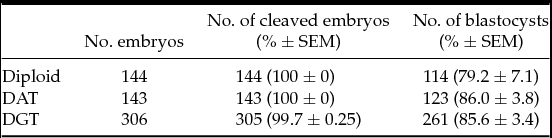Introduction
Polyploidy occurs relatively frequently among some animal groups, in particular in fish and amphibians (McKinnell, Reference McKinnell1964; Cuellar & Uyeno, Reference Cuellar and Uyeno1972). In mammals, triploids and tetraploids show severe developmental anomalies, resulting in lethality at midgestation (Triploid: embryonic day 11.5 (E11.5), Tetraploid: E14.5) (Kaufman et al., Reference Kaufman, Lee and Speirs1989a; Kaufman & Webb, Reference Kaufman and Webb1990; Henery & Kaufman, Reference Henery and Kaufman1992). As an exception, some human triploids could develop until the later stages of gestation, and rarely survive more than one month after birth (46–308 days), exhibiting neonatal mortality (Fryns et al., Reference Fryns, van de Kerckhove, Goddeeris and van den Berghe1977; Arvidsson et al., Reference Arvidsson, Hamberg, Johnsson, Myrdal, Anneren and Brun1986; Sherard et al., Reference Sherard, Bean, Bove, DelDuca, Esterly, Karcsh, Munshi, Reamer, Suazo and Wilmoth1986; Niemann-Seyde et al., Reference Niemann-Seyde, Rehder and Zoll1993; Hasegawa et al., Reference Hasegawa, Harada, Ikeda, Ishii, Hokuto, Kasai, Tanaka, Fukuzawa, Niikawa and Matsuo1999; Iliopoulos et al., Reference Iliopoulos, Vassiliou, Sekerli, Sidiropoulou, Tsiga, Dimopoulou and Voyiatzis2005). Triploidy has been estimated to occur in 1–3% of pregnancy cases in humans, which is one of primary causes of not only early miscarriage but also aberrations in a fetus or infant (Carr, Reference Carr1971a, Reference Carrb; Beatty, Reference Beatty1978; Niebuhr, Reference Niebuhr1974). The possible causes include dispermy or inhibited extrusion of the second polar body during fertilization (Jacobs et al., Reference Jacobs, Angell, Buchanan, Hassold, Matsuyama and Manuel1978). Hence, triploid embryos could be classified into two types according to whether the extra haploid set of chromosomes (in triploidy) was derived from a sperm or oocyte, namely diandric triploid (DAT) and digynic triploid (DGT) embryos, in which the extra haploid set is either paternal or maternal, respectively. Nonetheless, it is unknown how triploidy in mammals leads to lethality, which is not the case in other animal groups such as fish and amphibians.
Because the triploid human conceptuses at pre- and postimplantation stages are unavailable for experimental analysis, researchers have to use experimentally constructed mammalian triploid embryos that are not human. In mice, both DAT and DGT embryos show distinct phenotypes, particularly on E9.5 (Kaufman et al., Reference Kaufman, Lee and Speirs1989a). The DGT fetuses are significantly smaller than diploid fetuses with many morphological aberrations such as cardiac defects, craniofacial anomalies, and neural tube defects, although the frequency of these problems has not been determined after E9.5 precisely (Kaufman & Speirs, Reference Kaufman and Speirs1987; Kaufman et al., Reference Kaufman, Lee and Speirs1989a). In contrast, all of the DAT fetuses are relatively normal in terms of morphological characteristics. DAT fetuses had about 20–25 pairs of somites, of which the cephalic and cardiac regions were developmentally advanced as well as those of diploid fetuses at E9.5 (Kaufman et al., Reference Kaufman, Lee and Speirs1989a; Henery & Kaufman, Reference Henery and Kaufman1992). In both types of triploid embryos, the composition of sex chromosomes; i.e., XXX and XXY, appears not to cause significant phenotypical changes, although DAT fetuses with the XYY set have not been observed (Kaufman et al., Reference Kaufman, Speirs and Lee1989b; Speirs & Kaufman, Reference Speirs and Kaufman1989). The above-mentioned contrasting phenotypes of DAT and DGT embryos are evidence that the male and female genomes are not functionally equivalent, which is also the case for uniparental gynogenetic (parthenogenetic) and androgenetic diploids (McGrath & Solter, Reference McGrath and Solter1984; Surani et al., Reference Surani, Barton and Norris1984; Surani et al., Reference Surani, Barton and Norris1986). The functional and transcriptional asymmetry between parental genomes is explainable in terms of genomic imprinting. The latter is a mechanism for making one parental allele different from the other, and this process leads to parental-origin-dependent transcription in mammals.
