INTRODUCTION
The introduction of non-indigenous species (NIS) poses a significant ecological and economic threat to coastal marine communities (Cohen & Carlton, Reference Cohen and Carlton1998; Sakai et al., Reference Sakai, Allendorf, Holt, Lodge, Molofsky, With, Baughman, Cabin, Cohen, Ellstrand, McCauley, O'Neil, Parker, Thompson and Weller2001). Once introduced and established NIS proliferate and are one of the major threats to the world's biodiversity (Simberloff et al., Reference Simberloff, Martin, Genovesi, Maris, Wardle, Aronson, Courchamp, Galil, García-Berthou, Pascal, Pyšek, Sousa, Tabacchi and Vilà2013; Comtet et al., Reference Comtet, Sandionigi, Viard and Casiraghi2015). For this reason, the early detection of incipient invasions increases the possibilities of eradicating them or constraining their spread (Tidbury et al., Reference Tidbury, Taylor, Copp, Garnacho and Stebbing2016). Early detection has been recommended as a priority in management plans and programmes of biological invasions treatment (Vander Zanden et al., Reference Vander Zanden, Hansen, Higgins and Kornis2010; Xiong et al., Reference Xiong, Li and Zhan2016).
In these strategies of early detection of NIS DNA-based methods are playing an important role (Darling & Mahon, Reference Darling and Mahon2011; Darling & Piranio, Reference Darling and Piranio2015) as it has been successfully tested (Montoliu et al., Reference Montoliu, Miracle and Elías-Gutierrez2015; Brown et al., Reference Brown, Chain, Zhan, MacIsaac and Cristescu2016). However, DNA barcoding presents a bottleneck since it needs the previous knowledge of NIS DNA sequences deposited in accessible databases such as GenBank. However, recent technological advances, such as high-throughput sequencing (HTS) or a metabarcoding approach, are solving this problem (Brown et al., Reference Brown, Chain, Zhan, MacIsaac and Cristescu2016; Xiong et al., Reference Xiong, Li and Zhan2016).
Another way to detect NIS in aquatic ecosystems is the analysis of larval stages. An important number of aquatic taxa, including benthonic species, show a larval phase that differs in morphology and habitat to the adult. In many cases larval specimens are present in the plankton in higher numbers than the adult stage. Moreover, some species occupy cryptic habitats as adults and are easier to find as larval forms in the plankton, for example the trachelifer larvae of Jaxea spp. (Wear & Yaldwyn, Reference Wear and Yaldwyn1966).
There are cases where larvae of NIS were detected before the adults. For example, larvae of Palaemon macrodactylus Rathbun, 1902 were found in the Mediterranean in 2005 and 2010 surveys (Torres et al., Reference Torres, Dos Santos, Cuesta, Carbonell, Massutí, Alemany and Reglero2012), but the first adults were observed in 2012 and 2013 (Cuesta et al., Reference Cuesta, Bettoso, Comisso, Froglia, Mazza, Rinaldi, Rodriguez and Scovacricchi2014). In general, the use of DNA barcoding has allowed the identification of an important number of adults (Radulovici et al., Reference Radulovici, Saint-Marie and Dufresne2009) and planktonic larval stages of different taxa (Marco-Herrero et al., Reference Marco-Herrero, Torres, Cuesta, Guerao, Palero and Abelló2013).
In European marine waters there are four known species of the family Pinnotheridae. Becker & Türkey (Reference Becker and Türkay2010) reduced the number of European species from five to three, Nepinnotheres pinnotheres (Linnaeus, 1758), Pinnotheres pisum (Linnaeus, 1767) and Pinnotheres pectunculi Hesse, 1872, after synonymizing Pinnotheres ascidicola Hesse, 1872 and Pinnotheres marioni Gourret, 1888 to N. pinnotheres and validating the status of P. pectunculi. Later Subida et al. (Reference Subida, Arias, Drake, García Raso, Rodríguez and Cuesta2011) found an African species, Afropinnotheres monodi Manning, Reference Manning1993, in the Gulf of Cadiz, elevating to four the current number of European species of pinnotherids. For all these species there are larval data available. Atkins (Reference Atkins1954) described the complete larval development of N. pinnotheres (two zoeal and one megalopa stages), and P. pisum (four zoeal and one megalopa stages); only data of zoea I of P. pectunculi are included in Becker (Reference Becker2010), and Marco-Herrero et al. (Reference Marco-Herrero, Drake, Gonzalez-Gordillo and Cuesta2016) described the four zoeal stage and the megalopa of A. monodi. Also, with the exception of P. pectunculi, there are 16S mtDNA sequences available for all the European pinnotherids.
This previous information has allowed us to detect larvae of an unknown pinnotherid in the Gulf of Cadiz. In the present study these larval stages are described and DNA sequences are provided to help identify this new NIS for European waters and follow its spread, adding more value to the early detection of this aquatic invasion.
MATERIALS AND METHODS
Material studied
Larval stages: A total of 139 unidentified zoeae (stages of zoea II–IV) and 15 unidentified megalopae attributed to unknown pea crab species (Crustacea, Decapoda, Pinnotheridae), as well as eight zoeae (different stages) of Pinnotheres pisum, were collected in two plankton samples obtained in the context of the project ‘Ecology of the early stages of the Engraulis encrasicolus life cycle: the role of the coupled ecosystem ‘Guadalquivir estuary and its coastal influence area’ in the recruitment process of this species’ (ECOBOGE). The samples were collected with a suprabenthic sled equipped with two superimposed nets (200 µm mesh size) that sampled the motile fauna in the first metre near-bottom water layers. The two samples were obtained on 14 November 2013. The first one (PD10) was collected at 36,52900 (start) – 36,52978 (end) N – 6,28913 (start) – −6,29783 (end) W, 14 m depth, and the second one (MT10) at 36,57320 (start) – 36,57571 (end) N −6,3211 (start) – 6,32451 (end) W, 11 m depth (Figure 1). Both samples were fixed in ethanol (90%) for later morphological and molecular studies.
