Introduction
Henosepilachna vigintioctopunctata (Fabricius) (Coleoptera: Coccinellidae) is one of the most serious insect pests to a large number of nightshades and cucurbits in Asian countries (Wu et al., Reference Wu, Mu, Kang, Ze, Shen, Jin, Anjum and Li2021; Xu et al., Reference Xu, Ze, Kang, Wu, Jin, Ali and Li2020). At present, chemical pesticides are primarily used for controlling H. vigintioctopunctata (Guo et al., Reference Guo, Lü, Guo, Chen, Qiu, Sang, Yang, Zhang and Pan2019). However, these chemicals are hazardous to the environment and exert deleterious effects on human health (Zhu et al., Reference Zhu, Zhang, Li, Sun, Wang, Smagghe and Jiang2019). These issues provide compelling reasons to find safer and more species-specific alternatives to control the pest.
RNA interference (RNAi), a sequence-specific method of suppressing a targeted gene's expression mediated by double-stranded RNA (dsRNA), offers a reduced risk approach to insect pest control (Christiaens et al., Reference Christiaens, Whyard, Vélez and Smagghe2020; Liu et al., Reference Liu, Jaouannet, Dempsey, Imani, Coustau and Kogel2020; Zhu and Palli, Reference Zhu and Palli2020). Because each species is defined by the uniqueness of its genes' sequences, RNAi could be used selectively to kill pest insects without adversely affecting non-target species (Whyard et al., Reference Whyard, Singh and Wong2009). The first step to develop RNAi-based technologies for pest management is to identify amenable target genes (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019; Vogel et al., Reference Vogel, Santos, Mingels, Verdonckt and Broeck2019).
Vacuolar-type H+-ATPases (vATPases) are the complex membrane-bound rotary molecular motors in eukaryotes (Wieczorek et al., Reference Wieczorek, Beyenbach, Huss and Vitavska2009). By hydrolyzing ATP to ADP and phosphate, insect vATPases pump protons across membranes in nearly all epithelial tissues (Wieczorek et al., Reference Wieczorek, Brown, Grinstein, Ehrenfeld and Harvey1999; Wieczorek et al., Reference Wieczorek, Beyenbach, Huss and Vitavska2009). The structures of vATPases are conserved in eukaryote species (McGuire et al., Reference McGuire, Stransky, Cotter and Forgac2017; Song et al., Reference Song, Meng, Xu and Mao2020). A holoenzyme vATPase consists of two functional subcomplexes, cytoplasmic V1 and membrane V0. The V1 subcomplex is comprised of eight different subunits (A-H), with a stoichiometry of A3B3CDE3FG3H in Lepidopteran Manduca sexta and yeast Saccharomyces cerevisiae (Kitagawa et al., Reference Kitagawa, Mazon, Heck and Wilkens2008; Muench et al., Reference Muench, Huss, Song, Phillips, Wieczorek, Trinick and Harrison2009).
Depletion of vATPase subunits is lethal to insect species (Baum et al., Reference Baum, Bogaert, Clinton, Heck, Feldmann, Ilagan, Johnson, Plaetinck, Munyikwa, Pleau, Vaughn and Roberts2007; Fu et al., Reference Fu, Guo, Lü, Liu and Li2014). For instance, dsRNA targeting vATPaseE mRNA induces larval lethality in Coleopterans Diabrotica virgifera virgifera (Baum et al., Reference Baum, Bogaert, Clinton, Heck, Feldmann, Ilagan, Johnson, Plaetinck, Munyikwa, Pleau, Vaughn and Roberts2007), Diabrotica undecimpuctata howardii (Baum et al., Reference Baum, Bogaert, Clinton, Heck, Feldmann, Ilagan, Johnson, Plaetinck, Munyikwa, Pleau, Vaughn and Roberts2007), Leptinotarsa decemlineata (Zhu et al., Reference Zhu, Xu, Palli, Ferguson and Palli2011; Fu et al., Reference Fu, Guo, Lü, Liu and Li2014) and Tribolium castaneum (Whyard et al., Reference Whyard, Singh and Wong2009), in Lepidopteran M. sexta (Whyard et al., Reference Whyard, Singh and Wong2009), and causes an obvious lethal effect on the nymph in Hemipterans Acyrthosiphon pisum (Whyard et al., Reference Whyard, Singh and Wong2009) and Apolygus lucorum (Liu et al., Reference Liu, Yang, Zhang, Ding and Wang2019). Furthermore, ingestion of D. virgifera virgifera subunit vATPaseE dsRNA also kills approximately 40% of the D. undecimpunctata howardii larvae (Baum et al., Reference Baum, Bogaert, Clinton, Heck, Feldmann, Ilagan, Johnson, Plaetinck, Munyikwa, Pleau, Vaughn and Roberts2007).
The V1 subcomplex subunit C and H are among the most important components for a holoenzyme vATPase (Kitagawa et al., Reference Kitagawa, Mazon, Heck and Wilkens2008; Muench et al., Reference Muench, Huss, Song, Phillips, Wieczorek, Trinick and Harrison2009). Firstly, subunits C and H, along with subunits E and G, links V1 subcomplex to the membrane V0 subcomplex (Muench et al., Reference Muench, Huss, Song, Phillips, Wieczorek, Trinick and Harrison2009). Secondly, subunit C links the holoenzyme vATPase to actin filaments (Vitavska et al., Reference Vitavska, Wieczorek and Merzendorfer2003). Thirdly, subunit C dissociates from the V1 complex after disassembly of the vATPase, and binds not only to F actin but also to monomeric G actin (Vitavska et al., Reference Vitavska, Merzendorfer and Wieczorek2005).
However, only a few publications have documented the in vivo results of knockout/knockdown of vATPaseC and vATPaseH. In Drosophila melanogaster, a mutation in vha44 and vhaSFD (encoding the vATPaseC and vATPaseH subunits respectively) causes larval lethal (Allan et al., Reference Allan, Du, Davies and Dow2005). In non-Drosophilid insects, knockdown of vATPaseC in D. virgifera virgifera (Li et al., Reference Li, Khajuria, Rangasamy, Gandra, Woosely, Hasler, Schulenberg, Worden, Mcewan, Evans, Siegfried and Narva2015) and of vATPaseH in Locusta migratoria manilensis (Li and Xia, Reference Li and Xia2012) and Aphis gossypii (Rebijith et al., Reference Rebijith, Asokan, Ranjitha, Rajendra, Krishna and Kumar2015) leads to mortality. Moreover, ingestion of dsvATPaseC by the L. decemlineata first instar larvae reduces 75% of the target mRNA. However, the negative influence on the larvae has not been determined (Cappelle et al., Reference Cappelle, de Oliveira, Van Eynde, Christiaens and Smagghe2016). Does knockdown of vATPaseC or vATPaseH cause severe defective phenotypes in H. vigintioctopunctata?
Moreover, some genes are better RNAi targets than others (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019). For example, among 5000 random genes from iBeetle screened in T. castaneum, knockdown of only 100 genes lead to >90% mortality, 11 genes with 100% mortality by day 8 (Ulrich et al., Reference Ulrich, Dao, Majumdar, Schmitt-Engel, Schwirz, Schultheis, Ströhlein, Troelenberg, Grossmann, Richter, Dönitz, Gerischer, Leboulle, Vilcinskas, Stanke and Bucher2015). In Cylas puncticollis, of 24 genes examined by RNAi, silencing 12 genes induces almost 100% morality at 14 days post-injection (Prentice et al., Reference Prentice, Christiaens, Pertry, Bailey, Niblett, Ghislain, Gheysen and Smagghe2017). Are there any differences in RNAi efficiency among vATPase genes in H. vigintioctopunctata?
