Published online by Cambridge University Press: 13 May 2004
In order to improve our knowledge about the taxonomic status and the population structure of the causative agent of Human African Trypanosomiasis in the Central African subregion, 169 newly isolated stocks, of which 16 came from pigs, and 5 reference stocks, were characterized by multilocus enzyme electrophoresis, for 17 genetic loci. We identified 22 different isoenzyme profiles or zymodemes, many of which showed limited differences between them. These zymodemes were equated to multilocus genotypes. UPGMA dendrograms revealed one main group: Trypanosoma brucei gambiense group I and 3 T. brucei ‘non-gambiense’ stocks. T. b. gambiense group I zymodemes were very homogenous, grouping all the human stocks and 31% of the pig stocks. Two main zymodemes (Z1 and Z3) grouping 74% of the stocks were found in different remote countries. The genetic distances were relatively high in T. brucei ‘non-gambiense’ zymodemes, regrouping 69% of pig stocks. The analysis of linkage disequilibrium was in favour of a predominantly clonal population structure. This was supported by the ubiquitous occurrence of the main zymodemes, suggesting genetic stability in time and space of this parasite's natural clones. However, in some cases an epidemic population structure could not be ruled out. Our study also suggested that the domestic pig was a probable reservoir host for T. b. gambiense group I in Cameroon.
Human African Trypanosomiasis (HAT), formerly called sleeping sickness, remains a major health threat in most sub-Saharan African countries due to its alarming upward trend observed in this zone. WHO (1998) estimated that over 60 million people are at risk, less than 4 million of whom are presently under appropriate surveillance, and that about 300000 new cases occur each year. Moreover, current drugs especially those used for the treatment of the second phase are highly toxic and difficult to administer (Burri et al. 2000). In the Central African subregion, records of the evolution of this disease over the past 10 years show that it is strongly on an increase despite control measures deployed over the years (Cattand, 1994; Le Mardeley et al. 1995; Smith, Pepin & Stich, 1998), thereby confirming the observed general trend.
A clear identification of parasite stocks (type of multilocus genotype, and subspecies) and a better understanding of their distribution within and between foci is necessary for improving our surveillance of the disease and the evaluation of its pathogenicity or drug resistance. For such purposes, many studies on multilocus enzyme electrophoresis (MLEE) of human and animal stocks of trypanosomes from the West and the Central African sub-region have been carried out (Truc & Tibayrenc, 1993; Truc et al. 1997a,b; Nkinin et al. 1999, 2002; Jamonneau et al. 2000). The general outcomes of these studies were that the majority of human stocks were characterized as T. b. gambiense group I (Gibson, 1986), with one or two over-represented enzyme profiles or zymodemes while animal stocks were mainly T. brucei ‘non-gambiense’ or T. congolense. The debate on T. brucei population structure (Tait, 1980) has been recently reconsidered by MacLeod et al. (2000), who postulated that T. brucei has a flexible mode of reproduction, leading to a range of population structures among different transmission cycles. Although T. brucei population structure has been poorly analysed in this area, the widespread distribution of a few zymodemes suggested a predominant clonal mode of propagation rather than frequent sexual exchanges.
In this study, a MLEE analysis of Trypanosoma brucei s.l. stocks newly isolated from human and domestic pigs of the Central African subregion and an East African country (Uganda) has been carried out in order to identify the multilocus genotypes and the subspecies harboured by either humans or pigs and to contribute to the debate on T. brucei population structure.
Stocks were sampled from sites in the Central African Republic (RCA), Chad (TC), Equatorial Guinea (GE), the Republic of Congo (CG), Cameroon (CM) and an east African country state (Uganda), between 1997 and 2000 (Table 1). In Cameroon, 4 foci (Fontem, Bipindi, Campo, Doumé) were surveyed for HAT at different times. Domestic pigs were also sampled in 2 of them (Fontem and Campo). In the other countries, the study was limited to the human population. The KIVI (Kit for In Vitro Isolation – Aerts et al. 1992) was used to collect samples in the field as previously described (Truc et al. 1992; McNamara et al. 1995; Nkinin et al. 1999). After transport to the laboratory, the KIVI samples were maintained at 27 °C and checked for trypanosome growth at least twice a week by microscopical examination during 45 days. Stocks from positive KIVI and reference stocks were subsequently grown in Cunningham medium (Cunningham, 1977) until parasite harvest. The total collection of stocks analysed in the present study included 169 newly isolated stocks (16 from pigs and 153 from human) and 5 references stocks previously characterized as T. brucei gambiense group I (A005, Malounda and JUA), T. b. brucei (M253) and T. b. rhodesiense (TRPZ166).
Table 1. Origin and identification of the Trypanosoma brucei s.l. stocks surveyed in the present study (The localities names are given with the countries names in brackets: CM, Cameroon; CG, Popular Republic of Congo; UG, Uganda; TC, Chad; RCA, Central African Republic; GE, Equatorial Guinea; KE, Kenya; Zym, zymodeme; N stocks, number of stocks studied. The reference stocks lines are written in italic.)