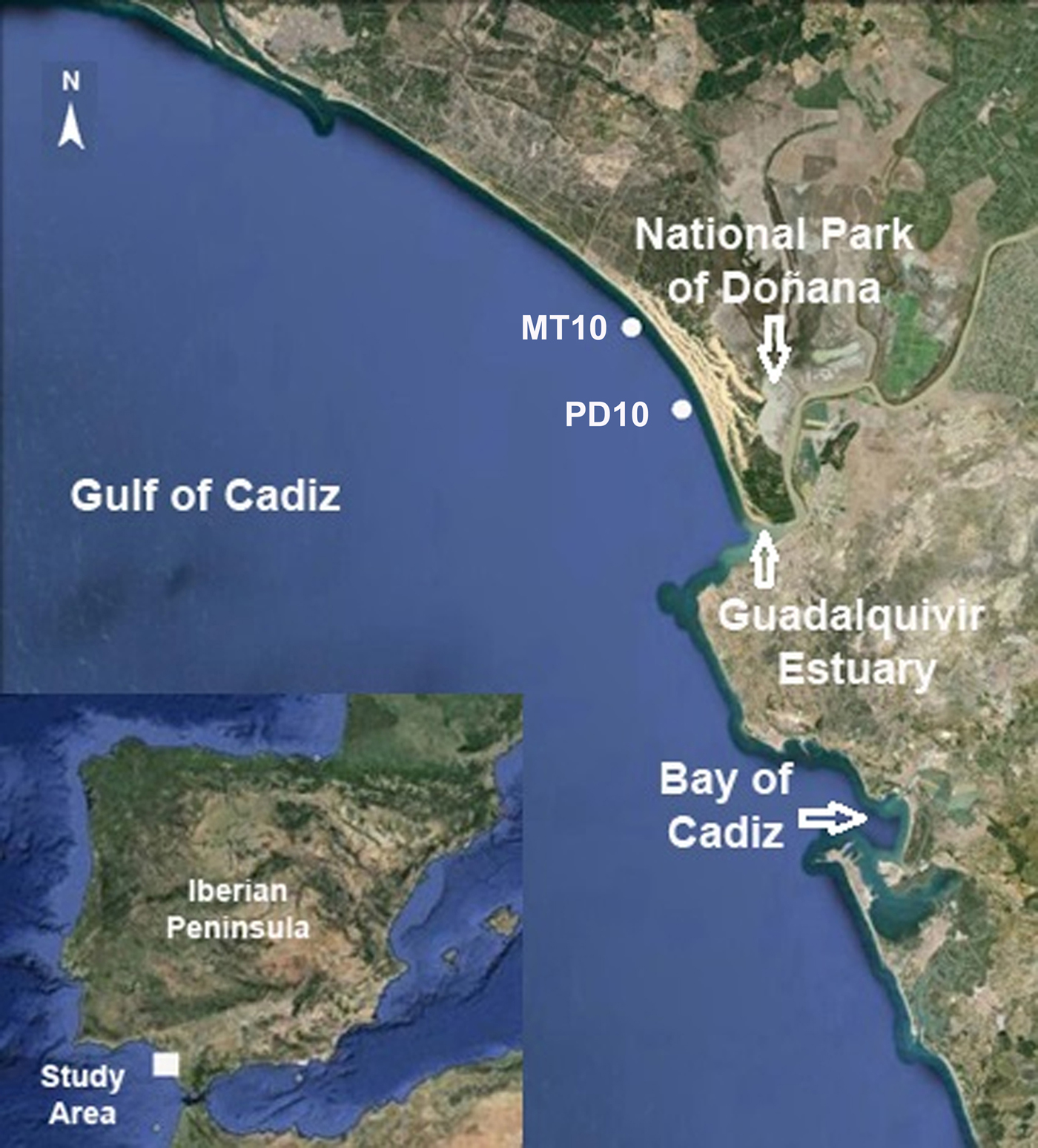
Fig. 1. Map of the Gulf of Cadiz (SW Iberian Peninsula) showing the sampling stations MT10 and PD10 where larvae of Pinnotheres pisum and Pinnotheres sp. were collected. Images from Google Earth, version 7.1.2.2041 ©2013.
Adult stages: two specimens of Pinnotheres pectunculi (one male SMF34383 and one female SMF34529 both from Roscoff, France) were borrowed as a loan from the Senckenberg Museum (Frankfurt, Germany) to obtain DNA sequences, since P. pectunculi was the only European Pinnotheridae species without gene sequences available in GenBank.
Identification of samples
Initially all zoeae and megalopae were undoubtedly identified as larval stages belonging to Pinnotheridae crabs. Eight zoeae were identified as zoeae II and III of Pinnotheres pisum, and the rest of the larval stages (zoeae II, III, IV and megalopae) shared characters that clearly placed them also in the subfamily Pinnotherinae, sensu Palacios-Theil et al. (Reference Palacios-Theil, Cuesta and Felder2016), and belonging to different larval stages of a single unknown species.
Molecular analysis
The DNA barcoding identification of larval stages was based on partial sequences of the 16S mtDNA gene, a molecular marker that has shown to be suitable for DNA barcoding in crustaceans (Schubart et al., Reference Schubart, Neigel and Felder2000). Cytochrome oxidase 1 (Cox1) was discarded as a marker because there are no Cox1 sequences available for all these species in GenBank, together with the difficulties found in the amplification of this gene for species of this family (Mantelatto et al., Reference Mantelatto, Carvalho, Simões, Negri, Souza-Carvalho and Terossi2016). Total genomic DNA was extracted from maxillipeds muscle tissue of three zoeal specimens and from pereiopods of one megalopa, following a modified Chelex 10% protocol by Estoup et al. (Reference Estoup, Largiadèr, Perrot and Chourrout1996).
Target mitochondrial DNA from the 16S mtDNA gene was amplified with polymerase chain reaction (PCR) using the following cycling conditions (with Taq polymerase): 2 min at 95°C, 40 cycles of 20 s at 95°C, 20 s at 45–48°C, 45 s at 72°C, and 5 min 72°C. Primer pairs 1472 (5′-AGA TAG AAA CCA ACC TGG-3′) (Crandall & Fitzpatrick, Reference Crandall and Fitzpatrick1996) and 16L2 (5′-TGC CTG TTT ATC AAA AAC AT-3′) (Schubart et al., Reference Schubart, Cuesta and Felder2002), and 16Sbr-H (5′-CCG GTC TGA ACT CAG ATC ACG T-3′) (Palumbi, Reference Palumbi, Hillis, Moritz and Mable1996) and L12 (5′-TGA CCG TGC AAA GGT AGG ATA A-3′) (Schubart et al., Reference Schubart, Diesel and Hedges1998) were used to amplify 420–540 bp of 16S mtDNA gene. PCR products were sent to Stab-Vida laboratories to be purified and then bidirectionally sequenced.
Sequences were edited using the free software Chromas Lite version 2.0. The obtained final DNA sequences were compared with those from adult specimens of the four Pinnotheridae species that inhabit European waters, obtained in the context of the MEGALOPADN and AFROBIV projects, as well as downloaded from GenBank databases. New partial sequences of 16S mtDNA obtained from larvae of Pinnotheres sp., zoea of Pinnotheres pisum and two specimens of Pinnotheres pectunculi were deposited in GenBank under accession numbers MF069147–MF069151.