Furthermore, RNAi efficiency is inconstant between life stages (Scott et al., Reference Scott, Michel, Bartholomay, Siegfried, Hunter, Smagghe, Zhu and Douglas2013; Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019). Higher RNAi efficiency is noted in the young juveniles than the elders in holometabolans, such as Bombyx mori, M. sexta and L. decemlineata (Guo et al., Reference Guo, Fu, Yang, Li and Li2015; Zhu and Palli, Reference Zhu and Palli2020), and hemimetabolans such as Rhodnius prolixus (Araujo et al., Reference Araujo, Santos, Pinto, Gontijo, Lehane and Pereira2006). Does RNAi efficiency vary among different larval instars in H. vigintioctopunctata?
In order to address the three issues mentioned above, we identified HvvATPaseC, HvvATPaseE and HvvATPaseH to knock down of them by RNAi, and to analyze their possible physiological roles in larval growth and development. We found that RNAi efficiency varied among different vATPase subunit genes and between various development stages. Our results imply the feasibility of RNAi as an alternative method for controlling this critical potato pest.
Materials and methods
Insect and rearing
H. vigintioctopunctata adults were collected from Solanum melongena L. in Nanjing city, Jiangsu Province, China, in the summer of 2018. The beetles were routinely maintained in an insectary at 28 ± 1 °C under a 16 h:8 h light−dark photoperiod and 50–60% relative humidity using foliage at the vegetative growth or young tuber stages in order to assure sufficient nutrition. At this feeding protocol, the larvae progressed through four distinct instars, with approximate periods of the first-, second-, third-, and fourth-instar stages of 3, 2, 2, and 3 days, respectively. Upon reaching full size, the fourth larval instars stopped feeding, fixed their abdomen ends to the substrate surface and entered the prepupal stage. The prepupae spent approximately 2 days to pupate. The pupae lasted about 4 days and the adults emerged.
Molecular cloning
The putative vATPaseC, vATPaseE and vATPaseH were obtained from the transcriptome data (Zhang et al., Reference Zhang, Wang, Guo, Deng, Chen and Lin2018). The correctness of the sequences was substantiated by polymerase chain reaction (PCR) using primers in table S1. The sequenced cDNAs were submitted to GenBank (accession numbers: vATPaseC, MW267249; vATPaseE, MW267251; vATPaseH, MW267254).
Preparation of dsRNAs
In order to improve the reliability of RNAi experiment, two cDNA fragments derived from HvvATPaseC, HvvATPaseE, or HvvATPaseH were selected. These targeted regions were further BLAST (BLASTN) searched against the H. vigintioctopunctata transcriptome data (Zhang et al., Reference Zhang, Wang, Guo, Deng, Chen and Lin2018) to identify any possible off-target sequences that had an identical match of 20 bp or more. A cDNA from the enhanced green fluorescent protein (egfp) gene from Aequorea victoria was used as control. Specific primers used to clone the fragments of dsRNAs were listed in table S1. These dsRNAs were individually expressed using Escherichia coli HT115 (DE3) competent cells lacking RNase III following the established method (Xu et al., Reference Xu, Deng, Li, Zhang, Mu, Fu, Guo and Li2019). Individual colonies were inoculated, and grown until cultures reached an OD600 value of 1.0. The colonies were then induced to express dsRNA by the addition of isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.1 mM. The expressed dsRNA was extracted and confirmed by electrophoresis on 1% agarose gel. Bacteria cells were centrifuged at 5000 × g for 10 min, and resuspended in an equal original culture volume of 0.05 M phosphate-buffered saline (PBS, pH 7.4). The bacterial solutions (at a dsRNA concentration of about 0.5 μg ml−1) were used for the experiment.
Dietary introduction of dsRNA
The similar method as previously reported in L. decemlineata (Xu et al., Reference Xu, Deng, Li, Zhang, Mu, Fu, Guo and Li2019) was used to introduce dsRNA into the H. vigintioctopunctata larvae. Potato leaves were immersed with a bacterial suspension containing a dsRNA (dsvATPaseC-1,2; dsvATPaseE-1,2;or dsvATPaseH-1,2) for 5 s, removed, and dried for 2 h under airflow on filter paper. The PBS- and dsegfp-dipped leaves were used as controls. Five treated leaves were then placed in Petri dishes (9 cm diameter and 1.5 cm height). The newly ecdysed third- and fourth-instar larvae were starved for at least 4 h prior to the experiment. Then, ten larvae were transferred to each dish as a repeat. For each treatment, 6 repeats were set. Three replicates were used to observe the survival, pupation and emergence by allowing the larvae to feed on treated leaves for 3 days (replaced with freshly treated ones each day), and on untreated foliage until reaching the prepupal stage. The resultant larvae were weighed daily using an analytical balance with an accuracy of 0.1 mg. The other three replicates were continuously fed on treated foliage for 3 days and were collected for extraction of total RNA.
Real-time quantitative PCR (qRT-PCR)
For temporal expression analysis, RNA templates were derived from eggs (day 3), the larvae from the first through the fourth instars, prepupae, pupae and adults. For analysis of the tissue expression patterns, RNA templates were from the foregut, midgut, hindgut, Malpighian tubules, epidermis and fat body of the day 1 final instar larvae. Each sample contained 20–30 individuals and repeated three times. For analysis of the effects of treatments, total RNA was extracted from treated larvae. Each sample contained ten individuals and repeated three times. The RNA was extracted using SV Total RNA Isolation System Kit (Promega). Purified RNA was subjected to DNase I to remove any residual DNA according to the manufacturer's instructions. Quantitative mRNA measurements were performed by qRT-PCR in technical triplicate, using 2 internal control genes (HvRPS18 and HvRPL13, the primers listed in table S1) according to the published results (Lü et al., Reference Lü, Chen, Guo, Ye, Qiu, Wu, Yang and Pan2018). An RT negative control (without reverse transcriptase) and a non-template negative control were included for each primer set to confirm the absence of genomic DNA and to check for primer-dimer or contamination in the reactions, respectively.
According to a previously described method (Bustin et al., Reference Bustin, Benes, Garson, Hellemans, Huggett, Kubista, Mueller, Nolan, Pfaffl, Shipley, Vandesompele and Wittwer2009), the generation of specific PCR products was confirmed by gel electrophoresis. The primer pair for each gene was tested with a 5-fold logarithmic dilution of a cDNA mixture to generate a linear standard curve (crossing point [CP] plotted vs. log of template concentration), which was used to calculate the primer pair efficiency. All primer pairs amplified a single PCR product with the expected sizes, showed a slope less than −3.0, and exhibited efficiency values ranging from 2.4 to 2.7. Data were analyzed by the 2−ΔCT method, using the geometric mean of the four internal control genes for normalization.
Data analysis
We used SPSS for Windows (Chicago, IL, USA) for statistical analyses. The averages (± SE) were submitted to analysis of variance with the Tukey−Kramer test. In both laboratory and greenhouse experiments, no significant differences between dsRNAs targeting two different regions of HvvATPaseC, HvvATPaseE, or HvvATPaseH genes (dsvATPaseC-1/dsvATPaseC-2, dsvATPaseE-1/dsvATPaseE-2, dsvATPaseH-1/dsvATPaseH-2) were found, the data of each gene were thus combined.
