference Cranston, Armitage, Cranston and Pinder1995). Chironomids vary in their tolerance to pollution (Lenat, Reference Lenat1993) and have become widely used in the assessment of water quality in Europe and North America (Johnson et al., Reference Johnson, Wiederholm, Rosenberg, Rosenberg and Resh1993). Particular genera and species of chironomids exhibit tolerance to nutrient enrichment (Sæther, Reference Sæther1979; Helson et al., Reference Helson, Williams and Turner2006), heavy metals (Warwick, Reference Warwick1989; Diggins, Reference Diggins2000), acid mine drainage (Heino et al., Reference Heino, Muotka, Paavola and Paasivirta2003; Waite et al., Reference Waite, Herlihy, Larsen, Urquhart and Klemm2004) and toxic organic compounds (Wright et al., Reference Wright, Ferrington and Crisp1996). Detailed taxonomic analysis at the genus or species level is required for bioassessment of the intensity of pollution impacts, as well as the identification of environmental stressors (Lenat & Resh, Reference Lenat and Resh2001). Unfortunately, the larval stage of chironomids, commonly collected in aquatic sampling surveys, possesses relatively few morphological characteristics useful for identification (Sharley et al., Reference Sharley, Pettigrew and Parsons2004). For example, discrimination of larval Cricotopus, Orthocladius and Paratrichocladius larvae is extremely difficult and depends on minute structures of the head capsule, e.g. the labral setae and premandibles (Epler, Reference Epler2001). Observation of such structures requires dissection, complex chemical clearing and preparation of permanent slide-mounts of larvae; but, depending on the orientation of specimens, crucial features may be obscured. Mature fourth instar larvae are required for effective use of larval keys; however, wear and damage of mouthpart structures may confuse identification. Phenotypic variation in pigmentation, another important character for distinguishing both Cricotopus species and Orthocladius species, can be highly variable (Gresens et al., Reference Gresens, Belt, Tang, Gwinn and Banks2007). Streams impacted by urbanization typically harbor large populations of Cricotopus and Orthocladius larvae; however, without individual associations of the larval stage with either pupa or adult, discrimination of these taxa is unreliable to impossible (Epler, Reference Epler2001; Gresens et al., Reference Gresens, Belt, Tang, Gwinn and Banks2007).
The time and expense of preparing and identifying chironomid larvae led Rabeni and Wang (Reference Rabeni and Wang2001) to suggest that it would be more economical to eliminate the Chironomidae from bioassessment programs. Fortunately, the cast pupal exoskeleton (exuviae) of chironomids provides the most efficient stage for identification to species level (Coffman, Reference Coffman, Armitage, Cranston and Pinder1995); exuviae present a wealth of morphological structures and require minimal dissection and no chemical processing prior to slide-mounting. In streams, exuviae are easily collected from the water surface in eddies and backwaters. Use of exuviae in bioassessment is cost effective in that samples are spatially integrated across microhabitats, and it takes less time to process exuviae than benthic samples (Ferrington et al., Reference Ferrington, Blackwood, Wright, Crisp, Kavanaugh, Schmidt, Peters and Walling1991). However, design of sampling surveys is complicated by the highly seasonal pattern of emergence of chironomid species (requiring multiple sample dates). Quantitative sampling of exuviae requires use of drift nets placed at comparable positions within study streams and must account for strong diurnal fluctuations in emergence rate (Wilson & Bright, Reference Wilson and Bright1973).
A final difficulty in species-level identification of chironomids, and its application to bioassessment, is the lack of qualified taxonomists (New, Reference New1996; Stribling et al., Reference Stribling, Moulton and Lester2003) and the limited availability of species-level keys for nearctic chironomids. Our knowledge of the taxonomy of nearctic Cricotopus and Orthocladius is particularly poor, and these genera are in need of revision (Epler, Reference Epler2001). Keys that have been used for identification of nearctic Cricotopus larvae and pupal exuviae (e.g. Simpson et al., Reference Simpson, Bode and Albu1983) draw heavily from palaearctic keys, even though several nearctic species do not fit this key, and it is not known whether nearctic and palaearctic species are indeed conspecific (Epler, Reference Epler2001).