An evolutionary distances analysis was carried out in MEGA6 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). An alignment of 16S sequences was built using the sequences obtained in the present study from the larval stages of Pinnotheres sp., one zoea of Pinnotheres pisum and two specimens of Pinnotheres pectunculi as well as the sequences of Afropinnotheres monodi, Nepinnotheres pinnotheres, Pinnotheres pisum, Orthotheres barbatus and Zaops ostreus downloaded from GenBank (http://www.ncbi.nlm.nih.gov). The phylogenetic reconstruction was inferred from neighbour-joining analyses using the P-distance method. The nodal confidence of the obtained topologies was assessed via 2000 bootstrap replicates.
Larval morphology description
Dissections, drawings and measurements of larval stages of Pinnotheres sp. were made using a Wild MZ6 and a Zeiss Axioskop compound microscope with Nomarski interference, both equipped with a camera lucida. For an easier observation of larvae structures and setation under the microscope, a digestion-stain procedure was carried out. First, entire specimens were placed for 10 min on a watch glass with 2 ml of heated lactic acid. Immediately after that, 3 drops of Clorazol Black stain (0.4 g Clorazol Black powder dissolved in 75 ml 70% ethanol) were added to the heated solution. After 5–10 min, the specimens were removed from the solution and placed on a slide with lactic acid, in order to proceed with the dissection of the appendages (for more details see Marco-Herrero et al., Reference Marco-Herrero, Rodríguez and Cuesta2012).
Drawings and description were based on examinations of three zoea II, six zoea III, seven zoea IV and nine megalopae. Measurements were made with an ocular micrometer, based on three zoea II, five zoea III, five zoea IV and five megalopae. In zoeal and megalopa stages the cephalothorax length (CL) was measured from the base of the rostrum to the posterior margin of the cephalothorax, and the cephalothorax width (CW) as the maximum width of the cephalothorax.
Descriptions and figures were arranged according to the standards proposed by Clark et al. (Reference Clark, Calazans and Pohle1998), and updated by Clark & Cuesta (Reference Clark, Cuesta, Castro, Davie, Guinot, Schram and von Vaupel Klein2015). Samples of larvae of Pinnotheres sp. and P. pisum are deposited at the Crustacean Decapod Collection from Cadiz Oceanographic Centre under the accession numbers IEO-CD-ECOB/1926–IEO-CD-ECOB/1928.
RESULTS
In the plankton samples were identified a total of eight zoeae (two zoea II, six zoea III) of Pinnotheres pisum, and three zoeae II, 97 zoeae III, 23 zoeae IV and 16 megalopae of Pinnotheres sp., but no zoea I attributed to this last species was found.
According to the morphology and size of the larvae identified, the larval development of Pinnotheres sp. consists of four zoeal stages and a megalopa.
The second zoea is described in detail, and only the main differences in subsequent stages are noted.
LARVAL DESCRIPTION
Order DECAPODA Latreille, 1802
Infraorder BRACHYURA Latreille, 1802
Family PINNOTHERIDAE De Haan, 1833
Genus Pinnotheres Bosc, 1801
Pinnotheres sp.
(Figures 2–5)
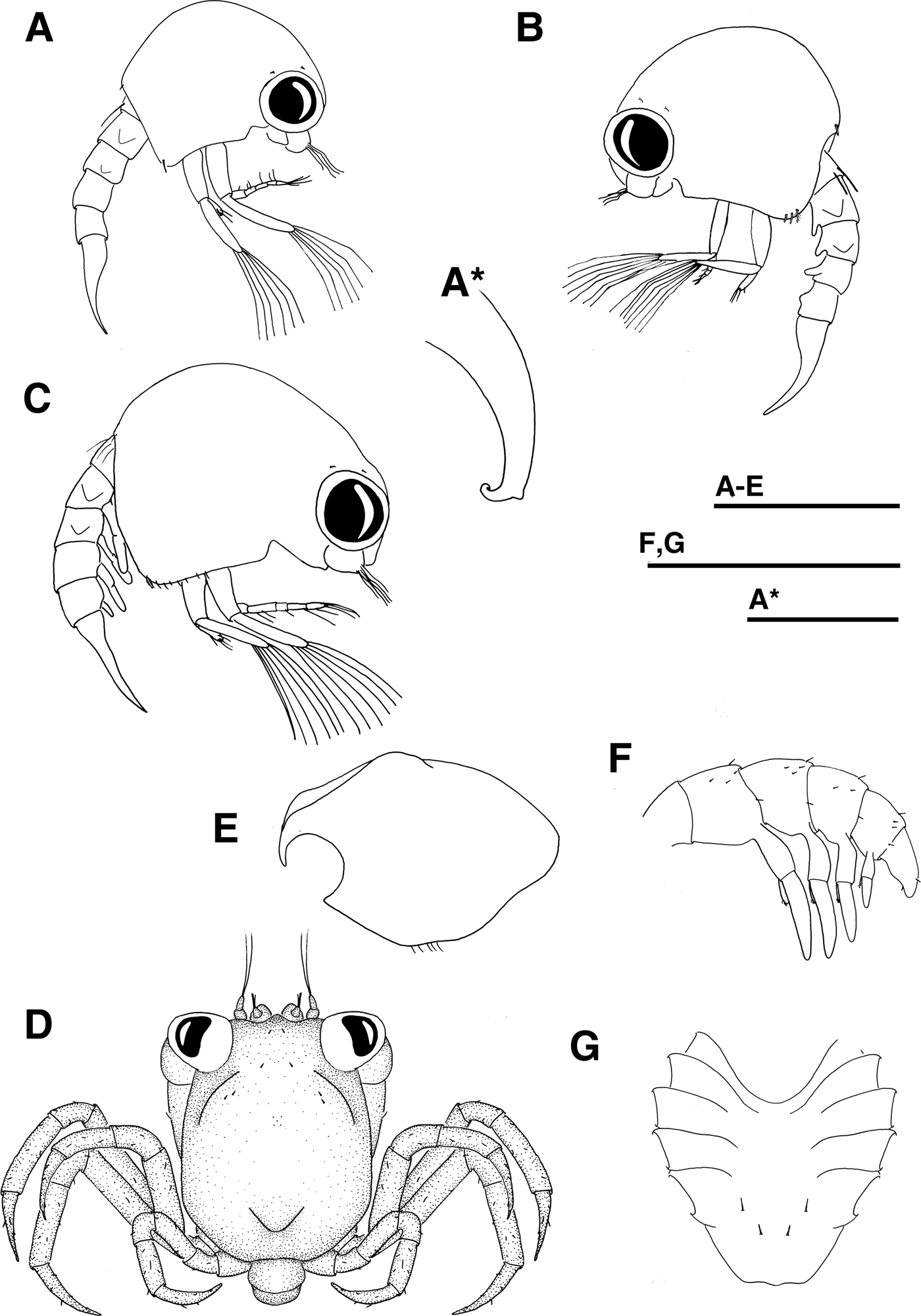
Fig. 2. Pinnotheres sp. Lateral view, (a) zoea II, (a*) detail of rostrum. (b) zoea III. (c) zoea IV. Megalopa, (d) dorsal view, (e, f) lateral view, (e) carapace, (f) pleon. (g) sternum. Scale bars, (a–g) = 0.5 mm, (a*) = 0.1 mm.
