Enterococci are gram-positive cocci responsible for many serious infectious syndromes including bacteremia and endocarditis. Out of concern for β-lactam resistance, vancomycin is often used for serious presumptive enterococcal infections until the results of antimicrobial susceptibility testing are available. However, particularly in the last 20 years, colonization with and infections due to vancomycin-resistant enterococci (VRE) have been increasing.Reference Ramsey and Zilberberg1
Patients with prolonged hospital stays, exposure to antibiotics,Reference Fridkin, Edwards and Courval2 hematological malignancies,Reference Suppola, Volin, Valtonen and Vaara3,Reference Mikulska, Del Bono and Raiola4 and solid-organ transplantationsReference Bedini, Codeluppi and Cocchi5 are at increased risk of VRE colonization. In these high-risk patients, the question of whether to add empiric VRE coverage for serious infections often arises. On one hand, failure to cover VRE may lead to clinically important delays in receipt of timely, appropriate therapy. On the other hand, the agents most commonly used to cover VRE, linezolid and daptomycin, have limitations. Although linezolid is the only antibiotic with FDA approval for VRE bacteremia, it can lead to hematological toxicity and important drug–drug interactions.Reference French6 The most commonly used alternative, off-label daptomycin, requires a higher dose than that quoted on the product monograph,Reference Foolad, Taylor, Shelburne, Arias and Aitken7 which may have implications on safety for acutely ill patients, particularly those with unstable renal function and those requiring longer-term therapy.Reference Chuang, Lin and Chen8 Widespread use anti-VRE coverage may also increase healthcare costs and promote resistance.Reference Bagga, Buckingham, Arnold, Nesbitt, Guimera and Lee9 However, failure to cover VRE at the onset may result in rapid worsening of an infection and may increase both the required treatment duration with these agentsReference Ye, Shie and Cheng10 and the risk of adverse events. The choice to add empiric VRE coverage entails a careful balancing of the harms and benefits of a therapy. Because VRE colonization is a major risk factor for infection, and based on previous work on MRSAReference Butler-Laporte, Cheng, Cheng, McDonald and Lee11,Reference Butler-Laporte, De L’Etoile-Morel, Cheng, McDonald and Lee12 and VREReference Brasg, Elligsen, MacFadden and Daneman13,Reference Webb, Healy and Majers14 prediction using screening swabs, we postulated that a patient’s VRE status could be used to identify those least and most likely to benefit from empiric VRE coverage in the context of enterococcal bacteremia.
Material and methods
We conducted a retrospective review of all consecutive adult enterococcal BSI from April 3, 2010, to March 24, 2017, at the McGill University Health Centre (2 hospitals, 770 beds, serving a population of 850,000). Only the first positive culture per patient was included. Antibiotic susceptibilities were determined using the VITEK-2 automated system (Biomérieux, Marcy-l’Étoile, France) and were interpreted in accordance with guidelines from the Clinical Laboratory Standards Institute.15 Patients with infections with Enterococcus spp that were intrinsically resistant to vancomycin (eg, E. gallinarum) were included.
Our hospital employed universal VRE screening on admission to the medical and surgical wards and the critical care units. Screening was repeated weekly while admitted to these units. Patients who were VRE colonized were placed in contact isolation, most often in private rooms, but they were occasionally placed in cohort groups. Screening swabs were collected from the rectum, stool, or colostomy, but clinical samples were also accepted if requested by the infection control team. In the laboratory, swabs were first incubated in a vancomycin and aztreonam selective broth. If there was growth within 48 hours, polymerase chain reaction (PCR) was performed using in-house primers for van A, -B, and -B-2/3 genes. If the PCR was positive, the broth was subcultured on a VRE specific chromogenic agar (Bio-Rad, Berkeley, CA) for confirmation. If either step was negative, the patient was considered not to be colonized with VRE. This protocol did not screen for other van phenotypes or for species with intrinsic vancomycin resistance. Local and national VRE epidemiology has been previously reported.Reference McCracken, Wong and Mitchell16,Reference McDonald, Dendukuri, Frenette and Lee17 The only change that occurred during the period of study was an institutional move for one of our study sites to a new facility with single patient rooms in 2015, which resulted in a 4-fold decrease in the VRE colonization incidence rate ratio.Reference McDonald, Dendukuri, Frenette and Lee17 Throughout the period described in our study, the infection control and laboratory protocols have remained unchanged and represent hospital standard operating procedures.
For our analysis, we categorized VRE colonization status in 3 ways. First, looking only at patients who were screened for VRE within the 30 days prior to the positive blood culture, a patient was considered positive for VRE colonization if there was any positive VRE swab within that period and was considered negative otherwise (30-day criteria). Second, we considered any patient with any prior positive VRE specimen as positive and patients with no positive and 1 or more negative VRE screens as negative (all-time criteria). Third, we included all patients who were not previously known to be colonized or infected as VRE negative, including those who had never been screened for VRE (inclusive criteria). The inclusive criteria simulates a “worse case” scenario in which all patients with unknown VRE carriage status are assumed to be negative, therefore increasing the number of false negatives and decreasing the sensitivity and negative predictive value (NPV) of the test.