Molecular-based approaches, such as DNA barcoding, are being used to supplement traditional taxonomic methods of species identification (Hebert et al., Reference Hebert, Cywinska, Ball and deWaard2003a; Savolainen et al., Reference Savolainen, Cowan, Vogler, Roderick and Lane2005; Witt et al., Reference Witt, Threloff and Hebert2006). DNA barcoding relies on sequence variation in short fragments of DNA to serve as a unique species identifier. Variation in the sequence of the mitochondrial gene, cytochrome oxidase I (COI), has proven informative for many animal taxa, including insects (Hebert et al., Reference Hebert, Penton, Burns, Janzen and Hallwachs2004a; Hogg & Hebert, Reference Hogg and Hebert2004; Ball et al., Reference Ball, Hebert, Burian and Webb2005; Monaghan et al., Reference Monaghan, Balke, Gregory and Vogler2005; Smith et al., Reference Smith, Fisher and Hebert2005; Hajibabaei et al., Reference Hajibabaei, Janzen, Burns, Hallwachs and Hebert2006; Meier et al., Reference Meier, Shiyang, Vaidya and Ng2006). DNA barcodes can be particularly useful when subtle variation in morphological features is used to distinguish species, when taxonomic keys describe only one life stage or when an investigator lacks the experience to identify taxa using morphology. One major advantage of DNA barcoding, particularly with insect identification, is the ability to identify the organism from any life stage or from a damaged specimen (Hebert et al., Reference Hebert, Cywinska, Ball and deWaard2003a; Ball et al., Reference Ball, Hebert, Burian and Webb2005; Savolainen et al., Reference Savolainen, Cowan, Vogler, Roderick and Lane2005). It is important to note that species boundaries must be determined through multiple molecular, morphological and behavioral characters prior to the use of only molecular data to identify taxa (Lee, Reference Lee2004).
In this study, we tested the efficacy of DNA barcoding for the identification of Cricotopus and Orthocladius collected from streams in Baltimore City and Baltimore County, Maryland. We compared COI sequence data with identifications based on the morphological characteristics of the pupal exuviae to create DNA sequence profiles specific to each species.
Materials and methods
Collection and rearing of larvae
Larval chironomids were collected from twelve streams in Baltimore City and Baltimore County, where previous sampling had indicated either high densities of Crictopus or the presence of species of particular interest. Larvae were collected during autumn 2004 and spring 2005, shortly before peaks in Cricotopus emergence were anticipated. Rocks covered with periphyton and larval chironomids and their retreats were removed from the streams, placed into small aerated aquaria covered with fine mesh netting and maintained in an incubator with controlled temperature and photoperiod, approximating the regime in their natural habitat. During this phase of rearing larvae to emergence, additional food was supplied as needed, in the form of cultured diatoms (i.e. no foreign larvae introduced). The enclosures were inspected twice daily, so that few adults should have emerged at any given period, thus allowing the floating pupal exuviae to be associated with the adult that produced it. The adult and its exuviae were placed in 70% ethanol and assigned an identification number based on collection site and order of emergence. For morphological identification of individuals, exuviae were dissected and slide mounted in euparal (Wiederholm, Reference Wiederholm1986). Species identifications were made using the taxonomic keys of Soponis (Reference Soponis1977), Simpson et al. (Reference Simpson, Bode and Albu1983) and Langton & Visser (Reference Langton and Visser2003). Slide-mounted exuviae have been deposited in the entomology collection of the Towson University Zoology Museum. For this initial study, the entire adult was used for extraction of DNA. In future studies, DNA will be purified from one or more legs of the individual or by enzymatic digestion of soft tissue (Ekrem & Willassen, Reference Ekrem and Willassen2004) to allow for the retention of adult vouchers.
DNA preparation
DNA was extracted from each adult following the protocol described in Guryev et al. (Reference Guryev, Makarevitch, Blinov and Martin2001). Briefly, adults were air dried then homogenized in extraction buffer (50 mm Tris-HCl pH 8.0, 400 mm NaCl, 20 mm EDTA, 0.5% SDS). Proteinase K was added to a concentration of 150 μg ml−1 and the solution incubated for 3 h at 55°C with shaking. After incubation, 5 m NaCl was added to a concentration of 1.1 m and the solution centrifuged to pellet debris. DNA was precipitated with the addition of one-half volume 4 m ammonium acetate and 2 volumes 100% ice-cold ethanol. Following centrifugation, the DNA pellet was washed with 70% ethanol, air-dried and resuspended in 15 μl TE pH 8.0.
Polymerase chain reaction (PCR) and sequencing
A 650-bp fragment of the COI mitochondrial gene was amplified with the primer pair 911 (5′-TTTCTACAAATCATAAAGATATTGG-3′) and 912 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994). DNA was amplified in the following 25 μl reaction: 2.5 μl of 10× PCR buffer pH 8.3 (10 mm Tris-HCl, 50 mm KCl, 1.5 mm MgCl2), 200 μm of each dNTP, 0.25 μm of each primer, 3.5 units of Taq polymerase and 75 ng of template DNA. The PCR thermocycling program consisted of 94°C for 3 min, 35 cycles of 94°C for 30 s, 50°C for 30 s and 75°C for 45 s followed by a final cycle at 75°C for 5 min. PCR products were verified by electrophoresis on a 1% agarose gel with ethidium bromide. Products were sequenced in both directions using the forward and reverse primers described above on an ABI 3100 at the Center of Marine Biotechnology in Baltimore, Maryland. Sequences for Cricotopus and Orthocladius COI have been deposited in Genbank, accession numbers DQ865173–DQ865180, DQ865182 and DQ865183.