Multilocus enzyme electrophoresis was done using cellulose acetate plates (Truc, Mathiau-Daude & Tibayrenc, 1991; Truc & Tibayrenc, 1993).
The following 14 enzyme systems were used: alanine aminotransferase (ALAT, EC 2.6.1.2), aspartate aminotransferase (ASAT, EC 2.6.1.1), glucose phosphate isomerase (GPI, EC 5.3.1.9), glucose-6-phosphate dehydrogenase (G6PDH, EC 1.1.1.49), isocitrate dehydrogenase (IDH, EC 1.1.1.42), malate dehydrogenase (MDH, EC 1.1.1.37), malic enzyme (ME, EC 1.1.1.40), peptidase (substrate L-leucyl-L-alanine) (PEP2, EC 3.4.11. or 13.-.), 6-phosphogluconate dehydrogenase (6PGDH, EC 1.1.1.44), phosphoglucomutase (PGM, EC 5.4.2.2), threonine dehydrogenase (TDH, EC 1.1.1.103), superoxide dismutase (SOD, EC 1.15.1.1) and nucleoside hydrolase (EC 3.2.2.1) using two substrates, inosine (NHi) and deoxyinosine (NHd).
The enzyme profiles were first interpreted phenotypically. Each reproducible band was considered as a separate character. Numbers were allocated to bands beginning with 1 for the most rapid. Then each distinct and reproducible pattern for a given identified genetic locus (see below) was equated to a distinct genotype, its allelic composition remaining hypothetical (Tibayrenc, Kjellberg & Ayala, 1990). The genetic diversity was analysed using the software ‘Genetics ToolBox’. The following indices were estimated: polymorphism rate (P95, number of polymorphic loci/total number of loci); mean genetic diversity index (Nei, 1972); genotypic diversity (number of different multilocus genotypes/number of stocks); total number of multilocus genotypes; number of multilocus genotypes per subsample; maximum, average and standard deviation of the Jaccard's distances observed between every pair of stocks in each subsample. The same software was used to check for linkage disequilibrium, or non random association of genotypes occuring at distinct loci (Tibayrenc et al. 1990; see Table 4 for detail of the tests). To test a potential Wahlund effect (departures from panmixia due to sampling of individuals from different populations or generations or both) in case of significant linkage disequilibrium, all the analyses were also performed on different subsamples composed according to the year of sampling of stocks, the host, the locality and the country, according to the recommendations of Tibayrenc et al. (1991). For the analysis of the genetic divergence between samples, the software ‘Genetics ToolBox’ was used to calculate matrices of Jaccard's genetic distances (Jaccard, 1908), and the software ‘neighbor’ from the ‘Phylip package’ (Felsenstein, 1989) was used to cluster the stocks from the distance matrice by the UPGMA method (Unweighted Pair Group Method with Arithmetic Averages) (Sneath & Sokal, 1973).
Three enzyme systems (NHi, ME and SOD) out of 14 exhibited 2 zones of activity, which were attributed to 2 separate loci, giving a total number of 17 loci. Five loci (6Pgdh, Mdh, Gpi, Nhi1 and G6pdh) did not show any variation. The other 12 loci exhibited at least 2 different electrophoretic patterns, which were attributed to distinct genotypes (Table 2). In total, 22 different enzyme profiles or zymodemes were recorded, with a majority of them differing at very few enzyme loci (Table 2). These zymodemes were equated to multilocus genotypes (MLG), and 74% of the stocks corresponded to 2 major zymodemes, Z1 (36%) and Z3 (38%). Two zymodemes were found on both humans and pigs whereas 15 were found only in humans and 2 were found only in pigs (Table 1). Three Ugandan stocks (R47UG, R56UG and R60UG) were considered resistant, since they were isolated from relapse cases after treatment with Arsobal®, and 13 Ugandan sensitive stocks were found to belong to Z1. The genetic variability indices are recorded in Table 3. For the whole sample, the polymorphism rate (P95) was 0·41 and ranged from 0·06 to 0·41 among the different subsamples. Nei's genetic index was low, ranging from 0·01 (for human RCA stocks) to 0·15 for Cameroonese stocks isolated in 1998. The genotypic diversity index was low for the whole sample, although it was relatively high for some subsamples (Equatorial Guinea, Campo, Campo, 1998 and Cameroon, 1998). All these indices showed relatively high differences between the different subsamples, by locality or by country (Table 3).
Table 2. Genotypes recorded at polymorphic isoenzyme loci surveyed (Full names of enzyme systems are given in the Materials and Methods section; Z, zymodeme. The number 0 recorded at loci ME2 for Z14 represents a null allele.)