Imprinted genes, where only one of the two parental chromosome copies is expressed, are the result of epigenetic modifications, such as DNA methylation (Li et al., Reference Li, Beard and Jaenisch1993). Acquisition of methylation of key controlling elements, i.e., imprinting of control regions (ICRs), occurs in parental germlines during male or female gametogenesis and these acquired modifications (methylation) are usually retained thereafter (Obata & Kono, Reference Obata and Kono2002). A number of imprinted genes are clustered with cis-acting ICRs carrying allele-specific methylation marks (Birger et al., Reference Birger, Shemer, Perk and Razin1999; Thorvaldsen et al., Reference Thorvaldsen, Duran and Bartolomei1998; Kim et al., Reference Kim, Ekram, Kim, Faisal, Frey, Huang, Tran, Kim and Yu2012; Lin et al., Reference Lin, Youngson, Takada, Seitz, Reik, Paulsen, Cavaille and Ferguson-Smith2003; Verona et al., Reference Verona, Thorvaldsen, Reese and Bartolomei2008; Shiura et al., Reference Shiura, Nakamura, Hikichi, Hino, Oda, Suzuki-Migishima, Kohda, Kaneko-ishino and Ishino2009). Because the dosage of imprinted genes is crucial for normal development, growth, and behavior in mammals, the loss or overexpression of those transcripts can cause dysontogenesis in some cases, resulting in death of uniparental mouse embryos (Sotomaru et al., Reference Sotomaru, Katsuzawa, Hatada, Obata, Sasaki and Kono2002; Ogawa et al., Reference Ogawa, Wu, Komiyama, Obata and Kono2006). In contrast, nothing is currently known about transcriptional dynamics of imprinted genes in triploid mouse embryos.
Uniparental diploid embryos in mice are thought to be a good experimental model for elucidation of the roles of parental genomes during early embryogenesis (Surani et al., Reference Surani, Barton and Norris1986). Recent studies have demonstrated that the parent-specific expression of imprinted genes is not always maintained in uniparental fetuses (Sotomaru et al., Reference Sotomaru, Katsuzawa, Hatada, Obata, Sasaki and Kono2002; Ogawa et al., Reference Ogawa, Wu, Komiyama, Obata and Kono2006). For instance, H19 and Igf2r, which are usually expressed from maternal alleles are transcribed in androgenetic fetuses consisting of paternal genomes only (Sotomaru et al., Reference Sotomaru, Katsuzawa, Hatada, Obata, Sasaki and Kono2002). Furthermore, the expression of U2af1, which is a paternally expressed gene was confirmed in parthenogenetic fetuses (Ogawa et al., Reference Ogawa, Wu, Komiyama, Obata and Kono2006). These results support the existence of yet unknown mechanism(s), such as those involving trans-acting parental alleles (act between parental genomes), that regulate the appropriate expression of imprinted genes (time and location). Because the imbalance of parental genomes in an embryo is one of the innate characteristics of triploid embryos per se, studies of expression of imprinted genes in both DAT and DGT embryos might be a promising way to elucidate why triploidy leads to mortality in mammals but not in other animal groups.