Sequence alignment and distance analysis
Nucleotide sequences of 550 bp were aligned using ClustalX (Thompson et al., Reference Thompson, Higgins and Gibson1994). Genetic distances were calculated using the Kimura-2-parameter (K2P) distance model (Kimura, Reference Kimura1980). Neighbour-joining and maximum parsimony trees of the K2P distances were created in PAUP*4.0b10 (Swofford, Reference Swofford2002). Bootstrap analysis was performed with 10,000 replicates for neighbour-joining and 1000 replicates for maximum parsimony. Intraspecific and interspecific sequence divergence based on K2P distances were calculated for all species. Pairwise divergences were calculated and plotted on a frequency histogram. Mean intraspecific and interspecific K2P divergences were calculated from the pairwise comparisons within each species and within each genus, respectively.
Results
A total of 93 adult chironomids and their associated pupae exuviae were analyzed, and seven species of Cricotopus, four species of Orthocladius and one species of Paratrichocladius were identified. One Cricotopus and two Orthocladius species were represented by single individuals. Paratrichocladius rufiventris exuviae superficially resembled those of C. tristis but differed distinctly in the size and form of the respiratory organs. In C. tristis, the respiratory organ is shorter (average 105 μm) and tapers evenly to a point; whereas, in P. rufiventris, the respiratory organ is longer (average 175 μm), slightly narrower at the base and is flattened apically. In addition, the hooklet row on the posterior of tergite II is shorter (ca. 0.3 times tergite width) in Paratrichocladius vs. Cricotopus. Only five exuviae could not be identified to species using the keys, and these are referred to as Cricotopus sp. 1. They resemble C. tristis in their lack of frontal setae and in the pattern of small points on tergite III; however, they are distinguished by conspicuous pedes spurii B (PSB) on both segments two and three, and the consistent presence of four lateral setae on segment eight. Unfortunately, since the entire adult body was used for DNA extractions, it is not possible to tell if this represents an undescribed species or merely a species for which the pupal exuviae have not yet been described. However, it will now be easier to identify additional pupal/adult associations which deserve comparison with known adults.
Two morphologically-defined Cricotopus species formed monophyletic groups in both the maximum parsimony and neighbour-joining profiles (figs 1 and 2, respectively). Bootstrap support for species nodes with multiple individuals was >99%. C. triannulatus formed a monophyletic group by parsimony analysis only (fig. 1). Orthocladius oliveri also formed a cohesive group. However, O. dorenus formed two distinctly separate genetic groups, which showed subtle but consistent differences in development of the enlarged cephalic tubercles and in degree of pigmentation of the exuviae. Individuals of Cricotopus sp. 1 were all placed in a group with C. tristis in both trees (figs 1 and 2), forming a mixed species group. However, one C. tristis individual was placed outside of this ‘mixed group’, sharing a node with a single specimen of C. triannulatus. COI sequences also located P. rufiventris individuals in very different parts of the parsimony and neighbor-joining trees (figs 1 and 2). For example, one individual of P. rufiventris shared a node with a group including all but one specimen of C. triannulatus, whereas another shared a node with C. trifascia, although C. trifascia formed a cohesive grouping after the node. Species represented by a single sequence: O. robacki, O. nigritis and one specimen of C. tristis, formed unique groups in both analyses.