Results
Molecular cloning and phylogenetic analyses
Three cDNAs, namely HvvATPaseC, HvvATPaseE and HvvATPaseH, were obtained from H. vigintioctopunctata transcriptome data. All the three cDNAs contained complete coding sequence. The correctness of the three sequences was substantiated.
To gain insight into evolutionary relationships of the vATPase subunits in relation to those from other insects, an unrooted phylogenetic tree was constructed with vATPaseC proteins from three Dipterans, two Lepidopterans, two Hymenopterans, three Coleopterans, four Hemipterans and a Siphonapteran. The result showed that vATPaseC sequences from the same order were clustered together. H. vigintioctopunctata vATPaseC sorted with the sequences from the other two Coleopteran species (fig. 1a).

Figure 1. Phylogenetic analyses of vATPaseC, vATPaseE and vATPaseH. (a) vATPase V1 subunit C proteins are derived from three Dipteran Ceratitis capitate, Drosophila melanogaster and Aedes aegypti, two Lepidopteran Helicoverpa armigera and Bombyx mori, two Hymenopteran Acromyrmex echinatior and Copidosoma floridanum, three Coleopteran Henosepilachna vigintioctopunctata, Leptinotarsa decemlineata and Anoplophora glabripennis, four Hemipteran Halyomorpha halys, Nilaparvata lugens, Myzus persicae and Melanaphis sacchari and a Siphonapteran Ctenocephalides felis. (b) vATPase V1 subunit E proteins are derived from four Coleopteran Henosepilachna vigintioctopunctata, Leptinotarsa decemlineata, Anoplophora glabripennis and Agrilus planipennis, a Thysanopteran Frankliniella occidentalis, three Hymenopteran Zootermopsis nevadensis, Orussus abietinus and Harpegnathos saltator, two Lepidopteran Trichoplusia ni and Bombyx mandarina, two Dipteran Ceratitis capitata and Aedes aegypti, and a Siphonapteran Ctenocephalides felis. (c) vATPase V1 subunit H proteins are derived from five Coleopteran Henosepilachna vigintioctopunctata, Leptinotarsa decemlineata, Tribolium castaneum, Anoplophora glabripennis and Coleomegilla maculata, two Hymenopteran Bombus terrestris and Harpegnathos saltator, two Dipteran Drosophila hydei and Aedes aegypti, a Siphonapteran Ctenocephalides felis, a Hemipteran Nilaparvata lugens and an Orthopteran Locusta migratoria manilensis. The trees are constructed using the neighbor-joining method based on the full-length protein sequence alignments. Bootstrap analyses of 1000 replications are carried out and bootstrap values >50% are shown on the tree.
Likewise, a phylogenetic tree was constructed on the basis of the amino acid sequences of selected vATPaseEs from four Coleopterans, a Thysanopteran, three Hymenopterans, two Lepidopterans, two Dipterans and a Siphonapteran. In the unrooted tree, the species from the same order were clustered together. Obviously, vATPaseE from H. vigintioctopunctata belonged to Coleopteran subclade (fig. 1b).
Similarly, vATPaseHs were selected from five Coleopterans, two Hymenopteran, two Dipterans, a Siphonapteran, a Hemipteran and an Orthopteran, and an unrooted phylogenetic tree was constructed on the basis of the amino acid sequences. The vATPaseH proteins formed order-specific branches. Out of 5 vATPaseH proteins from Coleopteran insect species, 2 from Coccinellidae species clustered together with 100% bootstrap support. Three from L. decemlineata and Anoplophora glabripennis were joined together with 94% bootstrap support. The two subclades then clustered together to form Coleopteran branch with 100% bootstrap support (fig. 1c).
Stage-specific expression patterns
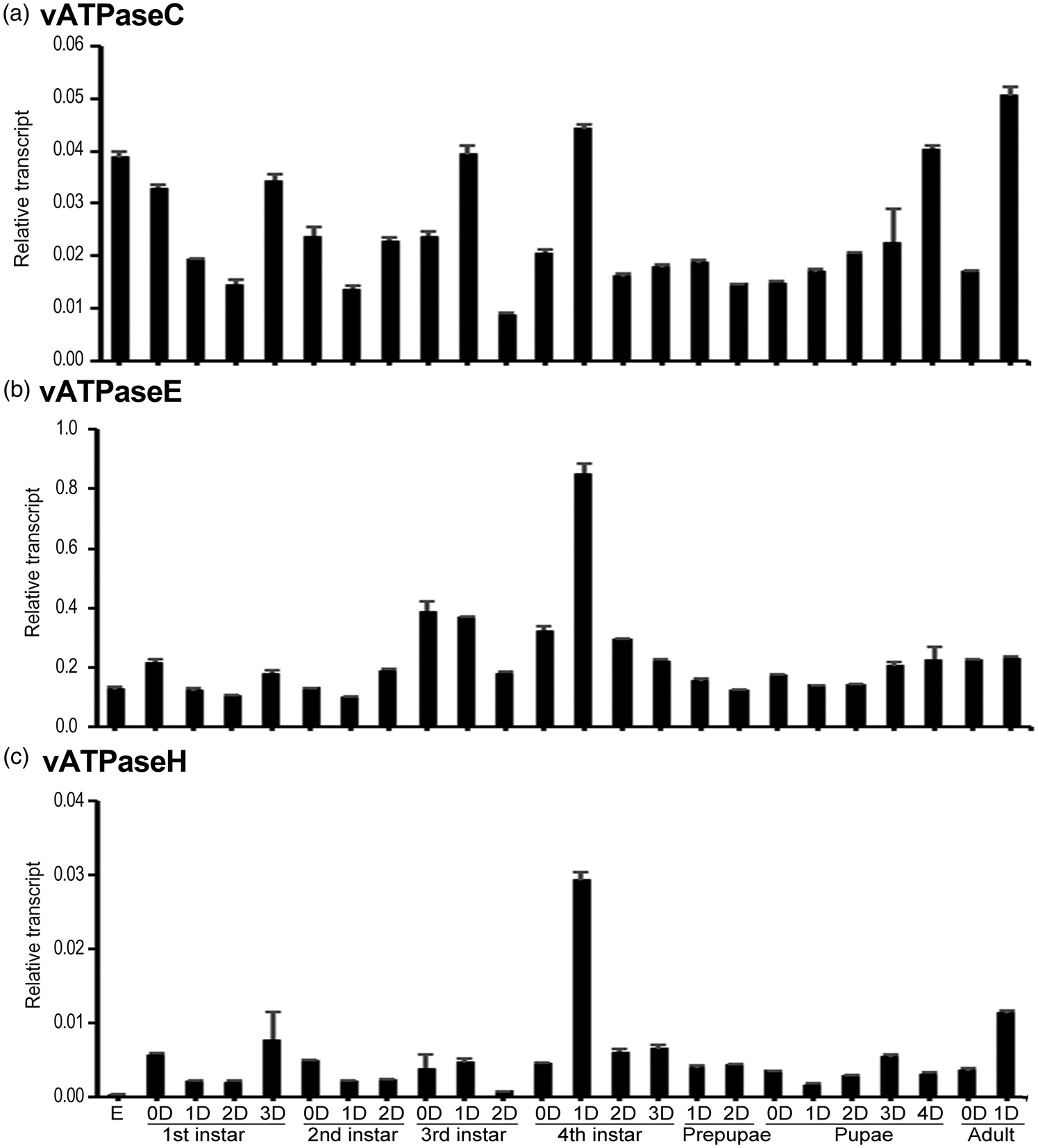
The temporal expression patterns of HvvATPaseC, HvvATPaseE and HvvATPaseH in whole body during different developmental stages were determined using real-time quantitative (qRT)-PCR. The results revealed that HvvATPaseC, HvvATPaseE, and HvvATPaseH were widely expressed in eggs, the first- to fourth-instar larvae, prepupae, pupae and adults. HvvATPaseC was steadily expressed throughout the whole development excursion. The highest and lowest levels were respectively noted at the day 1 adult and the day 2 third-instar larval stages, with the former being 5.7-fold of the latter (fig. 2a). In contrast, the expression of HvvATPaseH dramatically varied during development. The highest and lowest levels were respectively noted at the day 1 fourth-instar larval and the embryo stages, with the former being 104-fold of the latter (fig. 2c). The variation of the mRNA level of HvvATPaseE was intermediate. The variation range of the highest (at day 1 fourth instar larval stage) and lowest (at day 1 second instar larval stage) levels was 8.5 fold (fig. 2b).