Here, we report the morphological features of murine DAT and DGT embryos on E10.5 produced using the pronuclear transplantation technique. We further examined expression patterns of eight primary imprinted genes in each type of triploid fetus.
Materials and methods
Preparation of fertilized and parthenogenetic embryos
In order to create diploid fertilized embryos with an external haploid set from the other fertilized/parthenogenetic embryos by means of pronuclear transplantation, we carried out in vitro fertilization (IVF) and artificial oocyte activation. The oocyte donors were female B6D2F1 mice (C57BL/6N × DBA/2). They were superovulated with injections of 7.5 IU (international units) of equine chorionic gonadotropin (eCG; ASKA Pharmaceutical, Tokyo, Japan) and 7.5 IU of human chorionic gonadotropin (hCG; ASKA Pharmaceutical) given 48 h apart. Oocytes at the metaphase of the second meiosis (MII) were remove from the oviducts 16 h after hCG administration, and used for IVF and oocyte activation. All research and experimental protocols were approved by the Regulatory Committee for the Care and Use of Laboratory Animals, Hokkaido University.
IVF was performed using a previously described method (Kono et al., Reference Kono, Sotomaru, Sato and Nakahara1993). In brief, spermatozoa were collected from the cauda epididymis of mature B6D2F1 male mice and preincubated in the HTF medium (Quinn et al., Reference Quinn, Kerin and Warnes1985) in the humidified atmosphere containing 5% CO2 at 37°C for 1.5 h. MII oocytes that were collected as described above were transferred into 100-μl droplets of the HTF medium. Likewise, the spermatozoa after preincubation were added to the same HTF droplets. Six hours after insemination, the single-cell embryos were washed with the M2 medium (Quinn et al., Reference Quinn, Barros and Whittingham1982) and transferred into droplets of the M16 medium containing 0.1 mM ethylene diamine tetraacetic acid (EDTA; DOJINDO LABORATORIES, Kumamoto, Japan) (Whittingham, Reference Whittingham1971). The IVF embryos were used as donors and recipients of pronuclear transplantation.
For creation of parthenogenetic embryos, the cumulus cells of collected MII oocytes were removed by means of a brief incubation in hyaluronidase (300 units/ml; Sigma-Aldrich, Tokyo, Japan) in the M2 medium. After the hyaluronidase treatment, stripped oocytes were washed with the M2 medium several times, and transferred into droplets consisting of 50 μL of the Ca2+-free M16 medium containing 10 mM SrCl2 (KANTO CHEMICAL, Tokyo, Japan). After incubation for 4–6 h, activated haploid embryos containing both a second polar body and a female pronucleus were collected into M16 droplets at 37°C in the humidified atmosphere containing 5% CO2 until they were used for pronuclear transplant as a source of a maternal haploid set of chromosomes.
Pronuclear transplantation
Prior to micromanipulation, donor embryos; i.e., the IVF and activated embryos, were incubated for 1 h in the M16 medium and for 45 min in the same medium containing cytochalasin B (5 μg/ml; Sigma-Aldrich) and colcemid (0.1 μg/ml; Wako Pure Chemical Industries, Osaka, Japan). The male pronuclei were easily recognized within the IVF embryos because of the larger features compared to the female pronuclei. The male pronuclei were used as a source of an external haploid paternal genome for the production of DAT embryos (Fig. 1).

Figure 1 Creation of diandric and digynic triploid mouse embryos using the pronuclear transfer technique. To create a diandric triploid (DAT) embryo (3n), we fused a recipient IVF embryo (2n) with a male pronucleus (n) derived from another IVF embryo. Digynic triploid (DGT) embryos (3n) were created by transferring a female pronucleus derived from an activated haploid embryo (n) into a recipient IVF embryo (2n).
