Published online by Cambridge University Press: 05 December 2003
Biomphalaria glabrata embryonic (Bge) cells have been shown to be a valuable in vitro cellular model for the study of snail host–parasite interactions. They both promote the growth and differentiation of various trematode species including Schistosoma mansoni, and Echinostoma caproni and share some morphological and functional features with circulating haemocytes. As an approach to investigate snail genes potentially regulated following exposure to trematode excretory–secretory (ES) products, we compared gene expression profiles of Bge cells exposed to saline solution, or saline solution containing ES products from S. mansoni or E. caproni, two trematode species parasitizing B. glabrata. Following differential display RT-PCR analysis we characterized 23 differentially displayed cDNAs and we focussed on the 5 cDNAs showing sequence similarity to known genes for expression validation. Using RT-PCR, we confirmed that ES products from S. mansoni and E. caproni differentially affect the expression levels of 4 out of the 5 transcripts. These partial transcripts corresponded to novel B. glabrata sequences, and showed significant sequence similarity to genes coding for (i) cytochrome C, (ii) methyl-binding proteins, (iii) glutamine synthetases, and (iv) protease inhibitors from the Kunitz family. The possible significance of these gene expression changes in host–parasite molecular interactions is discussed.
Molecular studies of the interactions between the parasite Schistosoma mansoni and its molluscan host Biomphalaria glabrata have been facilitated by the recent use of the B. glabrata embryonic (Bge) cell line as an in vitro model. It is a feature of these cells to secrete soluble factors that promote growth and differentiation of various trematode species including S. mansoni (Yoshino & Laursen, 1995; Ataev, Fournier & Coustau, 1998; Laursen & Yoshino, 1999; Coustau & Yoshino, 2000). One major result of co-culture experiments in the presence of Bge cells is the achievement of the entire intra-molluscan development of S. mansoni (from mother sporocysts to cercariae) (Ivanchenko et al. 1999). A second feature of this cell line is that it shares some morphological and functional characteristics with circulating haemocytes (Yoshino et al. 1999). Both Bge cells and haemocytes possess a fibroblast-like appearance, substrate adhesive properties and are capable of encapsulating foreign material (including trematode sporocysts) under in vitro conditions (Yoshino et al. 1999). In addition, Bge cells and haemocytes present similar lysosomal enzyme contents (Yoshino et al. 1999). Bge cells therefore represent an invaluable tool for analysing host genes that may be involved both in general metabolic interactions (leading to parasite development) (Coustau & Yoshino, 2000), or in immunobiological interactions (i.e. adhesion, encapsulation response) (Duclermortier et al. 1999; Yoshino et al. 1999).
In the present study we investigated the effect, on Bge cell gene expression, of excretory–secretory (ES) products from 2 trematodes parasitizing B. glabrata. S. mansoni and E. caproni are known to interact quite differently with Bge cells. Co-cultivation experiments showed that Bge cells could support extended viability and growth of both parasites (Coustau & Yoshino, 2000). However, only S. mansoni mother sporocysts could differentiate and produce a second generation of larvae in the presence of these cells (Yoshino & Laursen, 1995; Ataev et al. 1998). In addition, Bge cells encapsulate S. mansoni, but not E. caproni sporocysts. Whether this differential adhesion results from different receptor/ligand combinations (Coustau & Yoshino, 2000) or from inhibitory effects of E. caproni ES products (Humbert & Coustau, 2001) is unknown. However, it seems likely that the molecular interactions between Bge cells and the 2 parasites involve different signalling pathways. Here, we used the technique of Differential Display RT-PCR (DDRT-PCR) to compare the transcription profiles of Bge cells under control culture conditions and after exposure to ES products from either S. mansoni or E. caproni sporocysts.