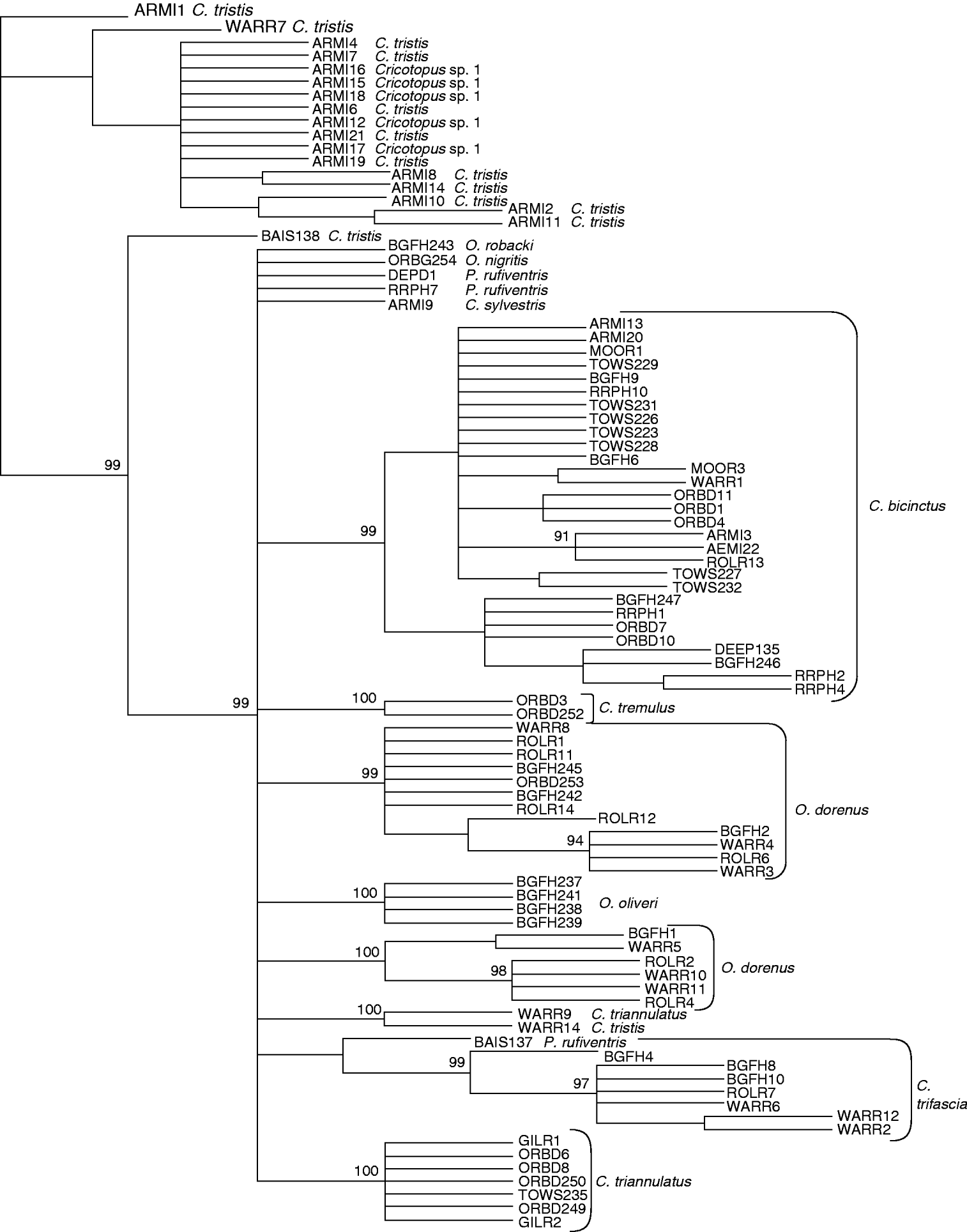
Fig. 1. Parsimony tree of COI sequence divergences (K2P) in Cricotopus, Orthocladius and Paratrichocladius species from Baltimore, Maryland. Numbers at nodes indicate bootstrap scores after 1000 replicates.
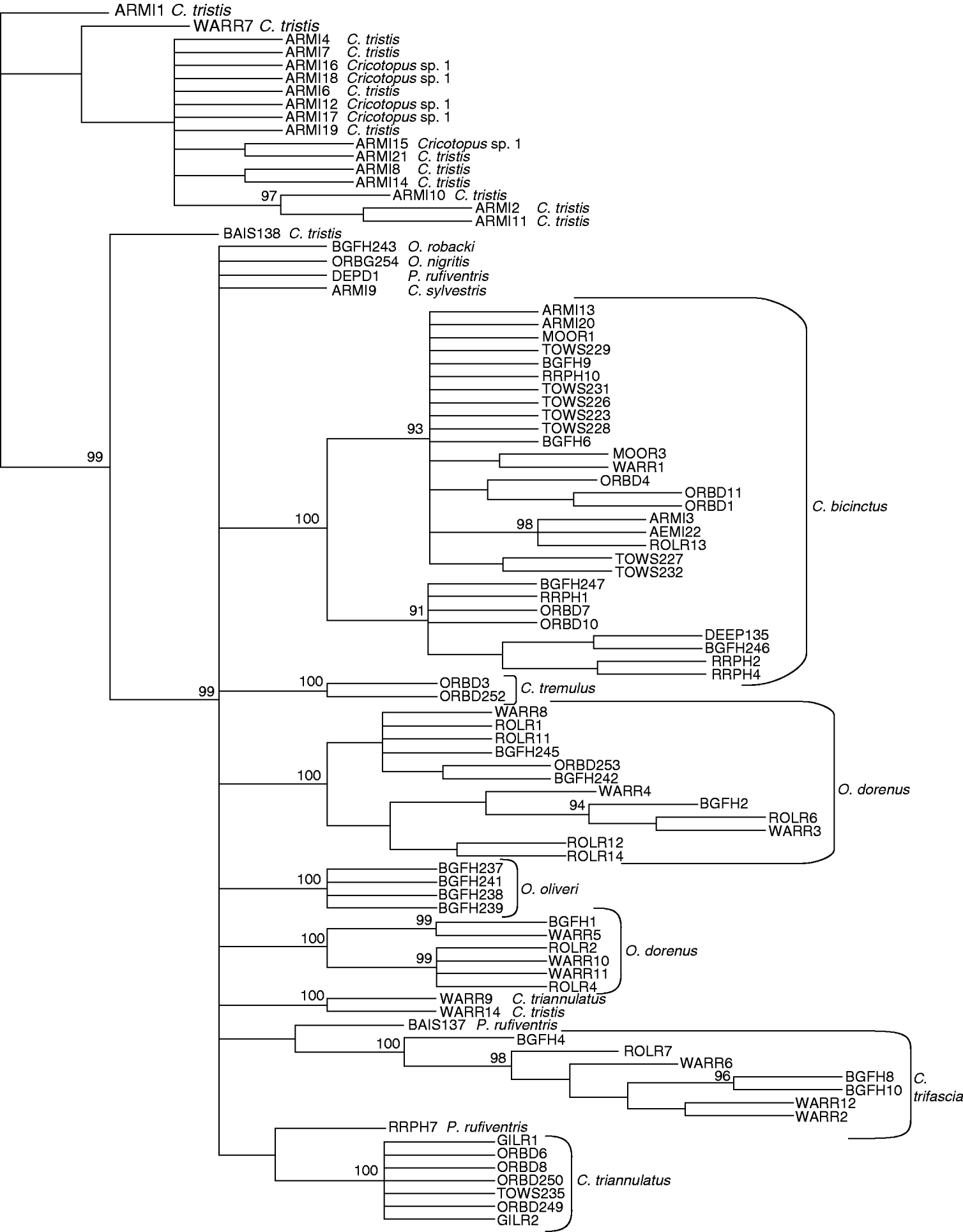
Fig. 2. Neighbour-joining tree of COI sequence divergences (K2P) in Cricotopus, Orthocladius and Paratrichocladius species from Baltimore, Maryland. Numbers at nodes indicate bootstrap scores after 10,000 replicates.
The mean intraspecific divergence for all species examined was 2.32% (range 0–22.21%) (fig. 