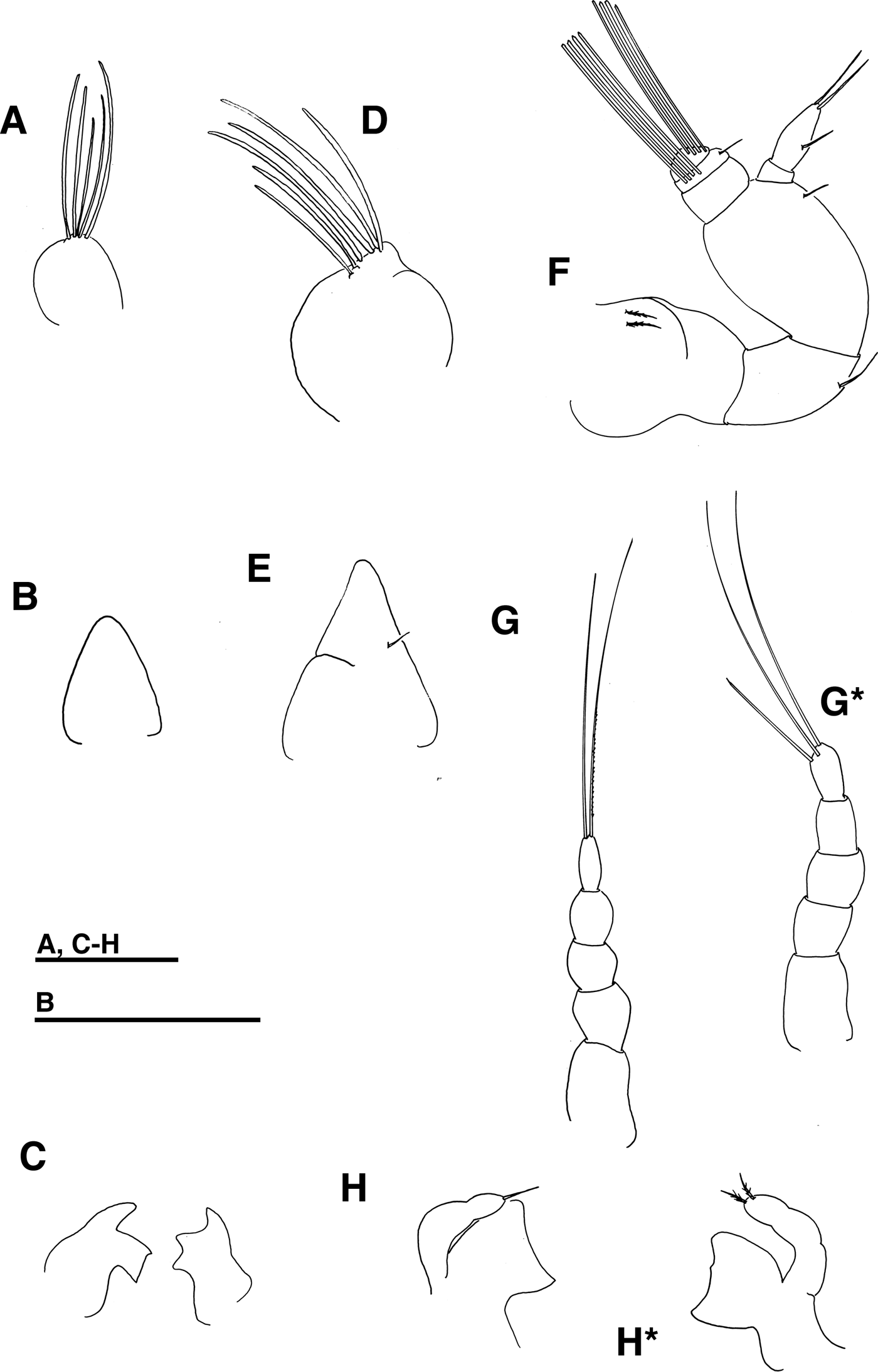
Fig. 3. Pinnotheres sp. Antennule, (a) zoea II, (d) zoea IV, (f) megalopa. Mandible, (c) zoea II, (h, h*) megalopa. Antenna, (b) zoea II, (e) zoea IV, (g, g*) megalopa. Scales bars = 0.1 mm.
