INTRODUCTION
Ciliated protozoa are a primary component of the microzooplankton communities in aquatic ecosystems and play important roles in the microbial food web (Finlay & Esteban, Reference Finlay and Esteban1998). With a short life cycle and more rapid response to environmental changes than many other eukaryotic organisms due to their delicate pellicles, special attention has been increasingly focused on ciliated protozoa as a useful bioindicator of water quality in many aquatic ecosystems in recent decades (Cairns et al., Reference Cairns, Lanza and Parker1972; Coppellotti & Matarazzo, Reference Coppellotti and Matarazzo2000; Corliss, Reference Corliss2002; Tan et al., Reference Tan, Shi, Liu, Xu and Nie2010; Jiang et al., Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a, Reference Jiang, Xu, Al-Rasheid, Warren and Songb; Xu et al., Reference Xu, Zhang, Jiang, Zhu, Al-Rasheid, Warren and Song2011a, Reference Xu, Zhang, Jiang, Min and Choib).
Our previous investigations have demonstrated that non-loricate ciliates, i.e. the forms without lorica or shell, are an important component of whole planktonic ciliate communities in terms of both abundance and occurrence in marine ecosystems (Jiang et al., Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a, Reference Jiang, Xu, Al-Rasheid, Warren and Songb; Xu et al., Reference Xu, Jiang, Zhang, Zhu and Al-Rasheid2011c). With regard to the contribution of the non-loricate ciliate assemblages to ecological patterns of total planktonic ciliate communities in response to water conditions, however, little information was available related to marine ecosystems (Jiang et al., Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a, Reference Jiang, Xu, Al-Rasheid, Warren and Songb; Xu et al., Reference Xu, Jiang, Zhang, Zhu and Al-Rasheid2011c).
In this study, the spatial patterns of non-loricate ciliate assemblages in Jiaozhou Bay, Qingdao, northern China were analysed based on 1-year dataset (June 2007–May 2008). Our aims were: (1) to reveal the contribution of non-loricate ciliate assemblages to total planktonic ciliate communities in ecological features; and (2) to discuss the availability of non-loricate ciliate assemblages being used as a potential bioindicator for bioassessment in marine ecosystems.
MATERIALS AND METHODS
Study sites
Five sampling sites were selected with a gradient of environmental stress in Jiaozhou Bay (Figure 1). Site A was slightly eutrophic due to tidal circulation of pollutants from inshore waters; Site B was selected as a severely eutrophic area due to organic pollutants and nutrients from domestic sewage and industrial discharges from two main rivers; Site C was heavily eutrophic due to pollutants from mariculture activities and the tidal circulation of inshore waters; Site D was moderately eutrophic and is subject to domestic pollutants; and site E was the cleanest area (Figure 1).