3). The mean intraspecific divergence for individuals forming cohesive groups by parsimony and neighbor-joining analysis was 1.80% (fig. 4). Intraspecific divergences for Cricotopus and Orthocladius ranged from 0–5.40% with the exception of C. tristis, C. triannulatus, O. dorenus and P. rufiventris (table 1). In C. tristis, two individuals varied by >8% from the other twelve which had ⩽3% variation. One individual, WARR9, was identified as C. triannulatus but varied more than 15% from the other seven C. triannulatus. The three samples identified morphologically as P. rufiventris vary genetically by 15–21% from one another.
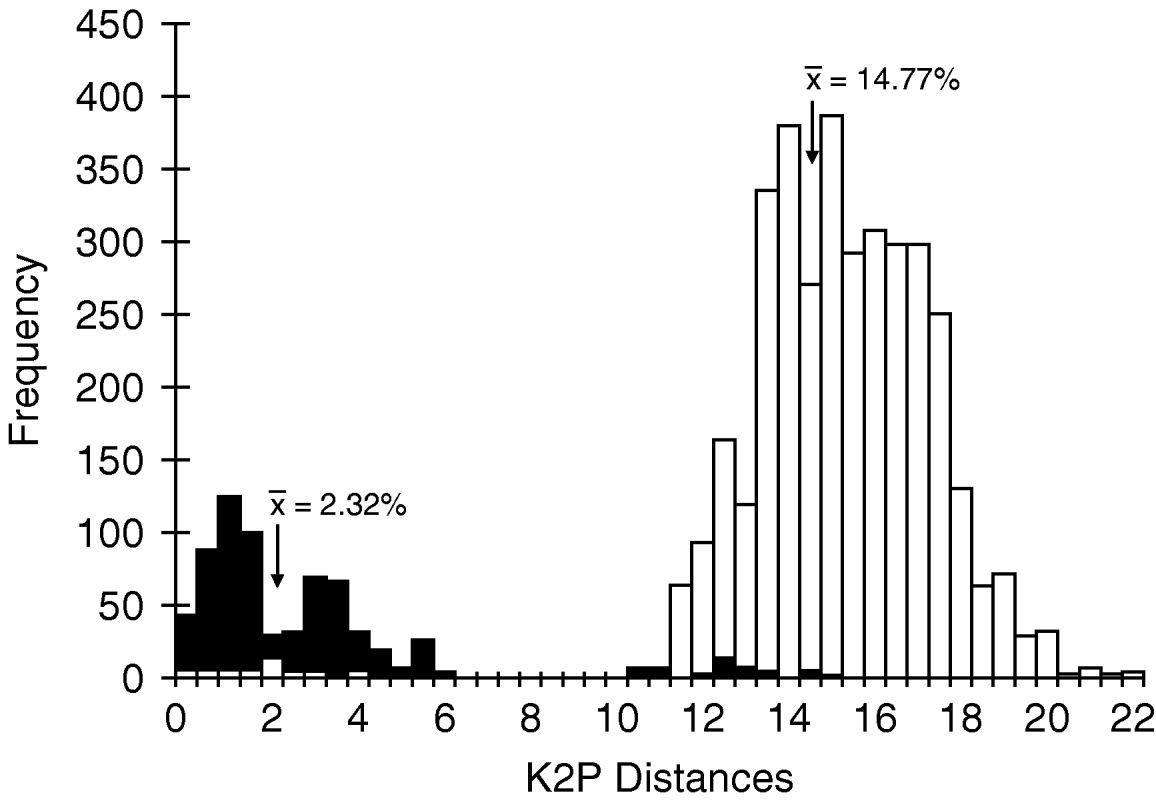
Fig. 3. Histogram of intraspecific and interspecific genetic Kimura-2-parameter (K2P) divergences. Values represent all 93 individuals analyzed in this study (■, Intraspecific; □, Interspecific).
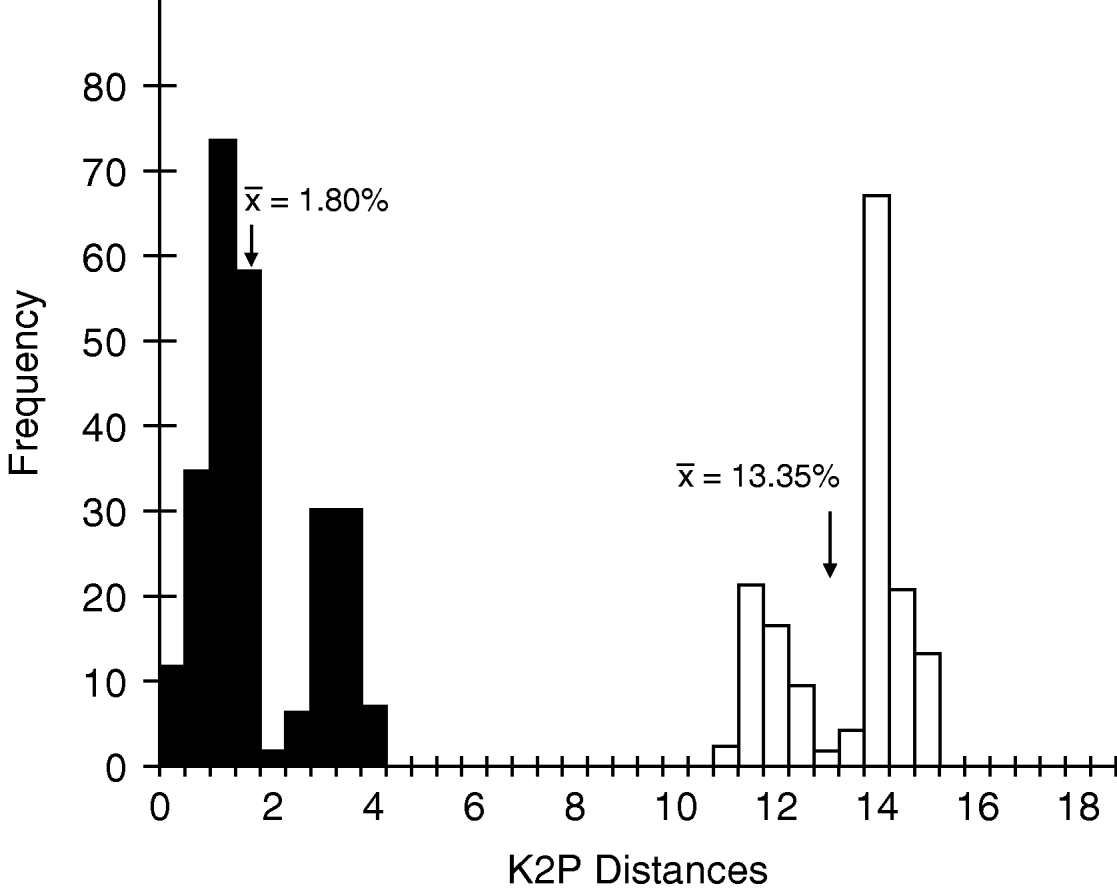
Fig. 4. Histogram of intraspecific and interspecific genetic Kimura-2-parameter (K2P) divergences. Values represent only those individuals forming cohesive groups by parsimony and neighbour-joining analysis, C. bicinctus, C. tremulus and O. oliveri (■, Intraspecific; □, Interspecific).