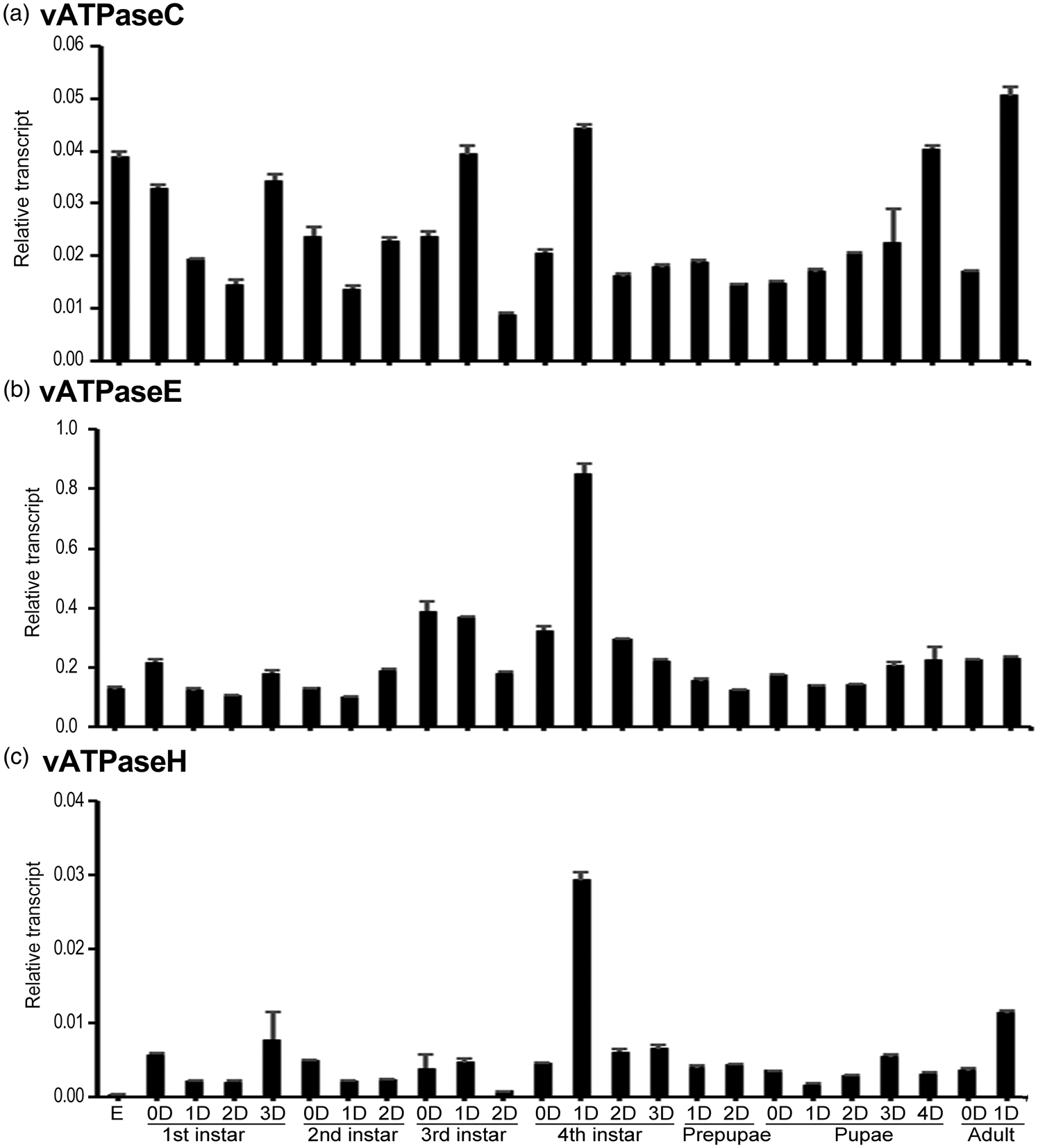
Figure 2. Temporal transcription patterns of HvvATPaseC, HvvATPaseE and HvvATPaseH in Henosepilachna vigintioctopunctata. Complementary DNA templates were derived from the eggs, first, second, third and fourth larval instars at an interval of 1 day (D0 indicates newly laid eggs or newly ecdysed larvae), the prepupae, pupae and adults. For each sample, 3 independent pools of 20–30 individuals were measured in technical triplicate using qRT-PCR. The values are calculated using the 2−ΔCT method. The columns represent averages with vertical lines indicating SE.
Based on the 2−ΔCT data, the mRNA abundance among HvvATPaseC, HvvATPaseE and HvvATPaseH can be compared. The average levels of HvvATPaseE at all development stages were higher than those of HvvATPaseC and HvvATPaseH (fig. 2b vs. 2a, 2c). The ratio of vATPaseE/vATPaseC averaged 10-fold, extending from 3-fold at the embryo stage to 20-fold at the day 2 third instar stage (fig. S1A). Similarly, the ratio of vATPaseE/vATPaseH averaged 75-fold, ranging from 20-fold at the day 1 adult phrase to 451-fold at the embryo stage (fig. S1B).
Tissue-specific expression profiles
To determine the tissue specificity of the expression of HvvATPaseC, HvvATPaseE and HvvATPaseH, six different tissues, including foregut, midgut, hindgut, Malpighian tubules, epidermis and fat body, were dissected from day 1 fourth-instar larvae. The expression levels were examined by qRT-PCR (fig. 3). HvvATPaseC, HvvATPaseE and HvvATPaseH were broadly expressed in these tissues. The three transcripts were highly expressed in the hindgut and Malpighian tubules, and lowly transcribed in the epidermis and fat body. Whereas HvvATPaseC and HvvATPaseE were also abundantly expressed in the foregut and midgut, the levels of HvvATPaseH were low in these two tissues (fig. 3).