For creation of digynic triploid (DGT) embryos, the female pronuclei in the activated embryos (that were produced without diploidization by means of a cytoskeleton inhibitor) were used as a source of external haploid maternal genome (Fig. 1). Either a male or female pronucleus was isolated with a small amount of the cytoplasm from donor embryos and in the presence of inactivated Sendai virus (hemagglutinating activity 2700 U/ml), was inserted into the perivitelline space of another IVF embryo. The IVF embryos that successfully fused with a karyoplast containing a male or female pronucleus were further cultured for 1 h.
In vitro culture and embryo transfer
Both DAT and DGT embryos were cultured in the M16 medium supplemented with 0.1 mM EDTA in the humidified atmosphere containing 5% CO2 at 37°C. The number of cleaved embryos and blastocysts was assessed after 1.0 and 3.5 days of culture, respectively. The resulting blastocysts were transferred into the uterine horns of female mice after 2.5 days of pseudopregnancy. To assess development, the recipients were euthanized on E10.5, and conceptuses were excised from the uterus. Fetuses with noticeable heartbeat were used for the following tests, including measurement of crown–rump length, assessment of embryonic turning, and quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) analysis of gene expression. The diploid embryos were also cultured and implanted in uterine horns of pseudopregnant females in the same manner, and the resulting fetuses served as developmentally matched controls.
Quantitative real-time PCR analysis
Total RNA from one fetus on E10.5 in each group, namely, the DAT, digynic triploid, or control diploid group, was prepared using the ReliaPrep RNA Cell Miniprep System (Promega, Madison, WI) according to the manufacturer's instructions. RNA concentrations were assessed using NanoDrop (Thermo Fisher Scientific, Wilmington, DE, USA), and then, quantity of the extracted RNA was normalized to the concentration of 1 μg among the number of samples necessary for one replicate. After normalization of RNA quantity, cDNA samples were synthesized using the ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan) in a reaction mixture (10 μl) containing 1 μg of the total RNA extracted from each fetus. Finally, the RNA samples from three fetuses in each group were prepared, and we performed quantitative analysis of the gene expression using real-time PCR (LightCycler; Roche Diagnostics, Basel, Switzerland) after preparing the reaction mixture using Thunderbird SYBR qPCR Mix (Toyobo) in triplicate as described previously (n = 3) (Nagatomo et al., Reference Nagatomo, Kagawa, Kishi, Takuma, Sada, Yamanaka, Abe, Wada, Takahashi, Kono and Kawahara2013). The primers are listed in the Appendix: Table A1. The transcript levels were calculated relative to the transcription of the housekeeping gene Gapdh in every sample.
Statistical analysis
We compared the means using Student's t-test. Data on the number of fetuses with embryonic turning were analyzed using the chi-squared test. All calculations were performed using the software Statview (Abacus Concepts, Inc., Berkeley, CA, USA). A P-value either < 0.05 or < 0.01 was assumed to denote statistical significance.
Grant support
This work was supported by a Grant-in-Aid for Research in Priority Areas to Young Scientists (B) to M.K. (No. 24780265) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Results
Developmental competence of triploid mouse embryos during pre- and postimplantation development
We first analyzed the developmental competence of triploid embryos until the blastocyst stage. Triploid mouse embryos were produced using pronuclear transplantation of one male and female pronucleus from IVF diploid and parthenogenetic haploid embryo into another IVF embryo, respectively: DAT and DGT embryos (Fig. 1). There were no significant differences in the rate of blastocyst formation among diploid, DAT, and DGT embryos (79.2 ± 7.1%, 86.0 ± 3.8%, and 85.6 ± 3.4%, respectively; mean ± standard error of the mean (SEM); Table 1). Next, we assessed the developmental characteristics of DAT and DGT embryos until E10.5. Compared to diploid conceptuses (85.2 ± 9.3%), the implantation rate of triploid conceptuses significantly decreased (P < 0.05; Table 2). In addition, the proportions of a live fetus (both DAT and DGT, 5.1 ± 2.4% and 5.2 ± 2.1%, respectively) were much smaller than in diploids (53.7 ± 5.0%; Table 2). The number of resorbed fetuses was 15 in diploid, 41 in DAT, and 68 in DGT. These results demonstrated that triploidy of the mouse embryos prevented them from developing beyond the early implantation stage.