Table 3. Level of genetic variability in newly isolated stocks (NS, number of stocks; P95, polymorphism rate at 95%; Nei, Nei's genetic index; GD, genotypic diversity; Freq, number of stocks expressing the most common zymodeme; NZ, number of zymodemes; MaD, maximal genetic distance between zymodemes; MD, mean genetic distance; SD, genetic distance standard deviation; CM, Cameroon; GE, Equatorial Guinea; RCA, Central African Republic; T. b. g. I, Trypanosoma brucei gambiense group I.)

The UPGMA dendrogram (Fig. 1) revealed only 1 cluster, the T. b. gambiense group I, that included all the human zymodemes, 5 zymodemes from pigs and 3 T. b. gambiense group I reference stocks (A005, Malounda and JUA). Genetic distances among this group of zymodemes were lower than 0·19 (Table 3). All the group had T. b. gambiense group I common Sod genotype and the 1.2.3 heterozygous genotype of Me2, except Z20 (Table 2). The other three zymodemes, T. b. brucei (M253) and T. b. rhodesiense (TRPZ166) reference stocks, and 1 newly isolated pig zymodeme (Z19, with 11 stocks) were all connected to T. b. gambiense group I, by high genetic distances. These stocks did not show specific Sod genotype, but none of them had the Me2 1.2.3 heterozygous genotype specific to T. b. gambiense group I (Table 2).

Fig. 1. UPGMA tree built from Jaccard's genetic distances observed between the 22 zymodemes (Z) or multilocus genotypes. A005, Malounda and JUA are Trypanosoma brucei gambiense group I reference stocks, M253 and TRPZ166 are respectively T. b. brucei and T. b. rhodesiense reference stocks. The number 36 corresponds to T. b. gambiense group I bootstrap value.
When we considered samples with stocks from distant areas or samples with both animal and human stocks (all stocks, all Cameroonese stocks, all human stocks and animal stocks) the tests were highly significant (Table 4), revealing high levels of linkage disequilibrium. Although RCA stocks showed limited genetic variability, which lowers the chances of obtaining positive tests (statistical type II error), they showed highly significant values for tests d2, e and f. These tests were also highly significant for 1998 and 1999 Cameroonese stocks showing that the linkage disequilibrium found in the pool sample was not due to the different sampling dates. For human stocks of Cameroon, Equatorial Guinea, Uganda, Campo and Bipindi, some tests were significant whereas others were not. When zymodemes (Multilocus Genotypes) were taken as the unit of analysis instead of the stocks, (see Table 4 ‘On zym’), no linkage disequilibrium was detected except for the whole set of zymodemes.
Table 4. Levels of significance of linkage disequilibrium tests among the various subsamples of Trypanosoma brucei s.l. newly isolated stocks (NR, number of repetition of the most common zymodeme; NZ, number of zymodemes; On zym, test f on zymodemes only; T. b. g. group I, Trypanosoma brucei gambiense group I. ****, P<10−4; —, test not done because of the low sample size. The linkage disequilibrium tests (Tibayrenc et al. 1990) are defined as: d1=probability of sampling the most common zymodeme as often or more than actually observed (threshold: 0·05); d2=probability of observing any zymodeme as often as or more often than the most common zymodeme in the sample (threshold: 0·05); e=probability of observing as few or fewer zymodemes in the population than observed in the sample (threshold: 0·05); f=probability of observing a linkage disequilibrium in the population as high or higher than observed in the sample (threshold: 0·01).)