Bge cells (ATCC CRL 1494) were cultured at 26 °C in complete Bge medium (Hansen, 1976) as previously described (Coustau et al. 1997). Miracidia from S. mansoni were hatched from eggs axenically recovered from 40-day infected hamster livers (Dissous, Dissous & Capron, 1981). Miracidia from E. caproni were obtained according to a previously described protocol (Humbert & Coustau, 2001). After 3 washes in sterile pond water containing an antibiotic/antimycotic (AB/AM) mixture (penicillin 100 units/ml, streptomycin 0·1 mg/ml, amphotericin 0·025 μg/ml (PSA); Sigma), miracidia from both species were placed into 50 ml tissue-culture plates containing sterile Chernin's balanced salt solution (CBSS; Chernin, 1963) plus the AB/AM mixture. When maintained in this solution at 26 °C under atmospheric conditions, miracidia from both species transformed into mother sporocysts within 24 h. CBSS containing the parasite ES products (including transformation products) was collected after 24 h and spun down (1000 g for 5 min) to eliminate ciliated plates. The supernatant fraction was filtered through a 0·22 μm membrane and protein content was determined using the micro BCA-protein kit assay (Pierce). Products were lyophilized, then solubilized in water and dialysed against CBSS in order to adjust the protein concentration to 90 μg/ml (Lodes & Yoshino, 1989; Humbert & Coustau, 2001). Prior to exposure to parasite ES products, cells were washed 3 times and incubated for 1 h in CBSS to allow the ‘shedding’ of residual Bge medium molecules potentially adsorbed onto their surface. CBSS was then removed and replaced either by fresh CBSS (control sample) or by CBSS containing S. mansoni or E. caproni ES products. After 4 h of incubation, cells were processed for RNA extraction. The entire procedure was carried out under sterile conditions in order to avoid any bacterial or fungal contamination of both ES products and Bge cells.
Differential display was based on the method described by Liang & Pardee (1992) and was carried out following the protocol supplied with the RNAimage Kit (GenHunter Corp.). Briefly, total RNA was prepared from control or exposed Bge cells using RNA now – TC reagent (Biogentex) according to the manufacturer's instructions. Residual contaminating DNA was systematically removed using the MessageClean kit (GenHunter). Integrity of total RNA was checked by electrophoresis of 1 μg RNA through a formaldehyde–agarose gel and staining with ethidium bromide according to standard procedures. Total RNA (0·2 μg) was then reverse-transcribed and 2 μl of the resulting cDNA was used for PCR. PCR reactions were performed in the presence of 2 μCi α-[33P]dATP (Amersham Pharmacia Biotech.) under the following cycling conditions: 30 s at 94 °C, 2 min at 40 °C, and 30 s at 72 °C for 40 cycles in a thermal cycler (MJ Research Inc., PTC-100). Each PCR reaction was performed in duplicate. The radio-isotope labelled PCR products were separated using a 6% denaturing polyacrylamide gel (60 W constant power; 4 h). Gels were dried and analysed by autoradiography after 24–48 h exposure onto X-ray film (Kodak).
The autoradiogram was aligned with the dried gel. Bands of interest, which were differentially represented between the duplicated samples, were cut out of the gel and rehydrated with 100 μl of water for 10 min. DNA was eluted from the gel by boiling for 15 min and recovered by ethanol precipitation. The precipitated cDNA was resuspended in 10 μl of water and a 4 μl aliquot was used to reamplify the DNA fragment, using the primers and conditions of the original PCR reaction. Cloning of the purified PCR products was performed using TOPO® TA Cloning Kit (Invitrogen™, Life Technologies, France), with the PCR® 2.1–TOPO® vector and chemically competent TOP10F′ One Shot® E. coli. Plasmid DNA of the positive recombinant clones was purified using QIAGEN® Plasmid Midipreps Kit (QIAGEN, Germany), and sequenced using ABI PRISM® Big Dye™ Terminator Kit (Applied Biosystems, France) on an automated sequencer (Applied Biosystems 373A DNA sequencer). Sequence analysis was performed using Sequencher™ software (Gene Codes Corporation). Consensus sequences from each set of clones were compared with the GenBank database using BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST/).