Table 1 Preimplantation development of triploid mouse embryos
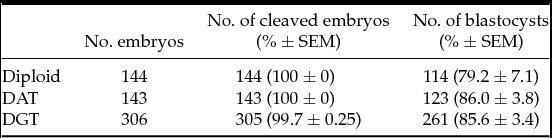
Diploid: IVF embryos. DAT: Diandric triploid embryos. DGT: Digynic triploid embryos. There were no significant differences in the rate of cleaved embryos and blastocyst formation to controls, or diploid. SEM, standard error of the mean.
Table 2 Postimplantation development of triploid embryos up to E10.5
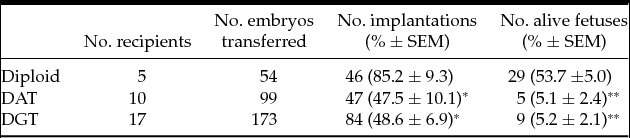
Diploid: IVF embryos. DAT: Diandric triploid embryos. DGT: Digynic triploid embryos.
Alive fetuses exhibited obvious heartbeat when recovered from uterine horns of recipients.
Asterisk means the significant difference to the developmentally matched controls (Diploid)
SEM, standard error of the mean. *P < 0.05; **P < 0.01.
Morphological characterization of triploid mouse conceptuses
DAT and DGT conceptuses on E10.5 showed contrasting phenotypes in relation to the entire size and vascularization on the surface of yolk sac (Fig. 2). The size of a DGT conceptus, particularly extraembryonic tissues, was smaller compared with DAT and diploid conceptuses, concomitant with impaired vasculogenesis on the surface of the yolk sac (Fig. 2 C). At the same time, there appeared to be no differences in both entire size and vasculogenesis between diploid and DAT conceptuses. In order to assess morphological characteristics more precisely, we analyzed the crown–rump length and embryonic turning of diploid, DAT, and DGT fetuses on E10.5 (Fig. 2 D–G). As expected, the crown–rump length of DGT fetus (1.87 ± 0.13 mm) was significantly smaller than that of diploids (4.81 ± 0.05 mm; P < 0.01; Fig. 2 H) and DAT (3.86 ± 0.43 mm; P < 0.01; Fig. 2 H).

Figure 2 Morphological characterization of triploid mouse conceptuses at E10.5. The whole diploid (A), DAT (B), and DGT (C) conceptuses are shown. Note that the DAT conceptus is equivalent in size to the diploid. In contrast, a DGT conceptus is much smaller than the other types, concomitantly with impaired vasculogenesis on the surface of the yolk sac (see the inset). Representative fetuses from diploid (D), DAT (E), and DGT (F, G) embryos are shown. (H) Assessment of the crown–rump length revealed that the length of DGT fetuses (1.87 ± 0.13 mm; n = 5) was much smaller than that of diploids (4.81 ± 0.05 mm; n = 3; P < 0.01) and DAT (3.86 ± 0.43 mm; n = 4; P < 0.01). Note that there was no obvious difference in the crown–rump length between diploid and DAT fetuses. (I) Evaluation of the number of fetuses with embryonic turning revealed that this percentage was significantly decreased in DGT fetuses (P < 0.01), although only two of nine DGT live fetuses with normal turning showed embryonic turning as shown in (G). The scale bar is 1 mm. The data are presented as mean ± standard error of the mean (SEM). *P < 0.05, **P < 0.01, respectively.