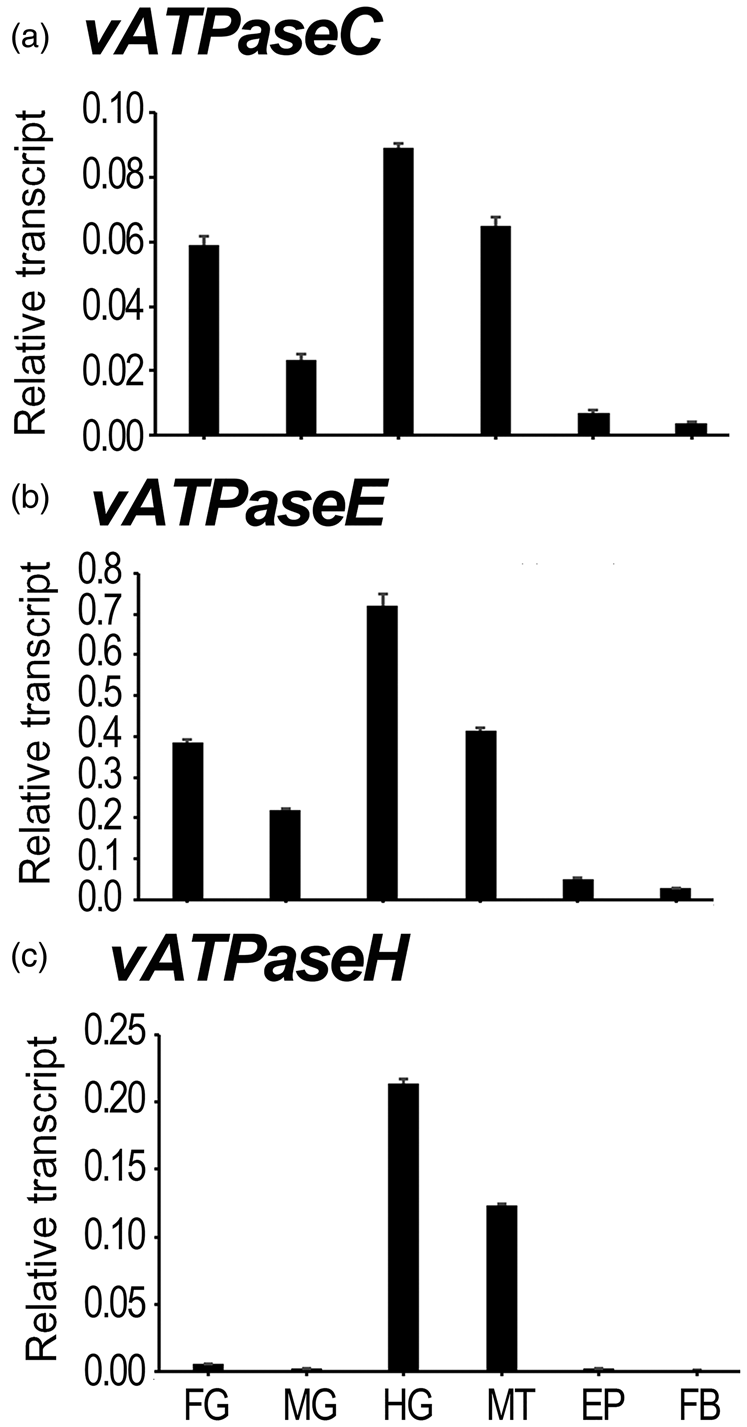
Figure 3. Tissue expression profiles of HvvATPaseC, HvvATPaseE and HvvATPaseH in Henosepilachna vigintioctopunctata. The relative transcripts were measured in the foregut (FG), midgut (MG), hindgut (HG), Malpighian tubules (MT), epidermis (EP) and fat body (FB) of the day 1 fourth instar larvae. For each sample, 3 independent pools of 20–30 individuals were measured in technical triplicate using qRT-PCR. The values were calculated using the 2−ΔCT method. The columns represent averages with vertical lines indicating SE.
The 2−ΔCT data of tissue expression in the day 1 fourth-instar larval stage also demonstrated that HvvATPaseE possessed the most abundant mRNA level, among the three vATPase genes (fig. 2b vs. 2a, 2c). The ratios of vATPaseE/vATPaseH and vATPaseE/vATPaseC averaged 7 and 42 folds, respectively. As for vATPaseE/vATPaseC, it extended from 6-fold in the Malpighian tubules to 9-fold in the midgut sample (fig. S2A). For vATPaseE/vATPaseH, it ranges from 3-fold in the hindgut specimen to 117-fold in the midgut sample (fig. S2B).
RNAi of vATPase subunit genes in the fourth instar larvae
Three days' ingestion of dsRNA-immersed foliage by the H. vigintioctopunctata final instar larvae significantly decreased the target mRNA level (fig. 4). The relative transcript levels of HvvATPaseC, HvvATPaseE and HvvATPaseH in dsRNA-fed larvae were greatly reduced by 90.1, 88.9 and 97.2%, respectively, compared with those in the PBS-fed larvae (fig. 4a–c).
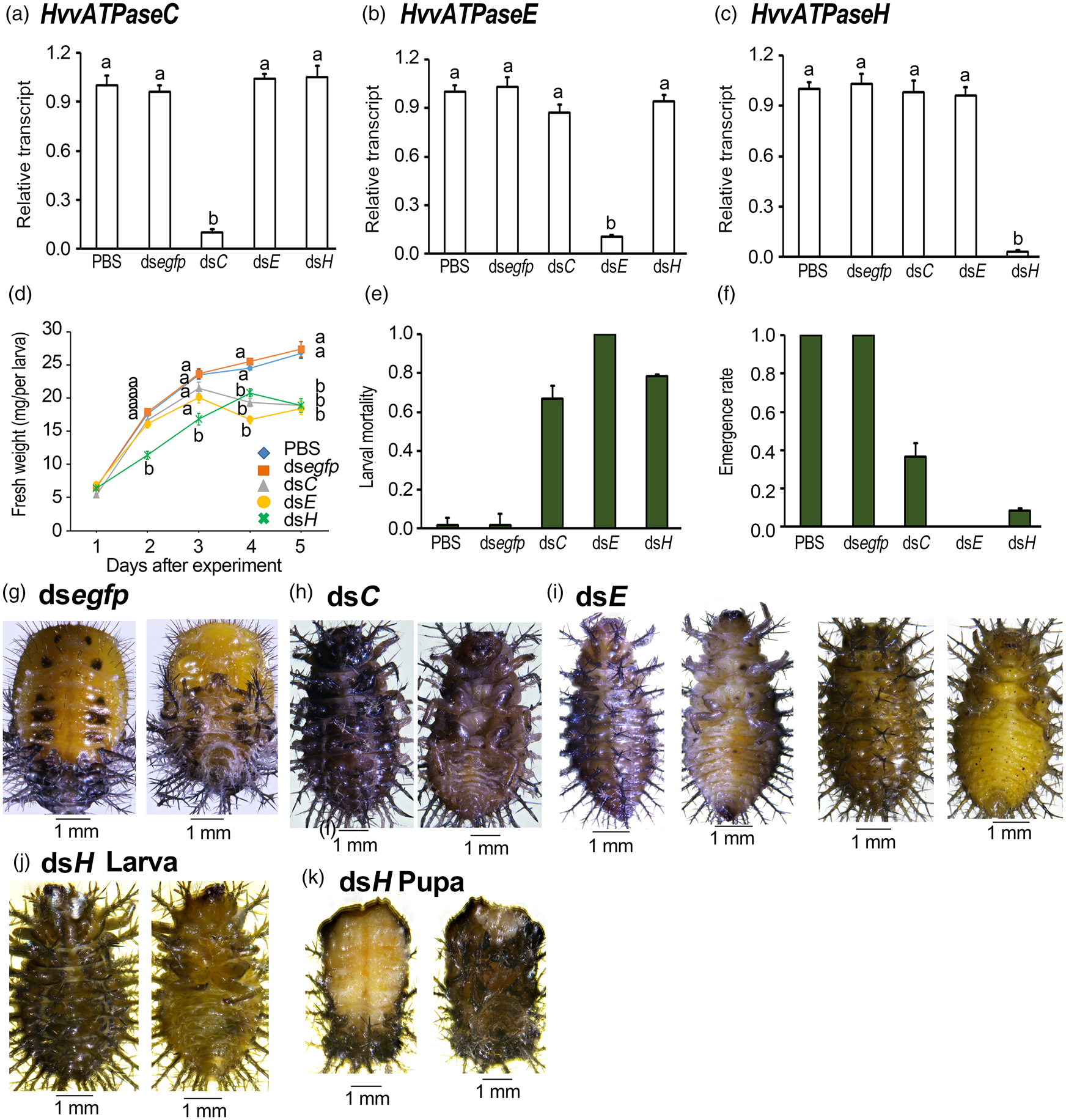
Figure 4. Ingestion of dsvATPaseC, dsvATPaseE or dsvATPaseH by the final instar larvae affects performance in Henosepilachna vigintioctopunctata. The newly ecdysed final instar larvae had ingested PBS-, dsegfp-, dsvATPaseC-, dsvATPaseE- and dsvATPaseH-dipped leaves for 3 days. The expression levels of HvvATPaseC, HvvATPaseE and HvvATPaseH were determined (a–c). Relative transcripts are the ratios of relative copy numbers in treated individuals to PBS-fed controls, which are set as 1. The fresh weights were measured 1–5 days after initiation of the experiment (d). The larval mortality and the emergence rate were recorded during a 3-week trial period (e, f). The bars represent values (± SE). Different letters indicate significant difference at P value < 0.05 using analysis of variance with the Tukey−Kramer test. While the CK larvae pupated 6 days after initiation of bioassay (g), most RNAi larvae remained as prepupae (h–j), a portion of dsvATPaseH-fed prepupae become deformed pupae (k).