Reverse transcription was performed with SuperScript™ II RNase H+ Reverse Transcriptase (Life Technologies, GIBCOBRL®, France), using 2 μg total RNA as substrate and an oligo dT primer (17-mer). Specific forward and reverse primers were designed using the LightCycler Probe Design software (Roche Applied Science), based on sequence data from the selected target genes. The primer sequences were: BgeA3aF 5′-AAGCTGGCGGAAAACA-3′; BgeA3aR 5′-GACCTGCAAACACCATC-3′; BgeA4aF 5′-GCTTCTCAACGAGTATGACA-3′; BgeA4aR 5′-AAAGAATTTTCTACGAGTGCC-3′; BgeMBDF 5′-CGGCAAGTCAT-GCAAT-3′; BgeMBDR 5′-TGCAGGCCAGATGGTA-3′; BgeG6cF 5′-CACCATGT-ACGACACATC-3′; BgeG6cR 5′-ACTAGCACTTCAGTGACG-3′; Bge.KIF 5′-TGGT5A-CAGTTCTCTATGCAAA-3′; Bge.KIR 5′-GGGAGGAACAAGTTGTCA-3′. Primers for the actin gene were designed from the actin sequence available in Genbank (Genbank Acc. no. Z72387): BgeACTF 5′-CACCTCAAACCCTAAA-GCCAACA-3′; BgeACTR 5′-TGAGAGCACA-GCTTG-GATGG-3′. PCR reactions were set up according to the LightCycler Manual (Roche Molecular Biochemicals, germany). A mastermix of the following reaction components was prepared as follows (final concentrations): 8·6 μl water, 2·4 μl MgCl2 (4 mM), 1 μl forward primer (0·4 μM), 1 μl reverse primer (0·4 μM), and 2 μl Lightcycler Fast Start DNA Master SYBR Green I (Roche Diagnostics). Lightcycler mastermix (15 μl) was filled in the Lightcycler glass capillaries and 5 μl cDNA was added as PCR template. All primers were highly purified and salt-free (Rajeevan et al. 2001). The following LightCycler run protocol was used: denaturation programme (95 °C, 10 min), amplification and quantification programmes repeated 40 times (95 °C for 15 s, annealing temperature for 5 s, 72 °C for 13 s), melting curve programme (60–95 °C with a heating rate of 0·1 °C/s and continuous fluorescence measurement), and a cooling step to 40 °C. Analysis of melting curves allowed optimization of annealing temperatures for each amplification product. Single highly specific amplification products were obtained using annealing temperatures ranging from 1 °C below to 1 °C above the Tm of primer pairs. For each reaction, the crossing point CP (defined as the cycle number at which the noise band intersects the fluorescent curves) was determined using the ‘Fit Point Method’ of the LightCycler Software 3.3 (Roche Diagnostics). PCR reactions were all set in triplicates and the mean value of the 3 CPs was calculated. Intra-assay variation (test precision) of CPs was assessed by the coefficient of variation CV=(standard deviation/mean)×100 (Pfaffl, 2001; Rajeevan et al. 2001). For each mRNA to be analysed, the absence of contaminating genomic DNA was verified by running a no-reverse transcription control using primers for actin. In addition, a no-reverse transcription control (H2O control) was analysed for each mastermix.