No significant difference was observed in the crown–rump length between diploid and DAT fetuses. Moreover, all DAT fetuses were morphologically normal, similar to the diploid one (Fig. 2 D, E). This result is consistent with the observation that all fetuses in both diploid and DAT groups exhibited normal embryonic turning on E10.5 (Fig. 2 I). In contrast, the number of fetuses (%) showing embryonic turning in the DGT group significantly decreased, although DGT fetuses with normal turning were found occasionally (Fig. 2 F, G, I). These results showed that the extra copy of a haploid genome had detrimental effects on fetal development during the early somite stage, ~E10.5. The extent of inhibitory effects on fetal growth depended on whether the extra copy of a haploid genome in a triploid fetus came from a male or female pronucleus.
Expression levels of imprinted genes in triploid mouse fetuses
We wanted to better understand the relationship of morphological characteristics in a triploid fetus on E10.5 with the expression pattern of imprinted genes. To this end, we performed quantitative expression analysis of eight imprinted genes (H19, Gtl2, Igf2r, and Grb10 among maternally expressed genes and Igf2, Dlk1, Ndn, and Peg3 among paternally expressed genes) in diploid, DAT and DGT groups by means of real-time RT-PCR (Fig. 3). As expected, the expression level of paternally expressed genes, Igf2, Dlk1, Ndn, and Peg3, remained significantly reduced, and that of maternally expressed genes, Igf2r and Grb10, increased in DGT fetuses (P < 0.05; Fig. 3). Additionally, in DAT fetuses, Igf2 mRNA expression was approximately two-fold higher than in diploid fetuses.

Figure 3 Graphical representation of the transcription levels of eight imprinted genes in the triploid mouse fetuses at E10.5. The expression of eight imprinted genes (H19, Gtl2, Igf2r, and Grb10 among maternally expressed genes and Igf2, Dlk1, Ndn, and Peg3 among paternally expressed genes) in diploid, DAT, and DGT fetuses (n = 3 each group) is shown. The values represent the levels of expression relative to that of an internal control gene (Gapdh). Diploid (n = 3, gray), DAT (n = 3, blue), and DGT (n = 3, pink). The data are presented as mean ± standard error of the mean (SEM). *P < 0.05.
Unexpectedly, DAT fetuses showed appropriate expression levels equivalent to that of diploid fetuses in the paternally expressed genes Dlk1, Ndn, and Peg3. In addition, among maternally expressed genes, the H19, Gtl2, Igf2r, and Grb10 mRNA expression levels in DAT fetuses expressed approximately normal levels seen in diploid fetuses. Even though DGT fetuses possessed the redundant maternal haploid set of chromosomes, the H19 mRNA expression level in DGT fetuses significantly decreased (P < 0.05; Fig. 3). These results serve as clear demonstration that DAT fetuses had an almost normal expression pattern of imprinted genes except for Igf2 mRNA expression, whereas in DGT fetuses, seven out of the eight imprinted genes tested exhibited aberrant expression patterns, consistent with the results of morphological analysis that DAT fetuses appeared to be relatively normal in contrast to DGT fetuses.
We further investigated a non-imprinted gene, Hoxc9, in DAT and DGT fetuses as well as imprinted genes (Appendix: Fig. A1). Hoxc9 is essential for fetal morphogenesis including formation of the vertebra, sternum and ribs (Suemori et al., Reference Suemori, Takahashi and Noguchi1995). There were no significant differences in Hoxc9 expression level between diploid and DAT fetuses. However, the expression level of DGT fetuses markedly reduced comparing diploids (P < 0.01). This result provided the possibility that expression levels of non-imprinted genes including Hoxc9 were disrupted in DGT fetuses, not but in DAT fetuses. This is consistent with our observation that DGT fetuses showed severer morphological aberrations than DAT fetuses (Fig. 2).