Table 1. Mean and range of intraspecific Kimura-2-parameter nucleotide divergences for Cricotopus, Orthocladius and Paratrichocladius species.
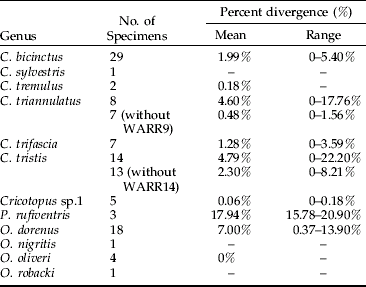
The mean interspecific divergence was 14.77% (range 0–21.90%) for all species (fig. 3) and 13.35% (range 10.61–15.40%) for the cohesive groups only (fig. 4). Interspecific variation within Cricotopus, with and without the mixed group, was 0–21.70% (mean 14.43%) and 11.04–21.70% (mean 15.20%), respectively. Interspecific variation within Orthocladius was 11.07–14.83% (mean 12.46%). Here, we note that specimens of two morphologically distinct species, C. tristis and Cricotopus sp. 1, had <3% divergence from one another with the exception of one C. tristis individual that was >15% divergent from the others.
Discussion
Our results indicate that DNA barcoding with the COI gene can be useful in identifying Cricotopus and Orthocladius species. Molecular classifications of three species of Cricotopus (C. bicinctus, C. sylvestris, C. tremulus) and three species of Orthocladius (O. nigritus, O. oliveri, O. robacki) from Baltimore-area streams produced monophyletic groups which were fully congruent with the morphological species identity. However, this COI sequence was not sufficient for accurate identification in all cases. For example, the ‘mixed group’ of Cricotopus sp. 1 and C. tristis had less than 3% COI sequence divergence, with the exception of two individuals, BAIS138 and WARR14, identified as C. tristis and having divergences of greater than 8% from all other individuals in the mixed group. Based on the suggested threshold values of 2–3% used to discern species (Hebert et al., Reference Hebert, Ratnasingham and deWaard2003b), this would suggest that the ‘mixed group’ is actually one species, despite obvious morphological distinctions. Perhaps Cricotopus sp. 1 may have been derived from the same parental species as C. tristis. Further analysis with additional mitochondrial or nuclear genes will be needed to clarify these two species.
In contrast to the ‘mixed group’, O. dorenus formed two separate groups, which nonetheless did show subtle but consistent morphological differences in development of the cephalic tubercles (an important species characteristic) and in pigmentation. Using COI sequence data, Ekrem et al. (Reference Ekrem, Willassen and Stur2007) similarly reported classification of Micropsectra notescens individuals into two monophyletic groups, although no morphological differences could be observed. Additional morphological, ecological or cytogentic data are needed before such patterns can be interpreted as cryptic species. Nevertheless, the feedback provided by the genetic analysis of O. dorenus was very helpful for correct interpretation of morphological variation within Orthocladius; Soponis (Reference Soponis1977) remarked that exuviae of O. dorenus, O. obumbratus, O. robacki and O. oliveri are difficult to distinguish. In this case, the genetic analyses discriminated morphological species quite well.
Overall, mean intraspecific and interspecific sequence divergences differed by an order of magnitude and did not overlap when only those individuals forming cohesive groups were evaluated. Three species, C. bicinctus, C. triannulatus and C. tristis, had greater than 3% intraspecific divergence due to one or two individuals. Two C. tristis, BAIS138 and WARR14, collected from a forested and a suburban stream, respectively, had greater than 4% divergence from the other C. tristis, which were from a polluted urban stream. This suggests a response to selection. However, this does not appear to be a factor with C. bicinctus, where <3% divergence was found among individuals from rural, suburban and highly urban streams.