Fig. 1. Sampling sites in Jiaozhou Bay, north China: (A) Site A near Huangdao; (B) Site B near the mouths of the Yang and Dagu Rivers; (C) Site C near mariculture area; (D) Site D near the mouths of the Haipo and Licun Rivers; (E) Site E at the mouth linking the bay with the Yellow Sea.
Sampling and sample processing
A total of 120 samples were collected biweekly at a depth of 1 m from five sampling sites during the period of June 2007–May 2008. For quantitative studies and for the species identification of ciliated protozoa, 1000 ml water samples were fixed with acid Lugol's iodine solution to a final concentration of 2% (volume/volume).
Salinity (Sal), pH, and dissolved oxygen concentration (DO) were measured in situ, using a multi-parameter sensor (MS5, HACH). Samples for nutrient analyses were preserved immediately upon collection by placing at –20°C in the dark. Concentrations of soluble reactive phosphorus (SRP), ammonium nitrogen (NH3-N), nitrate nitrogen (NO3-N) and nitrite nitrogen (NO2-N) were determined using a UV-visible spectrophotometer (DR-5000, HACH) according to the Standard Methods for the Examination of Water and Wastewater (APHA, 1992).
For the enumeration of ciliates a 0.1 ml aliquot of each concentrated sample was placed in a Perspex chamber and the ciliates were counted under a light microscope at a magnification 400×. Five aliquots of 0.1 ml from each sample were counted and yielded a standard error (SE) of <8% of the mean values of counts. Those individuals whose identity could not be ascertained following examination of Lugol's-fixed specimens were picked out with a micropipette and identified using protargol impregnation after re-fixing with Bouin's solution (Montagnes & Humphrey, Reference Montagnes and Humphrey1998). Species identification of ciliates was based on keys and guides such as Song et al. (Reference Song, Warren and Hu2009).
Biovolumes of ciliate cells were determined from measurements of their linear dimensions and using volume equations of appropriate geometric shape (Winberg, Reference Winberg1971). Conversion factors of carbon biomass were 0.14 pg C m−3 for non-loricate ciliates preserved with 2% vol:vol Lugol's iodine and 0.053 pg C m −3 for tintinnids (Putt & Stoecker, Reference Putt and Stoecker1989; Stoecker et al., Reference Stoecker, Sieracki, Verity, Michaels, Haugen, Burkill and Edwards1994).
Data analysis of samples
Shannon's diversity (H′), evenness (J′) and richness (D) of samples were computed following the equations:
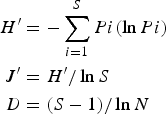
where H′ = observed diversity index; Pi = proportion of the total count arising from the ith species; S = total number of species; N = total number of individuals.
Multiple linear regression and Pearson correlation analysis between biotic and abiotic variables were carried out using the software SPSS v16.0. The best possible regression models were explored using the stepwise selection mode and the optimal model was estimated based on the statistical significance (P < 0.05). Original data were log-transformed in terms of the equation y = log (x + 1). Regression analysis between non-loricate ciliate assemblages and whole communities was performed using the statistical program SigmaPlot v10.0.
Multivariate analyses of spatial patterns of the ciliate communities were analysed using the PRIMER v6.1 package (Clarke & Gorley, Reference Clarke and Gorley2006) and the PERMANOVA+ for PRIMER (Anderson et al., Reference Anderson, Gorley and Clarke2008). The routine SIMPER was used to detect the dominant/common species with a cumulative contribution of 90% to the total ciliate communities (Xu et al., Reference Xu, Zhang, Jiang, Zhu, Al-Rasheid, Warren and Song2011a). Bray–Curtis similarity matrices were computed on log-transformed data. The separate clusters of biotic and abiotic samples were assigned by the routine CLUSTER, while the spatiotemporal differences of ciliate communities among the five sampling sites and four seasons were summarized using the submodule canonical analysis of principal coordinates (CAP) of PERMANOVA+ on Bray–Curtis similarities from log-transformed species-abundance data (Anderson et al., Reference Anderson, Gorley and Clarke2008; Jiang et al., Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a, Reference Jiang, Xu, Al-Rasheid, Warren and Songb). Differences between groups of samples were tested by the submodule ANOSIM (Clarke & Gorley, Reference Clarke and Gorley2006). The submodule BIOENV was used to reveal potential relationships between biotic parameters and the abiotic data. The significance of biota–environment correlations was tested using the routine RELATE (Clarke & Gorley, Reference Clarke and Gorley2006).
RESULTS
Contribution of non-loricate ciliates to spatio-temporal variations of the total ciliates
A total of 64 ciliate species were recorded in the dataset, 37 of which were non-loricate forms, accounting for 64%–79%, 66%–90% and 78%–95% in terms of species number, abundance and biomass from four seasons (Figure 2 A–C), while 64%–76%, 68%–88% and 69%–93% in terms of those from five sites (Figure 2 D–F).

Fig. 2. Seasonal and spatial variations in relative species number (A, D), relative abundance (B, E) and relative biomass (C, F) of planktonic ciliates in Jiaozhou Bay, northern China, during the study period.
The spatio-temporal distribution of the dominant/common ciliate species with a cumulative contribution of 90% to the total ciliate communities in each season is summarized in Figure 3. Of 24 dominant/common taxa, 20 species were non-loricate ciliates, while only 4 belonged to loricate ones which occurred in some seasons (Figure 3)