Discussion
Polyploidy exists across all major taxonomical groups of animals including fish and amphibians (Cuellar & Uyeno, Reference Cuellar and Uyeno1972; McKinnell, Reference McKinnell1964). Mammals also show the types of localized polyploidy, endopolyploidy, where only some tissues are polyploid, for example platelets, bone marrow, hepatic precursor cells, or trophoblast (Brodsky & Uryvaeva, Reference Brodsky and Uryvaeva1977; Carriere, Reference Carriere1969; Ilgren, Reference Ilgren1980; Saito et al., Reference Saito, Takada, Yamada and Fujimoto1996). In addition, the diploid–triploid and diploid–tetraploid mouse chimeras could show full term development (Lu & Markert, Reference Lu and Markert1980; Suwinska et al., Reference Suwinska, Ozdzenski, Waksmundzka and Tarkowski2005). Nevertheless, in mammals, polyploidy including triploidy is definitely nonviable (the latter means that one additional haploid set of chromosomes of either maternal or paternal origin is present in the karyotype) (Kaufman et al., Reference Kaufman, Lee and Speirs1989a; Kaufman & Webb, Reference Kaufman and Webb1990). Likewise, it is well known that uniparental diploidy (the genome consists of either two maternally or two paternally derived haploid sets of chromosomes) causes early embryonic mortality (E9.5), which can be explained by functional differences between parental genomes as a result of genomic imprinting (Surani et al., Reference Surani, Barton and Norris1986). Genomic imprinting leads to the parental-origin-dependent pattern of gene expression, which means that a group of genes, called imprinted genes, is transcribed from one parental allele (Surani et al., Reference Surani, Barton and Norris1984). Therefore, the contribution of these complementary parental genetic programs is essential for proper development in mammals (McGrath & Solter, Reference McGrath and Solter1984). Nonetheless, there are no data in the literature on the relationship between developmental characteristics of triploid embryos and their expression patterns of imprinted genes. In this work, we report morphological phenotypes of DAT and DGT embryos at E10.5, where the embryos were created using the pronuclear transfer technique. We examined the expression patterns of eight primary imprinted genes in each type of triploid fetus.
First, we determined the morphological characteristics of DAT and DGT fetuses at E10.5. From the standpoint of fetal growth, DGT fetuses were much smaller than the diploids; in contrast, DAT fetuses were similar to diploids in size. Additionally, the size of both types of triploids never exceeded that of diploids. These results point to two possible explanations as follows. One is that in triploids, the additional haploid set of chromosomes of either maternal or paternal origin has negative effects on postimplantation development (and there are mutual effects of reverse causation). The second explanation is that a triploid genome itself restricts fetal growth via some mechanism that detects the copy number of chromosomes. If we assume that the paternally and maternally derived genomes function for the benefit and detriment of fetal growth, respectively, then the morphological characteristics of triploids in this study are expected and logical results (Kaufman et al., Reference Kaufman, Lee and Speirs1989a). Furthermore, the morphological phenotypes of triploid conceptuses show clearer differences between these functions of two parental genomes compared to experimental models that involve parthenogenetic and androgenetic conceptuses. These differences are well pronounced in both fetal and extraembryonic development because androgenetic conceptuses show growth retardation compared to parthenogenetic ones (Surani et al., Reference Surani, Barton and Norris1986). In addition, a growing body of evidence points to the possibility that fetal growth is restricted by a triploid karyotype per se, regardless of which side (paternal or maternal) the triploidy is biased toward. This notion may be supported by the observation that DGT embryos show delayed cleavage during the preimplantation period, compared with developmentally matched controls (Takagi & Sasaki, Reference Takagi and Sasaki1976). Although this result was obtained with preimplantation embryos, there might be a delay in triploid embryos in the duration of the cell cycle during the postimplantation period.