In order to calculate amplification efficiencies (E) of each target cDNA, relative standard curves were generated using serial dilutions (1, 1[ratio ]10, 1[ratio ]50, 1[ratio ]100, 1[ratio ]500 and 1[ratio ]1000) of a unique Bge cell cDNA sample comprising a pool of the 3 cDNAs to be analysed (1[ratio ]1[ratio ]1). Standard curves were generated by the LightCycler software. They are based on the values of CPs and the log value of the standard cDNA dilution. RT-PCR efficiencies (E) were calculated from the given slope of the standard curve according to the equation E=10(−1/slope). For each sample, the level of expression of the target gene was compared to the expression of the constitutively expressed actin (act) gene (Lardans et al. 1997; Lockyer et al. 2000). The expression ratio (R) is calculated according to the formula R=(Etarget)CPtarget/(Eact)CPact. This ratio is calculated using the target and reference gene CP obtained with an identical cDNA dilution (1/20 in this case). However, because the expression levels of Bge.A4a and Bge.MBD were significantly lower than the expression level of actin, we increased the cDNA concentration to 1/10 and 1, respectively for these 2 target genes. The expression ratio was corrected by the concentration factor (×2 and ×20 respectively). Notice that because higher CP values correspond to weaker expressions, transcripts presenting expression levels lower than the reference gene will have R expression ratios greater than 1.
The relative quantification of expression (Rrel) of a target gene in 2 samples (control versus ES-exposed Bge cells) was calculated according to the mathematical model developed by Pfaffl (2001): Rrel=(Etarget)ΔCPtarget(control−sample)/(Eref)ΔCPref(control−sample), where ΔCP target is the deviation of control−sample of the target gene transcript and ΔCP ref is the deviation of control−sample of the reference gene transcript (actin). This equation actually corresponds to the control expression ratio divided by the sample expression ratio Rcontrol/Rsample (see Pfaffl, 2001, equation 3). In the present study, we used Rrel, or the reciprocal of this ratio 1/Rrel to indicate genes which are up- or downregulated, respectively (Rajeevan et al. 2001).
To analyse the effects of S. mansoni and E. caproni on Bge cell transcription profiles, the DDRT-PCR assay was performed using RNA from Bge cells exposed to CBSS (control) or CBSS containing parasite ES products. PCR reactions were performed using all possible combinations of 3 one-base anchored oligo(dT) primers and the eight 13-mers arbitrary primers available in the GenHunter RNAimage™ Kit 1. Each PCR reaction was performed in duplicate, and the duplicate reactions were loaded onto neighbour lanes of the gel in order to consider only the bands that were identical on both lanes (Ali, Markham & Isaacs, 2001). As an example, Fig. 1 shows defined areas of a DDRT-PCR gel with bands that are differentially displayed between or among duplicates. Complex patterns of amplification were observed for most of the primer combinations. Out of the 3000 fragments amplified in this study, 82 were observed to be differentially displayed (approx. 2·7%). Because DDRT-PCR is prone to generate both false positives and 3′-untranslated portion of transcripts (Kozian & Kirschbaum, 1999; Ali et al. 2001; Oura, Tait & Shiels, 2001), we selected 35 fragments based on the strongest differential amplification and largest size (0·2–1 kb). These fragments were reamplified, cloned and sequenced.

Fig. 1. Selected area of a DDRT-PCR gel showing consistent (black arrows) and inconsistent (white arrows) differences in the transcription profile of duplicated samples.