To allow for comparison with other studies, we also used the method from Brasg et alReference Brasg, Elligsen, MacFadden and Daneman13 in which only the last available screening swab result was considered (most recent criteria). We then pooled our results with theirs to obtain the overall sensitivity, specificity, likelihood ratios, and predictive values as functions of prevalence. These data can be found in the online supplement.
Sensitivity, specificity, likelihood ratios, positive predictive values (PPVs) and NPVs were calculated using standard formulas. Clopper-Pearson confidence intervals for the sensitivities and specificities were computed. The likelihood ratio and PPV/NPV confidence intervals were obtained by simulating the appropriate binomial random variables 1,000 times using the parametric bootstrap method.Reference Efron and Tibshirani18 These were plotted as a function of the proportion of vancomycin-resistant enterococcal bacteremia (the VRE proportion). Missing data were not inferred. Analyses were performed in R version 3.2.0 software (R Foundation for Statistical Computing, Vienna, Austria) with the ggplot2 version 1.0.1 package.
Results
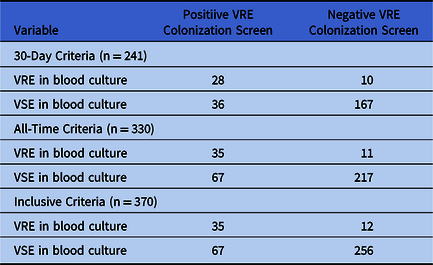
In total, our study included 370 enterococcal bacteremias. For 241 episodes, a screening swab was performed in the 30 days prior. The 30-day criteria yielded an NPV of 90% for a VRE proportion below 27.0%, and 95% for a VRE proportion below 15.0%. An additional 89 patients with VRE bacteremia had a prior screening swab dating back more than 30 days. The resulting all-time criteria had an NPV exceeding 90% and 95% at VRE proportions below 27.2% and 15.2%, respectively. Finally, 40 patients never had a VRE screening swab performed prior to their bacteremia. Assuming these patients had negative screening tests, the inclusive criteria yielded an NPV of 90% at a VRE proportion of 26.7% or less, and of 95% at VRE proportions 14.9% or less. In all cases, the PPV exceeded 50% at a VRE proportion exceeding 25%. Table 1 contains the full 2 × 2 tables for all 3 criteria. Sensitivity, specificity, and likelihood ratios are shown in Table 2. Figures 1–3 show the PPVs and NPVs values as a function of the VRE proportion. For context, at our center, the VRE proportion at the time this study was undertaken was 11% on surgical units, 14% on solid-organ transplant units, 15% on medical units, 29% in the intensive care unit (ICU), and 57% on the hematology-oncology units. Thus, at the time, outside the ICU and hematology-oncology units, a negative VRE screening swab had an excellent NPV for VRE bacteremia in suspected enterococcal infection and could help exclude the need for VRE coverage.
Table 1. Association Between Vancomycin-Resistant Enterococci Colonization and Concurrent Enterococcus Bacteremia Vancomycin Resistance, Obtained Using the 30-day, All-Time, and Inclusive Criteriaa
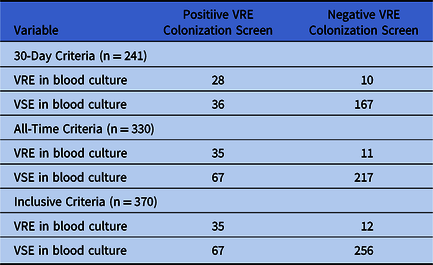
Note. VSE, vancomycin susceptible Enterococci; VRE, vancomycin-resistant Enterococci.
a Please refer to the Methods section for definitions of the 3 criteria and details on the colonization screening procedures.
Table 2. Diagnostic Properties for the 30-Day, All-Time, and Inclusive Criteria for VRE Colonization Screening in the Prediction of Concurrent Enterococcus Bacteremia Vancomycin Resistancea

Note. CI, confidence interval; LR+, positive likelihood ratio; LR−, negative likelihood ratio; NPV, negative predictive value; VRE, vancomycin-resistant Enterococcus.
a Please refer to the Methods section for definitions of the 3 criteria.

Fig. 1. Positive and negative predictive values (solid line: median, dashed lines: 95% confidence intervals) for the 30-day criteria, as a function of the proportion of Enterococcus bacteremias with vancomycin resistance.

Fig. 2. Positive and negative predictive values (solid line: median, dashed lines: 95% confidence intervals) for the all-time criteria, as a function of the proportion of Enterococcus bacteremias with vancomycin resistance.

Fig. 3. Positive and negative predictive values (solid line: median, dashed lines: 95% confidence intervals) for the inclusive criteria, as a function of the proportion of Enterococcus bacteremias with vancomycin resistance.