Fig. 3. The spatio-temporal distribution of the dominant/common ciliate species at five sampling sites in Jiaozhou Bay during the study period, plotted using group-average clustering on Bray–Curtis similarities from standardized/log-transformed species–abundance data. I–V, groups I–V.
Contribution of non-loricate ciliates to spatio-temporal patterns of total ciliate communities
Multivariate correlation (RELATE) analyses revealed that non-loricate ciliate assemblages were significantly correlated with those of the total ciliate communities (R = 0.892, P < 0.001). Discriminating among 120 samples was plotted by CAP on Bray–Curtis similarities from log-transformed species–abundance data, showed temporal and spatial patterns of non-loricate ciliate assemblages (Figure 4). In Figure 4A, the first canonical axis separated the non-loricate ciliate assemblages sampled in summer and autumn (on the left) from those in spring and winter (on the right), while the second canonical axis discriminated the samples in spring and summer (upper) from those in autumn and winter (lower) (Figure 4A). In Figure 4B, the first canonical axis separated the non-loricate ciliate assemblages sampled at Sites A and E (on the left) from those at Sites B and C (on the right), while the second canonical axis discriminated the samples at Sites D and A (upper) from those at the other three sites (lower) (Figure 4B). The ANOSIM test revealed that there were significant differences among the four seasons (R = 0.168, P < 0.001), the five sites (R = 0.120, P < 0.001) and between each pair of sites (P < 0.05) apart from Sites B and C, between which there were no significant differences (P > 0.05).

Fig. 4. Canonical analysis of principal coordinates on Bray–Curtis similarities from log-transformed non-loricate ciliate species–abundance data of four seasons (A) and five sampling sites (B) in Jiaozhou Bay during the study period.
Correlations between structural parameters of non-loricate ciliates and total ciliates
The scatter plots between species number, abundance and biomass of non-loricate ciliate assemblages and those of total ciliate communities are shown Figure 5A, B & C. Linear regression analyses revealed that the species number, abundance and biomass showed high regression coefficients (R2 > 0.88; P < 0.001).

Fig. 5. Scatter plots between species number (A), abundance (B), biomass (C), species richness (D), species evenness (E) and species diversity (F) of non-loricate ciliate assemblages and those of total planktonic ciliate communities at five sampling sites in Jiaozhou Bay during the study period. S, species number; N, abundance; B, biomass; D, species richness; J ′, species evenness; H′, species diversity.
The scatter plots between all three structural indices of non-loricate ciliate assemblages and those of total ciliate communities are shown in Figure 5D, E & F. Linear regression analyses revealed that the three indices showed high regression coefficients (R2 > 0.30; P < 0.001), especially the species richness index (R2 = 0.87; P < 0.001).
Relationships between non-loricate ciliate data and abiotic parameters
The RELATE analysis revealed that there was a significant correlation between spatial variations in planktonic non-loricate ciliate abundances and changes of environmental variables (R = 0.855; P < 0.01). BIOENV matching analysis showed that the best matching with the non-loricate ciliates occurred with the combination of salinity, DO, NO3-N and SRP (Table 1).
Table 1. Summary of results from biota-environment (BIOENV) analysis showing the 10 best matches of environmental variables with spatial variations in non-loricate ciliate abundances at five sampling sites in Jiaozhou Bay during the study period.

R, Spearman correlation coefficient; Sal, salinity; DO, dissolved oxygen; NO3-N, nitrate nitrogen; NO2-N, nitrite nitrogen; SRP, soluble reactive phosphate.
Spearman correlations between biodiversity indices and environmental variables are summarized in Table 2. Results indicated that the species number was positively correlated with DO and negatively with NO2-N, while the species richness represented a similar correlation with DO and NO2-N and a negative correlation with the nutrients NO3-N and SRP (P < 0.05). Species evenness was negatively correlated with NO3-N while species diversity positively correlated with DO, but negatively correlated with NO3-N, NO3-N, NO2-N and SRP (P < 0.05) (Table 2).
Table 2. Correlations (Spearman analysis) between environmental variables and species number (S), abundance (N), species richness (D), species evenness (J′) and species diversity (H′) of non-loricate ciliates at five sites in Jiaozhou Bay during the study period.

*, P < 0.05; **, P < 0.01; NH3-N, ammonium nitrogen; Chl a, chlorophyll-a; see Table 1 for other abbreviations.
Results obtained by multiple linear regression analysis between spatio-temporal abundance data and environmental variables indicated that a total of 19 dominant/common species were correlated with physico-chemical factors (Table 3). Nine non-loricate species (e.g. Strombidium capitatum, Strombidium globosaneum and Strombidium acutum) were found correlated with nutrients. However, only three tintinnids (Stenosemella nivalis, Tintinnopsis orientalis and Leprotintinnus bottnicus) were associated with nutrients (Table 3).
Table 3. Multiple linear regression analysis of abundance of 19 dominant/common ciliates related to environmental variables. F of the model is only shown when the model is multiple.