Depletion of HvvATPaseC, HvvATPaseE and HvvATPaseH delayed larval growth (fig. 4d). Two days after the initiation of the bioassay, the fresh weights of the larvae having fed on dsvATPaseH were significantly smaller than those fed on PBS and dsegfp. Five days after initiation of bioassay, the average fresh body weights of dsvATPaseC-, dsvATPaseE- and dsvATPaseH-fed beetles were 19.3, 18.8 and 18.9 mg larva−1 respectively, where those in PBS- and dsegfp-fed larvae were 26.8 and 27.4 mg larva−1 respectively. Analysis of variance with the Tukey−Kramer test revealed that the weights of the former three were significantly smaller than those of the latter two (fig. 4d).
RNAi of HvvATPaseC, HvvATPaseE, and HvvATPaseH caused larval lethality (fig. 4e). In contrast to PBS- and dsegfp-fed larvae that pupated 6 days after experiment (fig. 4g), 66.7, 100, and 78.7% of the HvvATPaseC, HvvATPaseE and HvvATPaseH hypomorphs stopped development (fig. 4e), and remained as prepupae (fig. 4h–j). These prepupae gradually became dried and finally died within 10 days.
A total of 33.3 and 21.3% of the HvvATPaseC and HvvATPaseH depleted larvae pupated. A portion of the HvvATPaseH depleted larvae become deformed pupae (fig. 4k). The emergence percentages of those HvvATPaseC and HvvATPaseH depleted pupae were 36.0 and 8.7%, respectively (fig. 4f). However, all the HvvATPaseC and HvvATPaseH RNAi adults eventually died within 1 week after molting.
Knockdown of HvvATPaseC in the third larval instars
We knocked down HvvATPaseC in the newly ecdysed third-instar larvae (fig. 5). Three days' exposure of the third instar larvae to dsvATPaseC-immersed foliage significantly decreased HvvATPaseC mRNA abundance by 89.5%, comparing with PBS-exposed larvae (fig. 5a).
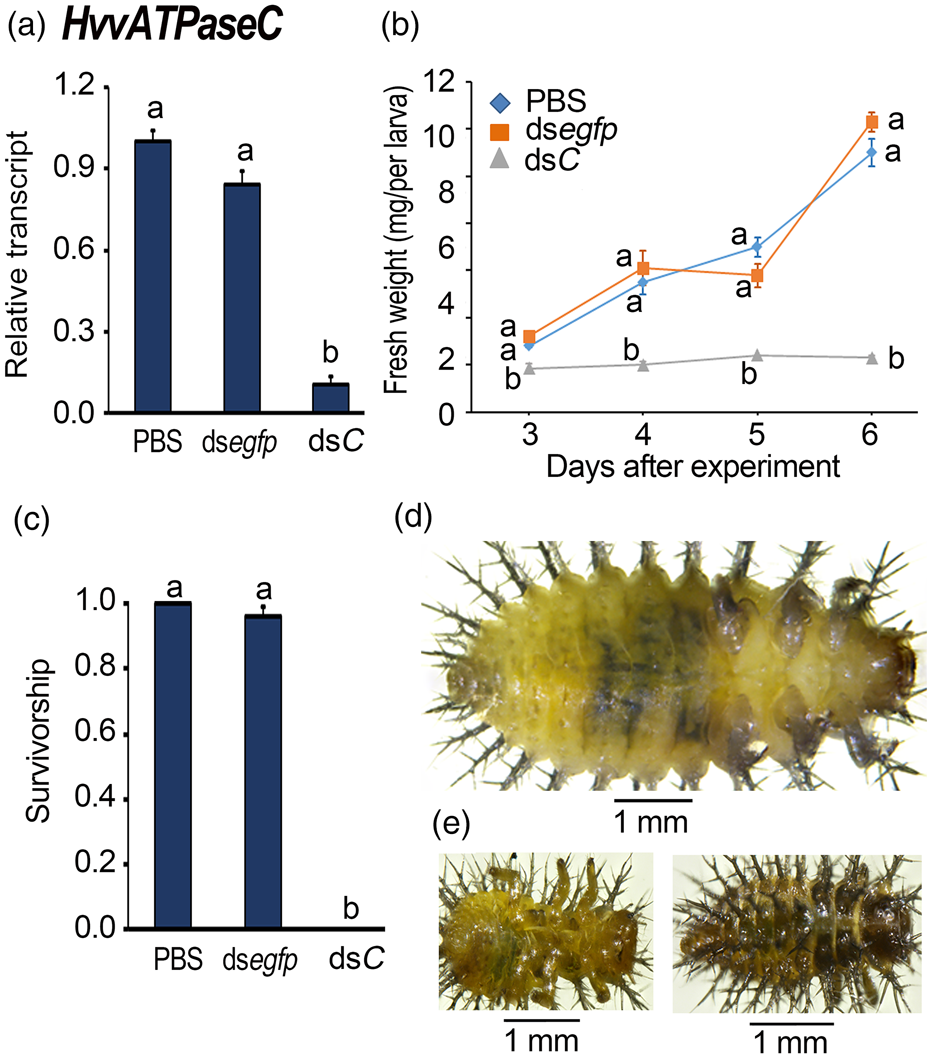
Figure 5. Ingestion of dsvATPaseC by the newly ecdysed penultimate instar larvae affects performance in Henosepilachna vigintioctopunctata. The newly ecdysed penultimate instar larvae had ingested PBS-, dsegfp-, dsvATPaseC-dipped leaves for 3 days. The expression levels of HvvATPaseC were determined (a). Relative transcripts are the ratios of relative copy numbers in treated individuals to PBS-fed controls, which is set as 1. The fresh weights were measured 3−6 days after initiation of the experiment (b). The larval survivorship was recorded during a 3-week trial period (c). The bars represent values (± SE). Different letters indicate a significant difference at P value < 0.05 using analysis of variance with the Tukey−Kramer test. While the PBS-fed larvae continuously grew 6 days after initiation of bioassay (d), the HvvATPaseC RNAi larvae remained stunting (e).