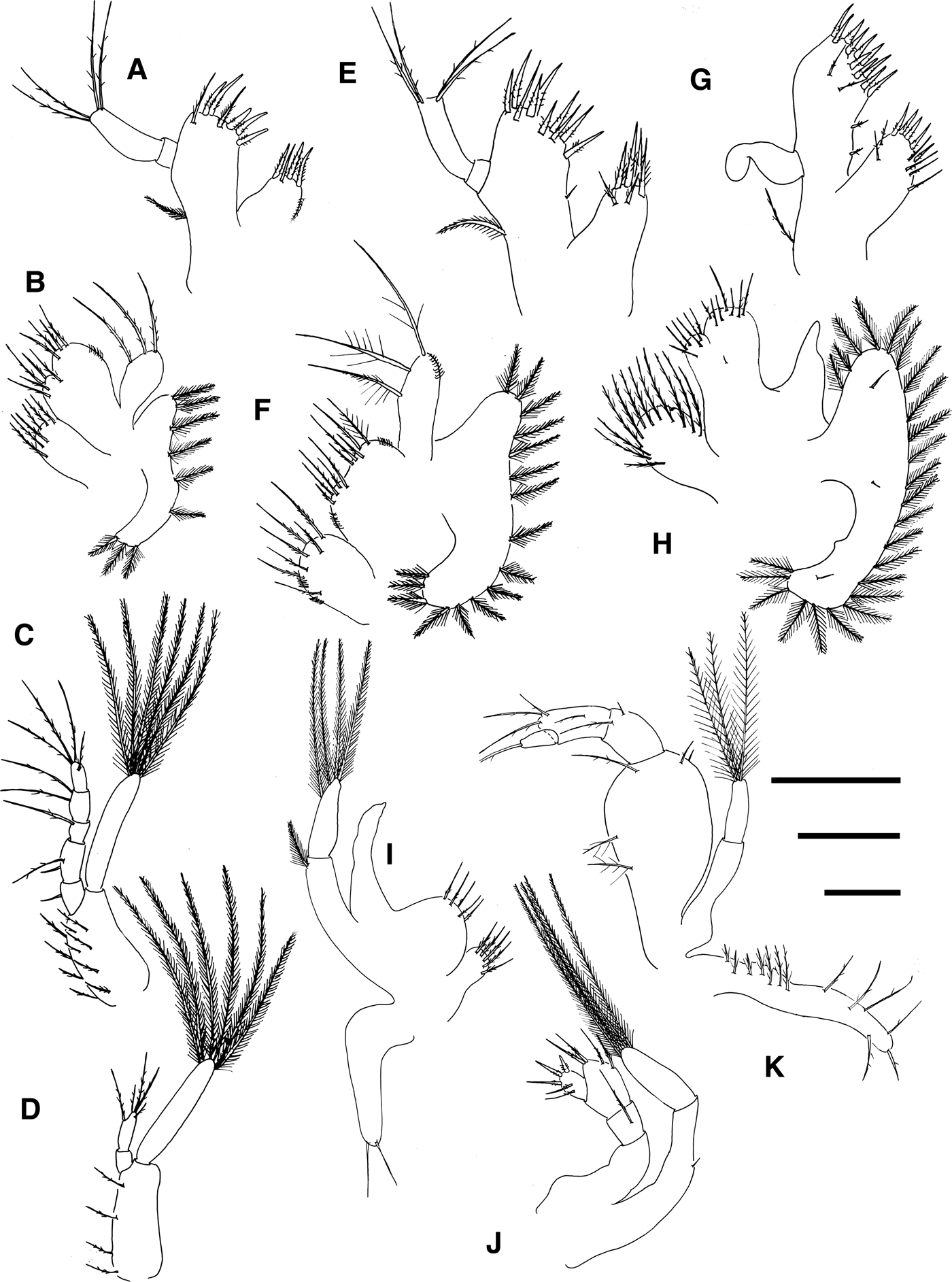
Fig. 4. Pinnotheres sp. Maxillule, (a) zoea II, (e) zoea IV, (g) megalopa. Maxilla, (b) zoea II, (f) zoea IV, (h) megalopa. First maxilliped, (c) zoea II, (i) megalopa. Second maxilliped, (d) zoea II, (j) megalopa. Third maxilliped, (k) megalopa. Scale bars = 0.1 mm.
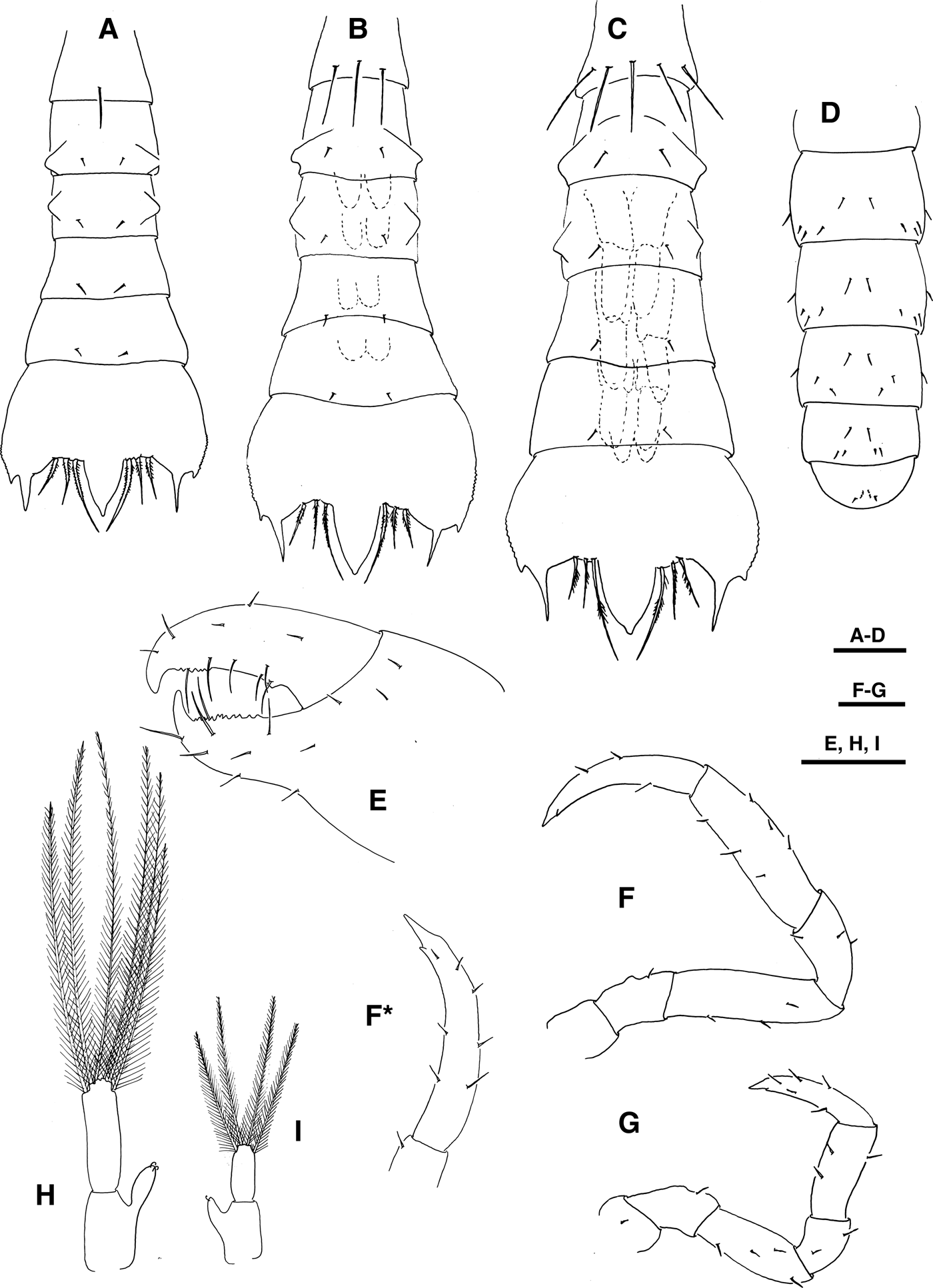
Fig. 5. Pinnotheres sp. Pleon, dorsal view, (a) zoea II, (b) zoea III, (c) zoea IV, (d) megalopa. Megalopa, (e) cheliped, (f, f*, g) pereiopods, (h) pleopod, (i) uropod. Scale bars = 0.1 mm.