In order to gain a further understanding of differences between DAT and DGT fetuses during postimplantation development, we analyzed mRNA expression patterns of eight imprinted genes including four paternally expressed genes and four maternally expressed genes in DAT and DGT fetuses. Among them, Igf2, Dlk1, Grb10, and Igf2r are directly responsible for growth and development of fetuses (Charalambous et al., Reference Charalambous, Smith, Bennett, Crew, Mackenzie and Ward2003; DeChiara et al., Reference DeChiara, Efstratiadis and Robertson1990; Ludwig et al., Reference Ludwig, Eggenschwiler, Fisher, D’Ercole, Davenport and Efstratiadis1996; Moon et al., Reference Moon, Smas, Lee, Villena, Kim, Yun and Sul2002). The first two genes in the list strongly promote the fetal size; in contrast, the last two genes, Grb10 and Igf2r, act as a potent growth inhibitor and a mediator, respectively, of insulin-like growth factor signaling, which promotes growth (DeChiara et al., Reference DeChiara, Efstratiadis and Robertson1990; Ludwig et al., Reference Ludwig, Eggenschwiler, Fisher, D’Ercole, Davenport and Efstratiadis1996; Moon et al., Reference Moon, Smas, Lee, Villena, Kim, Yun and Sul2002; Charalambous et al., Reference Charalambous, Smith, Bennett, Crew, Mackenzie and Ward2003). As expected, in DGT fetuses, the expression level of paternally expressed genes, Igf2, Dlk1, Ndn, and Peg3, remained significantly reduced, and that of maternally expressed genes, Igf2r and Grb10, increased (Fig. 3). These data are consistent with severe growth retardation of DGT fetuses (Fig. 2). We also found decreased expression of the Ndn and Peg3 genes that is responsible for the neurodevelopmental aberrations in DGT fetuses (Broad et al., Reference Broad, Curley and Keverne2009; Li et al., Reference Li, Keverne, Aparicio, Ishino, Barton and Surani1999; Pagliardini et al., Reference Pagliardini, Ren, Wevrick and Greer2005; Yang et al., Reference Yang, Adamson, Resnick, Leff, Wevrick, Francke, Jenkins, Copeland and Brannan1998). One early study showed craniofacial anomalies such as axial neural tube defects in DGT fetuses at E9.5 (Kaufman et al., Reference Kaufman, Lee and Speirs1989a), and in this study, we also observed severe morphological aberrations in DGT fetuses at E10.5 (Fig. 2). These problems might be due to the reduced expression of Ndn and Peg3 in DGT fetuses.
Contrary to overexpression of Igf2 in DAT fetuses that is particularly important for fetal growth during embryogenesis (DeChiara et al., Reference DeChiara, Robertson and Efstratiadis1991), the body size of DAT fetus tended to be smaller than that of diploid fetus. One possible reason is the polyploid states. Tetraploid fetuses possessing twice DNA contents also show smaller body size than developmentally matched controls (Kaufman & Webb, Reference Kaufman and Webb1990). This fact indicates that polyploidy per se has negative effect on fetal size for any reason. Therefore, it was possible that fetal-size-promoting effect by Igf2 mRNA overexpression in DAT fetus was compensated by polyploid state. Additionally, both DAT and DGT fetuses at E10.5 showed unregulated expression of the imprinted genes examined (Fig. 3). A likely explanation is that loss of methylation in these triploid fetuses happened during embryo manipulation or pregnancy, and then caused aberrant expressions of imprinted genes (Doherty et al., Reference Doherty, Mann, Tremblay, Bartolomei and Schultz2000; Sato et al., Reference Sato, Otsu, Negishi, Utsunomiya and Arima2007).
When taken together, our results demonstrate that imprinted genes in mouse triploid fetuses show distinctive expression patterns independent of the number of parental-origin haploid sets. These unregulated expression patterns of imprinted genes observed in triploids suggest that the appropriate number of parental alleles is essential for the maintenance of the parentally specific expression. Further research using triploid mouse embryos could provide a greater insight into the characteristics of mammalian ontogeny governed by both DNA ploidy and asymmetrical functions of parental genomes.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Supplementary Material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0967199414000550.