From the starting 35 DDRT-bands, 23 high-quality sequences have been retained. These sequences have been selected after the following validation steps: (1) validation of the expected size after gel excision and reamplification, and after cloning and digestion of the plasmid insert, (2) validation of the presence of a unique or highly predominant sequence (15 out of the 35 excised bands contained two or more different cDNA sequences) by sequencing of multiple clones (Rajeevan et al. 2001). These 23 sequences are available in Genbank under the accession numbers CB350562-CB350580, AY165005, AY161235, AY160953 and AY272040. DNA and protein database searches were conducted using BLAST search program (http://www.ncbi.nlm.nih.gov/BLAST/) in order to identify putative sequence similarities with known genes. Results showed that all sequences were new B. glabrata sequences and that 18 out the 23 sequences did not show significant matches to sequences in the databases. The deduced amino-acid sequence from 5 of the cDNAs showed similarities with known proteins (Table 1). The translated sequence of the BgeA3a fragment (GenBank Accession No. CB350562) showed high similarity to cytochrome c from various species including Homo sapiens (GenBank Accession No. BC009578), Manduca sexta (GenBank Accession No. M11382) and Drosophila melanogaster (GenBank Accession No. AC006214). A second fragment, Bge A4a (GenBank Accession No. AY161235) showed some similarity to both Dermatopontin from Suberites domuncula and the amoebocyte aggregation factor from Limulus polyphemus. However, based on the probability values (E-values >0·0001) obtained, the 52% and 58% values of sequence similarities were not significant (Table 1) (Anderson & Brass, 1998). The translated sequence of the Bge.MBD cDNA fragment (GenBank Accession No. AY165005) aligned significantly with the methyl binding domain of methyl CpG binding proteins from many different species including human (NM-003927; AF072242; AF120994; 67%), Xenopus laevis (AF170346; 64%), Mus musculus (AF072243; 64%) or Drosophila melanogaster (XM_081190; 85%). It also showed similarities with an unidentified EST from M-line Biomphalaria glabrata haemocytes (BI596282). The clone Bge.GS (GenBank Accession No. AY160953) aligned with glutamine synthetases (GS) from diverse species such as Xenopus laevis (Table 1), Panulirus argus (M96798; 86%); Paracentrotus lividus (L32699; 83%), or Drosophila melanogaster (AY060701; 84%). Finally, the Bge.KI fragment (GenBank Accession No. AY272040) significantly matched the Kunitz domain of various serine protease inhibitors of the Kunitz family including the putative secreted protease inhibitor from Ixodes scapularis (AF483726_1; 50%) (Table 1), the Kunitz inhibitor boophilin from Boophilus microplus (CAC82583·1, 57%) or the human tissue factor pathway inhibitor precursor (Q28864, 57%). Although the percentages of positive amino acids are moderate (around 50%), probability values are highly significant. In addition, as shown on Fig. 2, alignment of the Bge.KI translated sequence with Kunitz domains of known genes confirmed that Bge.KI presents the signature of the Kunitz domains consisting of 6 cysteine residues involved in disulfide bonds and located at conserved positions. The 459 base-pair Bge.KI cDNA contains both the 5′ ATG initiation codon, the stop codon, and the 3′ poly(A) tail suggesting that the complete coding region has been sequenced. This cDNA predicts a translated protein sequence of 82 amino acids.

Fig. 2. Translated amino acid sequence of Bge.KI (AY272040) and alignment with the Kunitz domain of other members of the Kunitz type family of serine proteases inhibitors. Listed are: AF483726, Ixodes scapularis putative secreted protease inhibitors; BMI304447, Boophilus microphilus mRNA for boophilin; S73337, tissue factor pathway inhibitor I from Rhesus monkeys; AF533590.1, Ancylostoma caninum Kunitz-like protease inhibitor precursor; NM_071009.1, Caenorhabditis elegans protease inhibitor; and SYNBOVTRYI, bovine synthetic colostrum trypsin inhibitor. The 6 conserved cysteine residues are shaded in light grey. The predicted inhibitory reactive site residue for each of the Kunitz inhibitors is underlined.

The 18 cDNA fragments that did not align with known sequences will be analysed in a future study consisting of both the confirmation of their differential expression, and the determination of the complete coding sequence and protein identification of the validated transcripts. In the present study, we focussed on the 4 cDNA fragments BgeA3a, Bge.MBD, Bge.GS and Bge.KI, for confirmation of their differential expression, since they showed significant sequence similarity to genes that may be potentially important in the host–parasite interaction. Bge.A4a, in spite of its non-significant alignment with Dermatopontin and the Limulus aggregation factor, also deserves further investigation as such proteins are respectively involved in cell–matrix and cell–cell interactions (Fujii et al. 1992; Forbes et al. 1994). We therefore included this transcript in the analysis of expression.