Within three to 6 days after the initiation of the bioassay, the fresh weights of the larvae having fed on dsvATPaseC were smaller than those fed on PBS and dsegfp (fig. 5b). When a PBS- or dsegfp-fed larva reached the fresh weights around 10 mg, a dsvATPaseC-fed larva was weighed only approximately 2 mg 6 days after initiation of bioassay (fig. 5b).
All the dsvATPaseC-fed larvae failed to shed their cuticle to become fourth-instar larvae. While the PBS- and dsegfp-exposed larvae became fourth-instar larvae and pupae (fig. 5c, d), all dsvATPaseC-fed beetles were completely wrapped in the old third-instar larval cuticles (fig. 5e). These stunting HvvATPaseC hypomorphs became withered, dried and darkened gradually, and finally died (fig. 5e).
Discussion
In this paper, we identified three vATPase subunit encoding genes HvvATPaseC, HvvATPaseE, and HvvATPaseH in H. vigintioctopunctata. We found that knockdown of each delayed growth and impaired development of the larvae, in a gene- and stage-dependent manner.
Transcription patterns
In the present paper, we found that HvvATPaseC, HvvATPaseE and HvvATPaseH were widely expressed in the eggs, first- to fourth-instar larvae, prepupae, pupae and adults (fig. 2). In agreement with our result, expressed sequence tags data in D. melanogaster reveal that vha44, vha26, vhaSFD (encoding C, E, H subunits, respectively) are expressed in the embryos, larvae, pupae and adults (Allan et al., Reference Allan, Du, Davies and Dow2005). The temporal expression data in H. vigintioctopunctata (this study) and those in other insects are compatible with the common idea that vATPase functions throughout the whole development excursion (Muench et al., Reference Muench, Scheres, Huss, Phillips, Vitavska, Wieczorek, Trinick and Harrison2014).
Our data revealed that HvvATPaseC, HvvATPaseE, and HvvATPaseH were highly expressed in the hindgut and Malpighian tubules, and lowly transcribed in the epidermis and fat body (fig. 3). In D. melanogaster, in situ hybridization shows that Vha44, Vha26, and VhaSFD are located in the Malpighian tubules, ovary, testes (including accessory gland, duct, and bulb), midgut, hindgut, and rectum (Allan et al., Reference Allan, Du, Davies and Dow2005). Moreover, insect vATPases have been found to be localized in the apical membranes of guts, Malpighian tubules, ovarioles, testes, salivary glands, labial glands and sensory sensilla in other insect species (Muench et al., Reference Muench, Scheres, Huss, Phillips, Vitavska, Wieczorek, Trinick and Harrison2014). The tissue expression data in H. vigintioctopunctata in the present paper and those in D. melanogaster (Allan et al., Reference Allan, Du, Davies and Dow2005) provide a piece of experimental evidence to support that vATPase functions in the apical membranes of nearly all epithelial tissues (Muench et al., Reference Muench, Scheres, Huss, Phillips, Vitavska, Wieczorek, Trinick and Harrison2014).
Gene-dependent RNAi efficiency
In the present paper, we found that 66.7, 100 and 78.7% of the HvvATPaseC, HvvATPaseE and HvvATPaseH hypomorphs stopped development and finally died (fig. 4). Our result is consistent with the fact that silencing effects among genes can differ considerably (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019; Zhu and Palli, Reference Zhu and Palli2020). The possible factors that influence the sensitivity of a target gene to RNAi include its function, tissue-bias expression, and the protein stability and turnover (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019; Christiaens et al., Reference Christiaens, Whyard, Vélez and Smagghe2020).
Gene function can significantly affect the outcome of an RNAi experiment (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019; Christiaens et al., Reference Christiaens, Whyard, Vélez and Smagghe2020; Liu et al., Reference Liu, Jaouannet, Dempsey, Imani, Coustau and Kogel2020; Zhu and Palli, Reference Zhu and Palli2020). Out of 130 lepidopteran genes that have been attempted to be knocked down by systemic RNAi, only 38% are silenced at high levels. Among these successfully silenced genes, immune genes occupy 80% (Zotti et al., Reference Zotti, Dos Santos, Cagliari, Christiaens, Taning and Smagghe2018; Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019). In M. sexta, for example, efficient silencing of immune genes can be achieved after injection of low to intermediate doses of dsRNA in the hemocoel (Eleftherianos et al., Reference Eleftherianos, Millichap, Ffrench-Constant and Reynolds2006a; Eleftherianos et al., Reference Eleftherianos, Marokhazi, Millichap, Hodgkinson, Sriboonlert, Ffrench-Constant and Reynolds2006b; Eleftherianos et al., Reference Eleftherianos, Xu, Yadi, Ffrench-Constant and Reynolds2009). Conversely, RNAi of nAChRs is more difficult than other genes in Nilaparvata lugens (Liu et al., Reference Liu, Ding, Zhang, Yang and Liu2010). In the present paper, however, all three genes encode vATPase subunits. Therefore, the difference of RNAi efficiency among HvvATPaseC, HvvATPaseE and HvvATPaseH is not related to gene function in H. vigintioctopunctata.
RNAi efficiency varies among different tissues (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019; Zhu and Palli, Reference Zhu and Palli2020). In Lepidoptera, epidermal tissues (larval epidermis and pupal wing) seem to be rather refractory to RNAi, even though RNAi successfully disrupts chitin synthesis during molting in S. exigua (Chen et al., Reference Chen, Tian, Zou, Tang, Hu and Zhang2008). Lepidopteran ovaries may be refractory to RNAi, although dsRNA injected into Hyalophora cecropia pupae and B. mori larvae can be absorbed by the developing oocytes in the ovaries (Bettencourt et al., Reference Bettencourt, Terenius and Faye2002; Mrinal and Nagaraju, Reference Mrinal and Nagaraju2008). In Caenorhabditis elegans, the brain is refractory to RNAi due to the expression of the eri-1 nuclease in this tissue (Kennedy et al., Reference Kennedy, Wang and Ruvkun2004). If a gene is specifically expressed in the tissues that impair dsRNA uptake, the gene is refractory to RNAi. In the present paper, we found that HvvATPaseC, HvvATPaseE and HvvATPaseH exhibited similar tissue expression profiles (fig. 3). Moreover, RNAi of the three vATPase genes in the fourth instar larvae reduced a comparable amount of target mRNA (fig. 4). Thus, tissue-biased expression of genes is not responsible for the difference of RNAi efficiency among the three vATPase genes in H. vigintioctopunctata.
Protein stability affects RNAi efficiency: the more stable of the protein, the less RNAi effect of its mRNA (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019; Zhu and Palli, Reference Zhu and Palli2020). In D. melanogaster and T. castaneum, α6 nicotinic acetylcholine receptors can be stable for more than 2 weeks (Lomazzo et al., Reference Lomazzo, Hussmann, Wolfe, Yasuda, Perry and Kellar2011). Consequently, RNAi of α6 gene does not result in changes in spinosad sensitivity (Rinkevich and Scott, Reference Rinkevich and Scott2013). In this survey, we found the average levels of HvvATPaseE at all development stages were higher than those of HvvATPaseC and HvvATPaseH (fig. 2). The ratios of vATPaseE/vATPaseH and vATPaseE/vATPaseC averaged 10 and 75 folds throughout development stages (fig. S1); they were 7 and 42 folds respectively among various tissues in the day 1 fourth-instar larvae (fig. S2). Higher expression levels of HvvATPaseE suggest lower protein stability and more efficient RNAi response. Consequently, knockdown of HvvATPaseE caused more severe defects in H. vigintioctopunctata.