The values reported here for intraspecific and interspecific divergence are comparable to similar studies in other organisms. Mean intraspecific divergences of 2.76% (Hebert et al., Reference Hebert, Penton, Burns, Janzen and Hallwachs2004a), 1.02% (Wiemers & Fiedler, Reference Wiemers and Fiedler2007) and less than 0.46% (Hajibabaei et al., Reference Hajibabaei, Janzen, Burns, Hallwachs and Hebert2006) in tropical Lepidopterans; 0.43% (Hebert et al., Reference Hebert, Stoeckle, Zemlak and Francis2004b) in birds; 0.17% (Smith et al., Reference Smith, Woodley, Janzen, Hallwachs and Hebert2006) in parasitoid flies; 1.1% in mayflies (Ball et al., Reference Ball, Hebert, Burian and Webb2005); and 0.9% (Ekrem et al., Reference Ekrem, Willassen and Stur2007) in Chironomidae have been reported. Mean interspecific divergences of 4.41–6.02% (Hajibabaei et al., Reference Hajibabaei, Janzen, Burns, Hallwachs and Hebert2006) and 9.38% (Wiemers & Fiedler, Reference Wiemers and Fiedler2007) in Lepidopterans; 7.93% (Hebert et al., Reference Hebert, Stoeckle, Zemlak and Francis2004b) in birds; 5.78% (Smith et al., Reference Smith, Woodley, Janzen, Hallwachs and Hebert2006) in parasitoid flies; 18.1% in mayflies (Ball et al., Reference Ball, Hebert, Burian and Webb2005); and 16.2% (Ekrem et al., Reference Ekrem, Willassen and Stur2007) in Chironomidae have been reported. Surprisingly, a mean intraspecific divergence of 0.048% and a mean interspecific divergence of 1.90% have been reported in coral (Shearer & Coffroth, Reference Shearer and Coffroth2007). These values clearly illustrate that a single threshold value for species identification is not feasible, and more studies will be needed to determine the best method for making these decisions.
Recently, Hebert et al. (Reference Hebert, Stoeckle, Zemlak and Francis2004b) suggested a threshold value of ten times the mean intraspecific divergence for species determination. Based on the mean intraspecific divergence reported here, 1.80%, that would give a threshold of 18.0% for species discernment. This value is greater than our mean interspecific value of 13.35%. More importantly, the value is greater than most observed interspecific divergences in our data set. The difficulty comes in separating the mixed group of Crictopus, which were identified by morphological characters as two distinct species yet they have nearly identical COI nucleotide sequence. In cases such as this, use of a nuclear marker may provide more accurate results. In order to get a good estimate of intraspecific (vs. interspecific) variation, it is necessary to collect a large number of representatives of each species from as wide a geographical range as possible (Zhou et al., Reference Zhou, Kjer and Morse2007). It must be kept in mind that the present study is biased in this regard, since only C. bicinctus, C. tristis and O. dorenus were represented by more than ten individuals each, and all collections were from one county in the Mid-Atlantic region. The disparate placement of a small number of C. tristis or C. triannulatus from otherwise ‘cohesive’ groupings may represent our underestimation of intraspecific genetic variation.
DNA barcoding has proven extremely useful for identifying organisms to species level and resolving taxonomic conflicts. Recently, these tools have been applied to the Chironomidae (Carew et al., Reference Carew, Pettigrove and Hoffmann2003; Ekrem et al., Reference Ekrem, Willassen and Stur2007; Pfenninger et al., Reference Pfenninger, Nowak, Kley, Steinke and Streit2007). Sequence data from the mitochondrial gene COI has been successful in monophyletic classifications that are largely congruent with morphological species in the genus Chironomus (Sharley et al., Reference Sharley, Pettigrew and Parsons2004; Carew et al., Reference Carew, Pettigrove and Hoffmann2005) and a large number of Tanytarsini genera (Carew et al., Reference Carew, Pettigrove, Cox and Hoffmann2007; Ekrem et al., Reference Ekrem, Willassen and Stur2007). Our study is novel in that it extends application of COI sequence analysis to three genera in the subfamily Orthocladiinae (Cricotopus, Orthocladius and Paratrichocladius), which have been difficult to discriminate in larval and even pupal life stages. A common conclusion of studies on chironomid taxa is that COI sequences work well for species identification in the majority of cases, but inclusion of sequence data from additional nuclear markers is highly recommended. Development of a standard set of genes/sequences is important for the construction of a chironomid barcode ‘library’, which would be effective in correct identification of larval life stages (Ekrem et al., Reference Ekrem, Willassen and Stur2007). Ultimately, such a library depends on a well-defined taxonomy based on morphological and other independent data. Nearctic species of Cricotopus are not as well described as those of the Western Palaearctic, and molecular analyses such as these are valuable in the correct interpretation of morphological variation in similar species and genera.
Acknowledgements
Funding for this project was provided by the Faculty Development and Research Committee of Towson University, Towson, Maryland. The authors would like to thank Dr Roland Roberts for his assistance with the parsimony and neighbour-joining analyses and manuscript review. The authors would also like to acknowledge the undergraduate students who assisted in the project, Chelsea Scott, Dan Cassilly and Wei Guo.