In order to verify that the selected differentially displayed fragments were effectively regulated following exposure to parasite ES products, we first conducted semi-quantitative RT-PCR assays and Northern blot analysis (results not shown). The level of sensitivity of the semi-quantitative RT-PCR technique only suggested weak differences in the expression of some transcripts but did not allow a precise conclusion about differential expression. Extensive efforts were also made to confirm the regulation of the different selected genes by Northern blot analysis. Conclusive results were only obtained for Bge.GS transcripts that were estimated to be upregulated by a factor 2.1 by S. mansoni ES products (result not shown). In the present work, we finally used the RT-PCR technique to confirm differential gene expression.
Analysis of serial dilutions of Bge cell cDNA showed that the 6 primer pairs amplified single specific PCR products, with amplification efficiencies ranging from 1·77 to 1·96 (Table 2) with high linearity (Pearson correlation coefficient r>0·95). This deviation from the expected efficiency of 2 may be explained by various factors including the high GC content of some sequences. We therefore used the ‘efficiency calibrated’ mathematical method (Pfaffl, 2001) rather than the 2−ΔΔCT method (Livak & Schmittgen, 2001) for the relative quantification of gene expression. Calculation of expression ratios (R) showed that, in control Bge cells, the investigated transcripts were from 142·8-fold less (Bge.MBD) to 2·77-fold more (Bge.GS, R=0·36) expressed than actin (Table 2). Analysis of the relative expression ratios (Rrel) revealed that exposure of Bge cells to parasite ES products differentially affected the expression levels of 4 of the transcripts (Table 2). Differentially expressed bands obtained by DDRT-PCR are generally considered validated if their relative expression ratio is equal or greater than 2 (Rajeevan et al. 2001). Using this criterion, expression of both Bge.A3a, Bge.GS and Bge.KI was shown to be significantly upregulated after exposure to S. mansoni but not to E. caproni ES products (Table 2). In contrast, expression levels of the Bge.MBD transcript was significantly downregulated by ES products of both parasites, with a stronger effect of E. caproni. Finally, the expression of Bge.A4a appeared weakly upregulated after exposure to S. mansoni ES and weakly downregulated after exposure to E. caproni ES products, but these differences cannot be considered as significant.

Differential display of mRNAs is one of the most commonly used methods for studying cellular responses to defined perturbations (Kagnoff & Eckmann, 2001), or for investigating constitutive gene expression differences between 2 or more samples (Kozian & Kirschbaum, 1999). Previous DDRT-PCR studies on Biomphalaria glabrata have investigated either the effect of S. mansoni infection on gene expression in various snail tissues (Lockyer et al. 2000) or in haemocytes (Miller et al. 2001), or the constitutive gene expression differences in haemocytes from S. mansoni-susceptible or -resistant snails (Schneider & Zelck, 2001). Using RT-PCR (Miller et al. 2001), or semi-quantitative RT-PCR (Lockyer et al. 2000) the validation of differential expression has been reported for 2 transcripts: a putative cytochrome P450 (Lockyer et al. 2000), and a transcript showing sequence similarity to E. coli transposase Tn5 (Miller et al. 2001).
In the present study, we used Bge cells as a host model to investigate genes potentially involved in molecular interactions between B. glabrata and the 2 trematodes S. mansoni and E. caproni. Among other interesting features, Bge cells (1) promote growth and differentiation of both parasites (Yoshino & Laursen, 1995; Ataev et al. 1998), (2) present functional characteristics of circulating haemocytes (Yoshino et al. 1999), and (3) show a differential encapsulation response in the presence of these parasites (Coustau & Yoshino, 2000). In addition, the use of this sterile cell line exposed only to sterile solutions provides us with a source of pure B. glabrata RNA. When comparing expression profiles of Bge cells after 4 h exposure to either CBSS, or CBSS containing S. mansoni or E. caproni ES products, it appeared that approximately 2% of the amplified fragments were differentially displayed. Following several selection and validation steps, 23 partial cDNAs were characterized. Sequence analysis of these cDNAs revealed that they all corresponded to novel B. glabrata cDNAs and that the translated sequence of 5 of them presented similarities with known genes. In the present study we focussed essentially on these 5 transcripts. Further studies will attempt to clarify both the identity and the potential involvement of the unknown transcripts in host–parasite interaction. Using quantitative RT-PCR, we validated the differential expression of 4 out of the 5 investigated transcripts, namely, Bge.A3a, Bge.MBD, Bge.GS and Bge.KI. It is interesting to note that the highest change in gene expression level was 5·6-fold. Such differences in expression may be biologically significant and are accurately measured using a quantitative PCR method, but our results have demonstrated that they are hardly or not detected using RT-PCR or semi-quantitative RT-PCR. Our data further support the idea that RT-PCR is a particularly appropriate method of validation of differentially expressed genes identified by DDRT-PCR (Rajeevan et al. 2001), and may explain why validation of DDRT-PCR approaches on Biomphalaria was extremely difficult in earlier studies that did not benefit from the RT-PCR technology.