Considering the 22 multilocus genotypes, 11 were unique (Table 1), whereas 11 were repeated from 2 to 64 times (60 and 64 times respectively for Z1 and Z3). Thus, besides the above statistical results showing the occurrence of strong linkage disequilibria in most of our subsamples, we noticed that 2 of the repeated genotypes (Z1 and Z3) occurred in several countries separated over wide geographical distances. Moreover, within Z1, A005, a T. b. gambiense group I reference stock isolated in 1988, and Menji-1F, Menji-2F, Menji-3F, all human stocks from Fontem isolated in 1999 showed a genetic permanency in time of this multilocus genotype. Interestingly, under the panmixia hypothesis (H0), the presence of Z1 60 times is expected (probability d1=0·343) while the presence of the other dominant zymodeme Z3 (64 times) is highly unexpected (probability d1=3·45 e-5). The probabilities of the other repeated zymodemes under H0 are: Z4 4 times p=6·38 e-2, Z5 3 times p=4·48 e-1, Z7 2 times p=1·13 e-2, Z10 2 times p=3·65 e-4, Z12 2 times p=2·35 e-5, Z19 11 times p <10 e-15, Z20 3 times p=8·91 e-1, Z23 4 times p=5·70 e-2 and Z24 6 times p=2·19 e-2. It is worth noting that the presence 11 times of Z19 is by far the most unexpected.
Data analysed have shown a relatively high polymorphism rate for the various subsamples surveyed here. However, the mean genetic diversity and the genotypic diversity indices were low, since the genetic variability was unequally distributed, often characterized by the occurrence of a few major alleles. T. b. gambiense group I is known to be very homogenous (Gibson, 1986) and the low level of polymorphism reported here for this group is consistent with the one found by Jamonneau et al. (2000) in a comparable study in West Africa.
Various authors have postulated a panmictic population structure in T. brucei because of the occurrence of genetic recombination (Tait, 1980; Jenni et al. 1986; Paindavoine et al. 1986). Later, T. brucei was included with several other major parasitic protozoa in the ‘clonal theory of parasitic protozoa’, stating that these species undergo predominant clonal evolution, with only rare bouts of genetic recombination (Tibayrenc et al. 1990, 1991; Tibayrenc, 1995). A panmictic population structure with occasional occurrence of epidemic clonality has been suggested (Cibulskis, 1992; Maynard Smith et al. 1993; Hide et al. 1994). More recently, Gibson & Stevens (1999) and MacLeod et al. (2000) postulated that T. brucei has a flexible mode of reproduction, leading to a range of population structures among different transmission cycles. In this study, high levels of linkage disequilibrium were recorded in most subsamples. This disequilibrium remained high within some groups of stocks isolated from the same geographical area or in the same year, suggesting that linkage disequilibrium was not due to a Wahlund effect, but rather to the mating system of the parasite itself. Maynard Smith et al. (1993) have proposed to take as the unit of analysis the genotypes instead of the individuals (stocks) to distinguish between true clonal evolution and epidemic clonality (occasional bouts of clonal propagation in a basically sexual species). If the linkage disequilibrium disappears, it is an indication for epidemic clonality rather than clonal evolution. Here, in most subsamples, the linkage disequilibrium becomes non-significant when applying this procedure. However, doing that, the sample size and the overall genetic variability become extremely low, which generates a high risk of statistical type II error (negative test, not because the working hypothesis is wrong but because the test lacks power). It is impossible therefore to know whether this result is an indication of epidemic clonality or a consequence of a statistical type II error. The permanency in space and time of Z3 (probability of apparition under panmixia condition=4·45 e-5), as well as the multi-occurrence of Z7, Z10, Z12, Z19 and Z24 was also a strong indication for predominant clonal evolution. In the light of these results, the multilocus genotypes represented by the various zymodemes recorded in the present study can be equated to natural clones, or rather, to ‘clonets’ (identical multilocus genotypes identified by a given set of genetic markers in a basically