ZOEA II
(Figures 2a, a*, 3a–c, 4a–d, 5a)
Size: CL = 0.472 ± 0.019 mm; CW = 0.375 ± 0.039 mm, N = 3.
Cephalothorax (Figures 2a, a*): Globular, without tubercles. Dorsal and lateral spine absent. Rostrum short, elephant trunk shape with a minute tubercle (Figure 2a*). One pair of posterodorsal and two pairs of anteromedian simple setae. Ventral margin with one plumose setae. Eyes stalked and movable.
Antennule (Figure 3a): Unsegmented and conical. Endopod absent. Exopod with 5 terminal aesthetascs (3 long, 2 shorter), without setae.
Antenna (Figure 3b): Endopod present as small bud. Protopod and exopod absent.
Mandible (Figure 3c): Incisor and molar processes developed. Palp absent.
Maxillule (Figure 4a): Coxal endite with 4 plumodenticulate setae. Basial endite with 7 setae (5 terminal cuspidate, 2 subterminal plumodenticulate). Endopod 2-segmented, proximal segment without setae, and 4 terminal (2 + 2) sparsely plumose setae on distal segment. Exopod seta present.
Maxilla (Figure 4b): Coxal endite with 6 plumodenticulate setae. Basial endite bilobed, with 5 + 4 plumodenticulate setae. Unsegmented endopod with 1 + 2 long plumodenticulate setae. Exopod (scaphognathite) with 9 plumose marginal setae.
First maxilliped (Figure 4c): Coxa without setae. Basis with 10 medial sparsely plumodenticulate setae arranged as 2 + 2 + 3 + 3. Endopod 5-segmented with 1,2,1,2,4 (1 subterminal + 3 terminal) sparsely plumodenticulate setae. Exopod unsegmented, with 6 terminal plumose natatory setae.
Second maxilliped (Figure 4d): Coxa without setae. Basis with 4 sparsely plumodenticulate setae arranged 1 + 1 + 1 + 1. Endopod 2-segmented with 0, 1 subterminal serrulate + 4 terminal setae (2 long plumodenticulate, 2 shorter: 1 plumodenticulate and 1 sparsely plumodenticulate). Exopod unsegmented with 6 terminal plumose natatory setae.
Third maxilliped: Absent.
Pleon (Figures 2a, 5a): 5 pleonites. Pleonite 1 with one middorsal simple seta. Pleonites 2–5 with a pair of minute simple setae on posterodorsal margin. Pleonites 2–3 with pair of laterally directed dorsolateral processes, those of pleonite 3 smaller.
Pleopods: Absent.
Telson (Figures 2a, 5a): Trilobed with 2 pairs of 3 serrulate setae on posterior margin, inner setae longest; each lateral lobes crenulated distally.
ZOEA III
(Figures 2b, 5b)
Size: CL = 0.521 ± 0.020 mm; CW = 0.442 ± 0.008 mm, N = 5.
Cephalothorax (Figure 2b): Ventral margin with 1 highly plumose and 2 sparsely setose setae.
Antennule: Exopod with 6 aesthetascs.
Antenna: Endopod enlarged. Exopod present as a simple seta.
Maxillule: Basial endite with 9 setae (3 subterminal plumodenticulate, 5 terminal cuspidate and 1 proximal simple seta).
Maxilla: Basial endite with 5 + 5 plumodenticulate setae. Scaphognathite with 12 plumose marginal setae.
First maxilliped: Exopod with 7–8 terminal plumose natatory setae.
Second maxilliped: Exopod with 7–8 terminal plumose natatory setae.
Third maxilliped: Present as undifferentiated buds.
Pereiopods: All present as buds, slightly segmented, first pair chelate.
Pleon (Figure 5b): Pleonite 1 with 3 middorsal simple setae.
Pleopods: Present on pleonites 2–5 as small buds, endopods absent.
ZOEA IV
(Figures 2c, 3d, e, 4e, f, 5c)
Size: CL = 0.626 ± 0.020 mm; CW = 0.510 ± 0.033 mm, N = 5.
Cephalothorax (Figure 2c): Ventral margin with 1 highly plumose and 7 sparsely setose setae.
Antennule (Figure 3d): Endopod bud present. Exopod with 6 aesthetascs (3 subterminal and 3 terminal).
Antenna (Figure 3e): Endopod more elongated than in zoea III.
Mandible: Palp present as unsegmented bud without setae.
Maxillule (Figure 4e): Coxal endite with 5–6 plumodenticulate setae. Basial endite with 10–12 setae (4–5 subterminal plumodenticulate, 5–6 terminal cuspidate, 1 proximal simple seta).
Maxilla (Figure 4f): Coxal endite with 7 plumodenticulate setae. Scaphognathite with 16 plumose marginal setae.
First maxilliped: Exopod with 7–8 terminal plumose natatory setae.
Second maxilliped: Exopod with 7–8 terminal plumose natatory setae.
Third maxilliped: Triramous. Endopod and exopod present as slightly segmented buds, without setae. Epipod bud present.
Pereiopods: Cheliped and pereiopods slightly segmented without setae.
Pleon (Figures 2c, 5c): Pleonite 1 with 5 middorsal simple setae.
Pleopods (Figures 2c, 5c): Biramous buds more elongated with endopod bud present.
MEGALOPA
(Figures 2d–g, 3f–h, h*, 4g–k, 5d–i)
Size: CL = 0.609 ± 0.042 mm; CW = 0.483 ± 0.016 mm; N = 9.
Cephalothorax (Figures 2d, e): Slightly longer than broad, subquadrate. Rostrum small, ventrally deflected (~70°), with median longitudinal depression. Protogastric, cardiac and mid-posterior region with tubercles. Setation as illustrated. Eyes stalked.
Antennule (Figure 3f): Peduncle 3-segmented, with 2 plumose, 1 simple, and 1 simple setae respectively. Endopod 2-segmented, without setae on proximal segment, and 1 subterminal and 2 terminal simple setae on distal one. Exopod 4-segmented, with 0, 0, 4, 4 aesthetascs and 0, 0, 1, 0 simple seta respectively.