Understanding the biological significance of the gene expression changes reported in the present study or in previous ones (Lockyer et al. 2000; Miller et al. 2001) clearly requires additional investigations. Expression profiles of any cell type may potentially be affected by the presence of exogenous molecules such as parasite nutrient mixture (proteins) or metabolic by-products for example. Therefore, part of the future studies should be aimed at clarifying how specific is the observed response. However, the validated changes in gene expression observed here are different according to the investigated transcripts (up- versus downregulations) and according to the nature of ES products (S. mansoni versus E. caproni). This result does not support the possibility of non-specific up- or downregulation of transcription and suggest that at least part of the regulation observed reflects a specific effect of some parasitic factors.
It is not surprising that ES products from the 2 parasites differentially affect Bge cell gene expression as these parasites are involved in quite different snail–trematode interactions (Adema & Loker, 1997). However, it is difficult, at this stage, to make assumptions on what role these changes in mRNA level may be playing in snail–parasite interactions. Indeed, the structural identities of the 4 differentially expressed proteins need to be confirmed as well as their functional activities, taking into account that they can be involved in various biochemical pathways or regulatory processes.
For example, the significant upregulation of the Bge.A3a and Bge.GS transcripts by S. mansoni ES products could reflect both a stimulatory effect on metabolism, or a response to the presence of toxic compounds. Although these 2 cDNAs are partial, alignments of their sequence with known genes are highly significant and suggest that they correspond to B. glabrata cytochrome c and glutamine synthetase (GS) genes, involved respectively in energy and general metabolism. Regarding cytochromes c, for a long time their sole function was assumed to be electron transfer from one to another of the mitochondrial electron transport complexes. Recently, they were also found to play a key role in the apoptotic and antioxidant cascades (Skulachev, 1998; Regula & Kirshenbaum, 2003). Similarly, glutamine synthetase (GS) enzymes, catalyse the conversion of ammonia and glutamate to glutamine and are therefore involved in multiple cellular functions related either to biosynthesis of glutamine for production of amino acids or sugars, or recycling of both the neurotransmitter glutamate and ammonia (detoxication) (Suarez, Bodega & Fernandez, 2002). In contrast to cytochromes c, the expression of glutamine synthetases is known to be highly regulated, at the transcriptional level, in a tissue-specific and developmentally controlled manner (Lie-Venema et al. 1998; Smartt et al. 2001). The rate of GS transcription is positively affected by glucocorticoids, growth hormone or insulin (Max, 1990; Lie-Venema et al. 1998). In the context of a host–parasite interaction, glutamine, which is an important precursor for protein and glucose synthesis, might represent one of the essential nutrients for parasite development. Interestingly, a recent study revealed that exposure of S. mansoni primary sporocysts to Bge cell products was associated with an increased level of transcription of the glutaminyl-tRNA synthetase (SmGlnRS) parasite gene (Coppin et al. 2003). Glutaminyl-tRNA synthetase catalyses the ligation of glutamine to its cognate tRNA, and its overexpression could be associated with an increase in protein synthesis (Coppin et al. 2003). The potential relationship between these 2 independent observations is currently being investigated in order to address the questions of the potential regulation of snail glutamine biosynthesis and parasite glutamine uptake during sporocyst development.