clonal species; Tibayrenc & Ayala, 1991). They represent convenient units of analysis for epidemiological tracking. This is especially true for the 2 dominant zymodemes, which have been already noted in previous studies (Godfrey et al. 1990; Truc & Tibayrenc, 1993), and represent a similar case than the ‘major clones’ identified in T. cruzi (Tibayrenc et al. 1986). Although T. b. gambiense group I was not supported by high bootstrap values it is clearly separated from Z19 and the two reference stocks of other species and it can be equated to ‘discrete typing units’ (discrete sets of genetically related genotypes; Tibayrenc, 1999). It could be the result of predominant clonal evolution, as is the case in T. cruzi (Tibayrenc et al. 1986; Tibayrenc, 1995; Barnabé, Brisse & Tibayrenc, 2000). However, one has to take into account that the range of hosts considered here (only humans and pigs) is quite limited, and therefore, that the set of T. brucei genotypes surveyed most probably represents only a limited subsample of all genotypes circulating in these areas (‘iceberg bias’; Tibayrenc, 1999). The present conclusions should therefore not be generalized to the whole species, and the hypothesis that T. brucei s.l. has a flexible population structure (Hide et al. 1994; Gibson & Stevens, 1999; MacLeod et al. 2000) is a parsimonious one.
The use of specific electrophoretic patterns, reference stocks and UPGMA tree easily enabled us to distinguish T. b. gambiense group I from T. brucei ‘non-gambiense’. Outside of T. b. gambiense group I, Z19 would likely be identified as T. b. brucei because of its animal origin and its location in the Central African subregion. Human stocks were almost all T. b. gambiense group I whereas 69% of pig stocks were T. brucei ‘non-gambiense’ and 31% were T. b. gambiense group I as previously found (Truc & Tibayrenc, 1993; Jamonneau et al. 2000).
Human parasites were represented by two major zymodemes found in many countries and some minor ones found in specific localities, questioning the spread of these clonal genotypes. The presence of Z3 in Batangafo (RCA) and Moundou (Chad) across the border may have resulted from displacement of infected flies and/or humans between the localities due to social, economic or political factors such as trade, hunting, and refugee migrations (Bureau et al. 2000). The same reasoning may explain the circulation of Z1 between Campo and Bipindi, neighbouring foci in Cameroon; and the cross-border foci of Campo (Cameroon) and Mbini (Equatorial Guinea). Apart from the widespread major zymodemes, minor ones have been sampled only from specific localities: Z6, Z7, Z11 and Z13 only from Campo; Z2 from Fontem; Z12 and Z15 unique to Equatorial Guinea and Z21, Z22 and Z23 unique to Uganda. These observations could reflect the ‘endemic’ nature of HAT in the various foci suggesting the occurence of particular alleles by mutation or by migration (Tibayrenc et al. 1991). The relation between such patchy distribution of the genetic variability and the clinical evolution of the HAT remains unknown. Lastly, 27% of stocks from domestic pigs in Fontem were identified as T. b. gambiense group I, pointing out their potential animal reservoir role (Gibson et al. 1978; Mehlitz et al. 1982; Nkinin et al. 2002).
This work was supported by a ‘Fonds d'aide à la Coopération’ grant by the Ministère Français des Affaires Etrangères and by the Institut de Recherche pour le Développement (IRD, France). We thank the OCEAC team for excellent technical assistance.

Table 1. Origin and identification of the Trypanosoma brucei s.l. stocks surveyed in the present study

Table 2. Genotypes recorded at polymorphic isoenzyme loci surveyed

Table 3. Level of genetic variability in newly isolated stocks
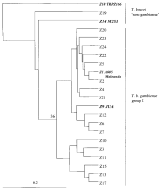
Fig. 1. UPGMA tree built from Jaccard's genetic distances observed between the 22 zymodemes (Z) or multilocus genotypes. A005, Malounda and JUA are Trypanosoma brucei gambiense group I reference stocks, M253 and TRPZ166 are respectively T. b. brucei and T. b. rhodesiense reference stocks. The number 36 corresponds to T. b. gambiense group I bootstrap value.

Table 4. Levels of significance of linkage disequilibrium tests among the various subsamples of Trypanosoma brucei s.l. newly isolated stocks