Discussion
We evaluated the sensitivity and specificity of VRE colonization status to predict the presence of VRE bacteremia in patients with enterococcal bloodstream infection and calculated estimates of the NPVs and PPVs based on the proportion of VRE among all Enterococcus bacteremia. We demonstrated that, for a given VRE proportion less than 27%, the NPV of a VRE screening test for predicting VRE bacteremia exceeded 90% and increased to 95% when the VRE proportion was below 15%. In stable patients for whom a 5%–10% risk of delaying VRE therapy is clinically acceptable, a negative screening test can guide a decision to forego empiric linezolid or daptomycin. As expected, the 30-day criterion was less sensitive but more specific than other criteria. However, the overall diagnostic properties were similar for all criteria and older results remain helpful depending on the local VRE proportion. Our findings suggest that even if screening practices vary, even a dated VRE colonization status can be helpful in choosing antimicrobials in the absence of high levels of nosocomial VRE transmission. We have provided PPV/NPV at different VRE proportions (Figs. 1–3) to allow other institutions to evaluate the operating characteristics of VRE screening swabs in their local context.
One limitation of our study was our lack of access to individual patient-level data. Nevertheless, in general, colonization likely precedes infection, and we have previously shown that, for MRSA, colonization status is the single most important risk factor for infection.Reference Butler-Laporte, Cheng, McDonald and Lee19 There is no reason a priori to suspect that it might be different for VRE. Another limitation is our focus on bloodstream infections. We chose this subset of infections due to the relative ease and specificity of case finding and because it provides both the worst case for the NPV and the best case for the PPV. Using a negative VRE colonization status to withhold anti-VRE therapy seems justified in most patients, especially for infections in which Enterococcus is unlikely to be contributory. At the same time, using a positive colonization status to drive empiric anti-VRE therapy when an Enterococcus infection is unlikely would be unjustified. However, in situations in which an Enterococcus spp is justifiably suspected, such as hepatobiliary infections or when susceptibility data from a confirmed gram-positive cocci infection is pending, empiric VRE-directed therapy could be justified. Essentially, clinicians would need to refer to local microbiology and clinical data when using VRE colonization for treatment guidance. Additionally, because second-line antimicrobials for Enterococcus are usually not tested during first-line susceptibility testing, a positive VRE screen might be used to alert the microbiology laboratory that early second-line susceptibility testing could be warranted for presumed enterococci to avoid delays.Reference Rubinstein, Hirsch and Bandyopadhyay20
Previously, most studies involving the use of VRE colonization status in predicting vancomycin resistance in clinical isolates have involved the role of VRE colonization as a risk factor for eventual VRE infection.Reference Bossaer, Hall and Garrett-Mayer21,Reference Ford, Lopansri and Haydoura22 To our knowledge, apart from Brasg et alReference Brasg, Elligsen, MacFadden and Daneman13 and Webb et al,Reference Webb, Healy and Majers14 no other study has analyzed the predictive accuracy of VRE screening tests to guide empiric therapy when an enterococcal infection is known or suspected. Although the conclusions of other studies were similar, their study designs differed in multiple ways, which significantly limits their generalizability. First, we screened all patients on admission to medical, surgical, and critical care wards, regardless of medical comorbidities, although the other studies restricted their patient population either by limiting screening to specific patient groups at high risk of VRE colonization or by only including patients undergoing stem cell transplantation or leukemia induction chemotherapy. Second, the overall proportion of VRE in their institutions was much lower than ours. Third, their VRE screening protocols differed greatly. In Brasg et al,Reference Brasg, Elligsen, MacFadden and Daneman13 the protocol began with VRE specific agar with confirmation of vanA and/or vanB gene carriage by PCR. By performing PCR first, our protocol may have achieved higher sensitivity. In Webb et al,Reference Webb, Healy and Majers14 no PCR was used, and the screening agar also differed, which also most likely reduced their sensitivity. We suspect that these methodological differences contribute to any discordant results.
Although the retrospective nature of our study does not provide proof of clinical benefit, we contend that knowledge of previous VRE screening test results can help guide appropriate empiric antibiotic therapy. Our research adds to the growing number of studies reporting on the importance of a patient’s bacterial flora on subsequent resistance in infection. In addition to MRSAReference Butler-Laporte, De L’Etoile-Morel, Cheng, McDonald and Lee12 and VRE, a similar predictive value has been observed in gram-negative bacterial infections.Reference MacFadden, Coburn and Shah23 We suggest that we are moving toward an era in which empiric therapy can be personalized based on an individual’s known colonization status and previous microbiology, local epidemiology, likely source of infection, and severity of the illness, to improve outcomes and reduce adverse events.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2019.380
Acknowledgments
We thank Selma, Darius, and Artémis for their unconditional support.
Financial support
Support for this study was provided by the Clinical Practice Assessment Unit, McGill University Health Centre.
Conflicts of interest
Emily G. McDonald receives salary support from the Fonds de Recherche Santé Québec and research funding from the Canadian Institutes of Health Research. Todd C. Lee receives salary support from the Fonds de Recherche Santé Québec and research funding from the Canadian Institutes of Health Research. All other authors report no conflicts of interest relevant to this article.