The downregulation of a cDNA fragment (Bge.MBD) showing sequence similarity to the methyl-binding domain of various methyl binding proteins (MBPs) is also intriguing. MBPs are important mediators of the methylated DNA silencing process. Their binding to methylated CpG dinucleotides generally present in promoter regions of vertebrate genes (Roder et al. 2000) prevents transcription of the corresponding genes (Meehan et al. 1992; Razin, 1998). This silencing process has been extensively studied in vertebrate species since it is involved in the regulation of tissue- or developmental stage-specific gene expression, X-chromosome inactivation, genomic imprinting or carcinogenesis (Robertson & Jones, 2000; Meehan et al. 1992). However, the existence of an equivalent process in invertebrate species was unsuspected until recently, and is still poorly documented (Shimizu, Takahashi & Tomita, 1997; Tweedie et al. 1997; Roder et al. 2000; Lyko, 2001). The potential existence of an MBP in a snail species therefore clearly deserves further investigations.
Finally, the upregulation of a Kunitz protease inhibitor could potentially be related to immunobiological interactions. Serine protease inhibitors of the Kunitz family can participate in any biological process involving neutralization of serine proteases including endogenous or exogenous proteases (Armstrong, 2001). In invertebrate species, they have been characterized as haemolymph or haemocyte proteins, and are suspected to function either in protecting their hosts from infection or in regulating endogenous proteases involved in immune cascades (Kanost, 1999). On the another hand, proteolytic enzymes are known to be essential virulence factors for most parasite species including helminths (Monroy & Dresden, 1996; Armstrong, 2001; Maizels et al. 2001). Therefore, it would be of particular interest to characterize the serine proteases potentially present in the ES products from S. mansoni and E. caproni, and to identify the molecular target of this B. glabrata Kunitz inhibitor.
In conclusion, our DDRT-PCR and RT-PCR approaches showed that S. mansoni and E. caproni secrete soluble factors that differentially affect the gene expression profile of Bge cells. In order to make progress in the understanding of snail–trematode molecular interactions, future studies should include, not only the characterization of host genes regulated following exposure to the parasites, but the purification and identification of the parasitic factors acting on these genes.
The authors are grateful to N. Dinguirard, N. Lallement, J.-R. Pagès and L. Plaisance for their help in the laboratory, and to anonymous referees for their critical comments and suggestions on an earlier version of this work. This work was supported by the CNRS and by a grant (PRFMMIP) from the Ministère de l'Education, de la Recherche et de la Technologie.
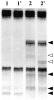
Fig. 1. Selected area of a DDRT-PCR gel showing consistent (black arrows) and inconsistent (white arrows) differences in the transcription profile of duplicated samples.

Fig. 2. Translated amino acid sequence of Bge.KI (AY272040) and alignment with the Kunitz domain of other members of the Kunitz type family of serine proteases inhibitors. Listed are: AF483726, Ixodes scapularis putative secreted protease inhibitors; BMI304447, Boophilus microphilus mRNA for boophilin; S73337, tissue factor pathway inhibitor I from Rhesus monkeys; AF533590.1, Ancylostoma caninum Kunitz-like protease inhibitor precursor; NM_071009.1, Caenorhabditis elegans protease inhibitor; and SYNBOVTRYI, bovine synthetic colostrum trypsin inhibitor. The 6 conserved cysteine residues are shaded in light grey. The predicted inhibitory reactive site residue for each of the Kunitz inhibitors is underlined.

Table 1. Summary of the sequence alignment results

Table 2. Relative quantification of target cDNA expression levels in Schistosoma mansoni (Bge S.m) or Echinostoma caproni (Bge E.c) exposed versus CBSS-exposed (Bge ctl) Bge cells