Tem, temperature; see Table 1 for other abbreviations.
DISCUSSION
So far, a number of studies have demonstrated that ciliate assemblages are strongly related to eutrophication status in a range of aquatic ecosystems (Beaver & Crisman, Reference Beaver and Crisman1982, Reference Beaver and Crisman1989; Jiang et al., Reference Jiang, Wu and Shen2007, Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a, b; Xu et al., Reference Xu, Jiang, Zhang, Zhu and Al-Rasheid2011c, Reference Xu, Jiang, Al-Rasheid, Al-Farraj and Songd). Our previous investigations have demonstrated that the ecological features of planktonic ciliate assemblages are significantly associated with environmental conditions in both temporal and spatial scales (Jiang et al., Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a; Xu et al., Reference Xu, Jiang, Al-Rasheid, Al-Farraj and Song2011d), and that some dominant species (mainly non-loricate species) are significantly correlated with concentrations of nutrients (Jiang et al., Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a, Reference Jiang, Xu, Al-Rasheid, Warren and Songb).
In the present study, the non-loricate ciliate assemblages represented a similar pattern to that of the total ciliate communities. Linear regression analyses showed that the species number, abundance and three biodiversity indices of non-loricate ciliate assemblages showed high regression coefficients with those of total ciliate communities. Linkage analyses to abiotic data demonstrated that the spatial variations in non-loricate ciliate community patterns were significantly correlated with environmental variables, especially nutrients (NO3-N, NO2-N and SRP), either alone or in combination with salinity and DO. In addition, more non-loricate ciliate species were correlated with abiotic parameters especially nutrients rather than tintinnids. These findings are consistent with our previous reports on marine bioassessment using total ciliate communities in Jiaozhou Bay, northern China (Jiang et al., Reference Jiang, Xu, Hu, Zhu, Al-Rasheid and Warren2011a, Reference Jiang, Xu, Al-Rasheid, Warren and Songb; Xu et al., Reference Xu, Jiang, Al-Rasheid, Al-Farraj and Song2011d).
Biodiversity indices (e.g. richness, diversity and evenness of species) are commonly used to simplify statistical analyses in community investigations (Connell, Reference Connell1978; Huston, Reference Huston1979; Magurran, Reference Magurran1991; Ismael & Dorgham, Reference Ismael and Dorgham2003). Generally, the higher the values are, the higher the water quality is (Ismael & Dorgham, Reference Ismael and Dorgham2003). In our study, species richness and diversity indices had higher values in the samples from less stressed sites than those from more stressed sites, and were significantly correlated with NO3-N, NO2-N and SRP (P < 0.05). Thus, we propose that the non-loricate ciliate assemblages might be used as a useful bioindicator for assessing water quality of marine ecosystems.
In summary, the non-loricate ciliate assemblages were the primary components and significantly correlated with the total ciliate communities in terms of species number, abundance and biomass. The spatial pattern of non-loricate ciliate assemblages was significantly related to that of both total ciliate communities and variations in environmental variables. Spatial variations in biodiversity (richness, diversity and evenness of species) indices of non-loricate ciliate assemblages were significantly correlated with those of total ciliate and the environmental conditions, especially nutrients NO3-N, NO2-N and SRP. These results suggest that the non-loricate ciliate assemblages represent an important contribution to the ecological pattern of total ciliate communities and might be used as a useful bioindicator of water quality in marine ecosystems. However, further studies on a range of marine environments and over long time periods are needed in order to verify this conclusion.
ACKNOWLEDGEMENTS
This work was supported by ‘The Natural Science Foundation of China' (Project No. 41076089), the Darwin Initiative Programme (Project No. 14-015) which is funded by the UK Department for Environment, Food and Rural Affairs and partially supported by the Center of Biodiversity Research, King Saud University, Saudi Arabia. We thank Professor Weibo Song, Laboratory of Protozoology, Ocean University of China (OUC), China, for his helpful discussions and Dr Xinpeng Fan, Dr Jiamei Jiang and Dr Xumiao Chen, Laboratory of Protozoology, OUC, China, for their help with sampling and sample processing.