Antenna (Figures 3g, g*): Peduncle 3-segmented without setae and flagellum 2-segmented without setae on first segment, and 2 long terminal setae (and sometimes 1 shorter subterminal) on distal segment.
Mandible (Figures 3h, h*): Palp 3-segmented with 1 or 2 terminal simple or sparsely plumodenticulate setae on distal segment.
Maxillule (Figure 4g): Coxal endite with 11 plumodenticulate setae. Basial endite with 15 setae (2 subterminal plumodenticulate + 5 subterminal cuspidate + 6 terminal cuspidate + 2 proximal plumodenticulate). Endopod unsegmented without seta.
Maxilla (Figure 4h): Coxal endite with 12 plumose setae. Basial endite bilobed with 6 + 7–8 plumodenticulate setae. Endopod unsegmented without setae. Scaphognathite with 23–26 marginal plumose setae plus 3 small simple setae, on the same lateral surface.
First maxilliped (Figure 4i): Epipodite with 2 terminal simple setae. Coxal endite with 4–5 plumodenticulate setae. Basial endite with 4–5 plumodenticulate setae. Endopod unsegmented without setae. Exopod 2-segmented with one terminal plumose setae on proximal segment, and 4 terminal plumose setae on distal segment.
Second maxilliped (Figure 4j): Epipodite absent. Coxa and basis undifferentiated without setae. Endopod 4-segmented with 0, 1 long sparsely setose, 4 plumodenticulate, 5 (2 subterminal sparsely setae + 3 plumodenticulate) setae respectively, dactylus inserted subterminally on propodus. Exopod 2-segmented with one medial simple setae on proximal segment and 4 terminal plumose setae on distal segment.
Third maxilliped (Figure 4k): Epipodite well developed with 7 proximal plumodenticulate setae and 6 long terminal setae. Protopod without setae. Endopod 4-segmented, ischium and merus fused with 5 setae (3 long sparsely setose + 2 small simple setae), carpus with 2 setae (1 plumodenticulate, 1 simple), propodus with 4 plumodenticulate setae (3 terminal, 1 subterminal), dactylus inserted subterminally on propodus with 2 terminal simple setae. Exopod 2-segmented, proximal segment without setae and 3 terminal plumose setae on distal segment.
Pereiopods (Figures 2d, 5e–g): All segments well differentiated. Cheliped sparsely setose as shown. Pereiopods 2–5 thin and setose, with a small spine subteminal in dactylus (Figure 5f, f*).
Sternum (Figure 2g): Maxillipeds sternites fused with 2 simple setae. Chelipeds sternites with 2 simple setae. Sternites of pereiopods 2–5 without setae.
Pleon (Figures 2f, 5d): Sixth pleonite absent, setation as shown.
Pleopods (Figures 2f, 5h, i): Biramous, present on pleonites 2–5. Endopod of pleonites 2–4 with 3 cincinuli and exopod with 6 long terminal plumose natatory setae. Endopod of pleonite 5 with 1–2 cincinuli and exopod with 2–5 long terminal plumose natatory setae. Uropods absent.
Telson (Figures 2f, 5d): Rounded with one pair of simple setae on dorsal margin and one pair on ventral margin.
DNA BARCODING IDENTIFICATION
The 16S mtDNA (439 bp) of the zoea initially identified as belonging to Pinnotheres pisum show a 100% match with the sequences of P. pisum from Germany and France obtained by Schubart et al. (Reference Schubart, Vannini, Cannicci and Fratini2006) and Bui et al. (Reference Bui, Casse, Leignel, Nicolas and Chenais2008) respectively. However, the sequences of larval stages (zoea and megalopa) of Pinnotheres sp. do not match with any sequence deposited in GenBank, or with any other European pinnotherid. The sequences obtained for zoea and megalopa are 100% identical to each other and in the obtained tree both cluster together. A major clade joins them to Pinnotheres pisum and P. pectunculi, clearly separated from N. pinnotheres and A. monodi (Figure 6).
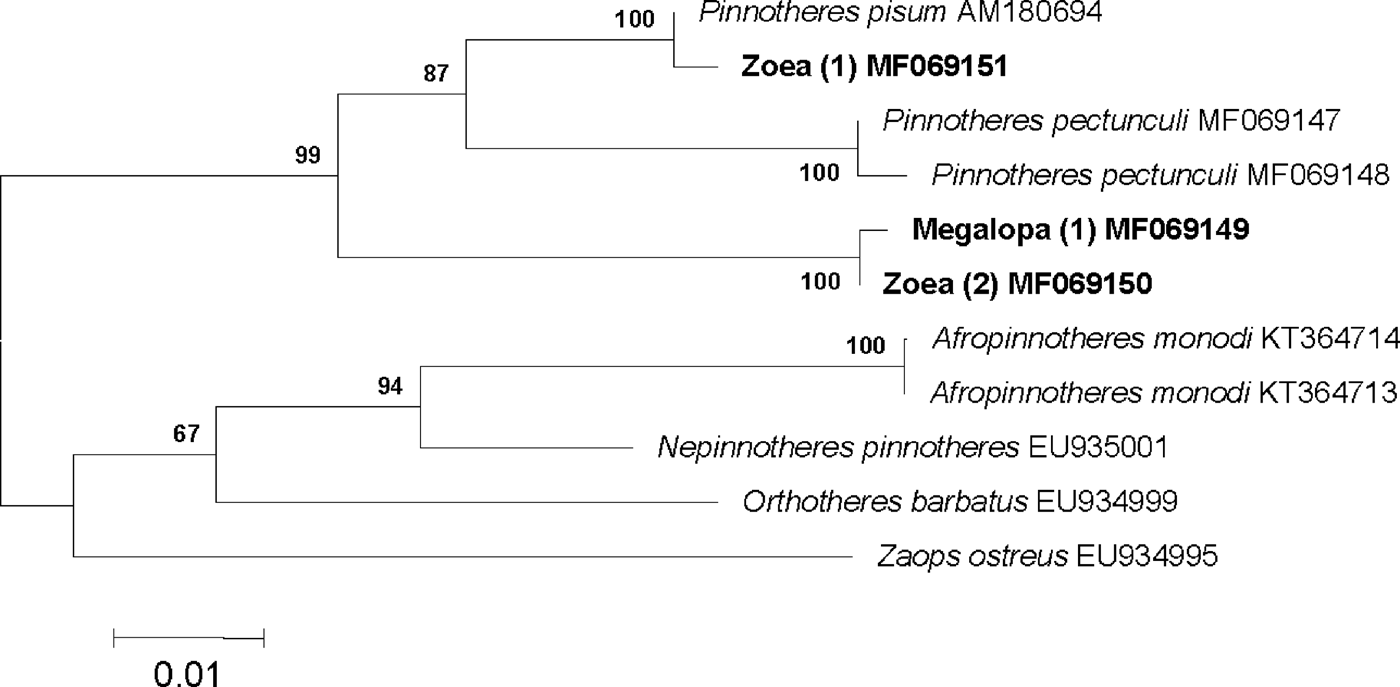
Fig. 6. Topology of neighbour-joining tree based on 540 bp of the 16S mtDNA gene sequences, showing inferred phylogenetic relationships within the European representatives of the family Pinnotheridae with Zaops ostreus and Orthotheres barbatus as outgroups. Numbers close to nodes indicate bootstrap support (only values above 60% shown). GenBank accession numbers are shown after name of species.