RNAi efficiency differs between different development stages
Our results showed that ingestion of dsvATPaseC caused more severe negative effects at the third-instar stage, compared with those at the fourth-instar stages (fig. 4 vs. 5). In agreement with our results, RNAi efficiency is inconstant between life stages (Scott et al., Reference Scott, Michel, Bartholomay, Siegfried, Hunter, Smagghe, Zhu and Douglas2013; Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019). In R. prolixus, for instance, injection of dsRNA into the second instar nymphs causes higher RNAi efficiency than that into the fourth instar bugs (Araujo et al., Reference Araujo, Santos, Pinto, Gontijo, Lehane and Pereira2006). In some lepidopterans, such as B. mori and M. sexta, the dsRNA triggers RNAi more efficiently during the non-feeding wandering stage of the final instar larvae than during the feeding stages of the larvae (Zhu and Palli, Reference Zhu and Palli2020). In L. decemlineata, ingestion of dsLdSAHase targeting S-adenosyl-L-homocysteine hydrolase kills more young larvae than the old ones (Guo et al., Reference Guo, Fu, Yang, Li and Li2015). In order to improve RNAi efficiency, the appropriate timing for dsRNA treatment is the young larval instars.
An interesting question then arises: what causes the differences in RNAi activity among different larval instars? Several factors may affect RNAi effects in different development stages after a dsRNA has been orally introduced into the midgut (Christiaens et al., Reference Christiaens, Whyard, Vélez and Smagghe2020). Firstly, the availability of the dsRNA affects RNAi efficacy. After ingestion, the dsRNA may bind to food molecules that interfere with cellular uptake. In honey bee larvae, the dsRNA can be bound to the main ingredient of the larval diet, royal jelly. The binding reduces RNAi efficacy (Christiaens et al., Reference Christiaens, Whyard, Vélez and Smagghe2020). Similarly, no mortality was observed when D. virgifera virgifera adults are fed with a mixture of royal jelly and a lethal concentration of dsvATPaseA (Vélez et al., Reference Vélez, Jurzenski, Matz, Zhou, Wang, Ellis and Siegfried2016). Most importantly, dsRNA degradation ability dramatically varies among different insect stages. Nucleases, such as dsRNases, can degrade dsRNA (Arimatsu et al., Reference Arimatsu, Kotani, Sugimura and Furusawa2007; Wynant et al., Reference Wynant, Santos, Verdonck, Spit, Van Wielendaele and Broeck2014; Song et al., Reference Song, Zhang, Li, Cooper, Silver, Li, Liu, Ma, Zhu and Zhang2017). Silencing of genes encoding dsRNases improved RNAi efficiency in L. migratoria (Song et al., Reference Song, Zhang, Li, Cooper, Silver, Li, Liu, Ma, Zhu and Zhang2017), L. decemlineata (Spit et al., Reference Spit, Philips, Wynant, Santos, Plaetinck and Broeck2017), Bemisia tabaci (Luo et al., Reference Luo, Chen, Luan, Chung, Van Eck, Turgeon and Douglas2017) and A. pisum (Chung et al., Reference Chung, Jing, Luo and Douglas2018). Generally, Lepidopterans and Hemipterans showed higher nuclease activity compared to that in other insects (Christiaens et al., Reference Christiaens, Swevers and Smagghe2014; Wang et al., Reference Wang, Peng, Pu, Fu, Wang and Han2016; Singh et al., Reference Singh, Singh, Mogilicherla, Shukla and Palli2017; Song et al., Reference Song, Zhang, Li, Cooper, Silver, Li, Liu, Ma, Zhu and Zhang2017). For example, the dsRNA-degrading nucleases are much more active in Heliothis virescens (inefficient RNAi) than in L. decemlineata (highly efficient systemic RNAi) (Shukla et al., Reference Shukla, Kalsi, Sethi, Narva, Fishilevich, Singh, Mogilicherla and Palli2016). As a result, the Lepidopteran and Hemipteran insect species are more refractory to RNAi. The variability of RNAi efficacy in different insect stages can also be influenced by different physiological pH. For example, ssRNA is most stable at pH 4.0–5.0, while it is susceptible to hydrolysis at pH >6.0 and <2.0, and to depurination at <3.0 (Cooper et al., Reference Cooper, Silver, Zhang, Park and Zhu2019). All these factors may be different between H. vigintioctopunctata young larvae and old larvae, and may result in higher susceptibility of young larvae to dsRNA.
The uptake of the dsRNA by target tissues is the next barrier to successfully RNAi (Christiaens et al., Reference Christiaens, Whyard, Vélez and Smagghe2020). The machinery components involved in cellular uptake of dsRNA such as Sid-like and clathrin genes (Xiao et al., Reference Xiao, Gao, Xu, Liang, Li, Yao and Zhu2015; Cappelle et al., Reference Cappelle, de Oliveira, Van Eynde, Christiaens and Smagghe2016) may be lowly expressed in the old larval instar. As a result, the cellular membrane transport of the dsRNA is suppressed and the efficiency of RNAi is lowered in the old larvae in H. vigintioctopunctata.
Thirdly, the dsRNA needs to be released from the endosome to get in contact with the RNAi machinery once it enters the target cells. In Spodoptera frugiperda, lack of endosomal release of the dsRNA leads to low sensitivity to RNAi (Shukla et al., Reference Shukla, Kalsi, Sethi, Narva, Fishilevich, Singh, Mogilicherla and Palli2016; Yoon et al., Reference Yoon, Shukla, Gong, Mogilicherla and Palli2016). Whether the release of dsRNA from the endosomes differs between young vs. old larvae need further research to address in H. vigintioctopunctata.
Finally, the dsRNA is processed by the RNAi core machinery to generate gene knockdown after the release of the dsRNA from the endosomes. Resting expression levels of core RNAi genes and their responses to dsRNA may be involved in RNAi efficiency (Christiaens et al., Reference Christiaens, Whyard, Vélez and Smagghe2020). In L. decemlineata, ago1, ago2, and aubergine are essential for RNAi (Yoon et al., Reference Yoon, Shukla, Gong, Mogilicherla and Palli2016). These core RNAi genes are highly expressed at the first and second instar stages and relatively lowly expressed at the third and fourth instar stages. Moreover, the levels of dcr2 and ago2 are significantly elevated after 6 h of dsegfp exposure (Guo et al., Reference Guo, Fu, Yang, Li and Li2015). The different expression levels of these core RNAi genes may be responsible for the different RNAi efficiency between H. vigintioctopunctata young larvae and old larvae.
In summary, knockdown of three vATPase genes, especially HvvATPaseE, killed a large proportion of larvae. Young larvae are more susceptible to dsRNA. Therefore, the appropriate timing for dsRNA treatment is the young larval instars in order to improve RNAi efficiency. Our data imply the feasibility of RNAi as an alternative method for controlling this critical potato pest.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485321000420
Acknowledgments
This research was supported by the National Key R & D Program of China (2017YFD0200900), and China Agriculture Research System (CARS-09-P22).
Conflict of interest
The authors have declared that no competing interest exists.