DISCUSSION
Eradication and control of invasive species are often possible only if populations are detected when they are small and localized (Inglis et al., Reference Inglis, Hurren, Oldman and Haskew2006). For early detection accurate surveillance and monitoring programmes must be established, and these management programmes must take into account the last DNA-based techniques (Darling, Reference Darling2015).
For successful early detection of NIS a good knowledge of the native fauna is required. This knowledge should include both adult/larval morphology and DNA barcodes. In the present study concerning European Pinnotheridae this knowledge has been the key for the detection of an unknown species.
Firstly, the morphology of these larvae caught our attention. Pinnotherid zoea are clearly distinguishable from the rest of the brachyuran larvae due to a typical trilobate telson (see Figures 5a–c), but when comparing them to known European pinnotherid (Atkins, Reference Atkins1954; Becker, Reference Becker2010; Marco-Herrero et al., Reference Marco-Herrero, Drake, Gonzalez-Gordillo and Cuesta2016) these zoeae of Pinnotheres sp. did not exhibit dorsal and lateral spines in the cephalothorax (see Figures 3a–c). In Pinnotheres pisum and P. pectunculi, lateral spines are present, and N. pinnotheres and A. monodi show both dorsal and lateral spines. With these findings, a more accurate study of the megalopae from the same samples also indicated that these megalopae were different from those of P. pisum (for example in the number of antennal flagellum segments, 3 in P. pisum and only 2 in Pinnotheres sp.), and N. pinnotheres and A. monodi (pleon with 6 pleonites and only 5 pleonites in Pinnotheres sp.). However, due to the lack of data for the megalopa stage of P. pectunculi, this species cannot be discarded.
The results of comparing the fragments of 16S mtDNA confirmed that the zoeae and the megalopae here analysed belong to the same species (Figure 6) and that this species is not any of the four previously recognized European pinnotherids, or any of the Pinnotheridae currently included in the GenBank databases.
The lack of cephalothoracic dorsal and lateral spines in zoeal stages of Pinnotheres sp. allow to distinguish it from the rest of European pinnotherids, but this feature does not place it close to other species with these characters, for example Zaops ostreus (Say, 1817) (see Sandoz & Hopkins, Reference Sandoz and Hopkins1947) or Orthotheres barbatus (Desbonne, 1867) (see Bolaños et al., Reference Bolaños, Rivero, Hernández, Magán, Hernández, Cuesta and Felder2005) (Figure 3).
From the DNA analysis it was clear that Pinnotheres sp. is closer to species of the genus Pinnotheres than any other Pinnotheridae genera, but currently Pinnotheres comprises 64 species (Palacios-Theil, Reference Palacios-Theil2014) and there are DNA sequences available for only two species of this genus, P. pisum and P. pectunculi.
Regarding the origin of this species there are two main possible explanations, (1) it could have been introduced within ballast water or as part of ship hull fouling. Samples were collected in an area close to the Guadalquivir estuary, a place where several species have been introduced as a result of the transport by one or more of the many vessels entering and leaving the Sevilla Harbour, which experiences considerable international ship traffic (Cuesta et al., Reference Cuesta, González-Ortegón, Drake and Rodríguez2004). Taking into account that Pinnotheres species are mainly hosted by bivalve and ascidian species common in fouling communities and allowing alive transport for long time periods, Pinnotheres sp. could have come from anywhere in the world. (2) Considering that Afropinnotheres monodi was detected in 2010 and the first samples of this species were collected in 1995 (López de la Rosa, Reference López de la Rosa, García Raso and Rodríguez2002), this species could be another African species just in an initial stage of establishment (taking into account that larvae are a sign of reproduction). It could be arriving in southern Europe in a natural northward expansion of this species. One of the effects of global warming is the northward expansion of African species, and this could be another example. Manning & Holthuis (Reference Manning and Holthuis1981) listed a total of 11 Pinnotheres species in West Africa, including five Pinnotheres spp. More recently Manning (Reference Manning1993), studying West African Pinnotheres, erected six new genera (Afropinnotheres, Alainotheres, Ernestotheres, Hospitotheres, Nepinnotheres and Waldotheres) to remove 14 species from this genus. Except for Afropinnotheres monodi and Nepinnotheres pinnotheres no DNA data are available for any of these 14 species of African pinnotherid species.
The information provided here (larval morphology and DNA sequences) will help in the near future to identify this unknown species of Pinnotheridae (when adults are collected), and to follow its spread in European waters through the presence of larvae in plankton samples.
ACKNOWLEDGEMENTS
We are grateful to César Vilas (project leader) and the rest of researchers of the ECOBOGUE project for their participation in the campaigns where samples were obtained, and Carlos Sánchez Nieto for his laboratory work. Emma Palacios-Theil and one anonymous reviewer provided useful comments and corrections that clearly improved this paper. Thanks are also due to Dr Michael Türkay, curator of Senckenberg Museum for loan and allowing DNA extraction of the P. pectunculi specimens.
FINANCIAL SUPPORT
This study was partially funded by the Andalusian ‘Consejería de Economía, Innovación y Ciencia’ with ‘Junta de Andalucía’ and European FEDER funds through Project ECOBOGUE (RNM-7467), the Spanish ‘Ministerio de Ciencia e Innovación, Plan Nacional I + D’ and the European FEDER funds through project MEGALOPADN (CGL2009–11225) and the Spanish ‘Ministerio de Economía y Competitividad, Plan Nacional I + D’ and the European FEDER funds through project AFROBIV (CGL2014-53557-P). The ‘Ministerio de Ciencia e Innovación’ also supported a FPI PhD grant to EMH (BES-2010-033297).








