Introduction
Aspidogastrea is a minor group of flatworms that infect poikilothermic animals, including molluscs, fish and reptiles, and, as an exception, some crustacean species in marine and freshwater environments (Alves et al., Reference Alves, Viera, Santos, Scholz and Luque2015). According to the most popular opinion, this group is considered as a subclass within the class Trematoda (Skrjabin, Reference Skrjabin and Skryabin1952; Dollfus, Reference Dollfus1958; Rhode, Reference Rohde, Jones, Bray and Gibson2002). In contrast to members of the other subclasses, Aspidogastrea have a simple life cycle with no parthenogenetic stages. For this reason, and due to a number of morphological features, few authors have considered aspidogastreans as a distinct class of flatworms (Timofeeva, Reference Timofeeva1975). A number of freshwater aspidogastreans have a rather wide geographical distribution (Achmerov, Reference Achmerov1956; Vosnesenskaya, Reference Vosnesenskaya1968; Stromberg, Reference Stromberg1970; Nagibina & Timofeeva, Reference Nagibina and Timofeeva1971; Strelkov, Reference Strelkov1971; Shimazu, Reference Shimazu, Otsuru, Kamegai and Hayashi2003; Schludermann et al., Reference Schludermann, Laimgruber, Konecny and Schabuss2005; Zhang, Reference Zhang2006; Popiolek et al., Reference Popiolek, Luczynski and Jarnecki2007; Yuryshynets & Krasutska, Reference Yuryshynets and Krasutska2009; Shedko et al., Reference Shedko, Sokolov, Koschelev, Evteshina, Mikheev and Litovchekno2010; Alves et al., Reference Alves, Viera, Santos, Scholz and Luque2015). However, there is still no molecular evidence of conspecificity of species from European and Asian territories. Underestimation of this fact has led to inappropriate conclusions about the phylogeny and distribution of endemic aspidogastrean species (Chen et al., Reference Chen, Zhang, Wen, Sun and Gao2010). Molecular studies of trematode phylogeny are commonly based on ribosomal DNA sequences, including 18S, 28S rDNA and internal transcribed spacer (ITS) regions (Jousson et al., Reference Jousson, Bartoli and Pawlowski2000; Lockyer et al., Reference Lockyer, Olson and Littlewood2003; Olson et al., Reference Olson, Cribb, Tkach, Bray and Littlewood2003; M.-X. Chen et al., Reference Chen, Wang, Yao and Nie2007; Petkevičiūtė et al., Reference Petkevičiūtė, Stunzenas, Staneviciute and Sokolov2010).
Nucleotide sequences of the ITS1–5.8S–ITS2 fragment of the ribosomal cluster were used in our study to evaluate the phylogenetic relationships of European and Far Eastern representatives of the genus Aspidogaster Baer, 1827: A. conchicola Baer, 1827; A. limacoides Diesing, 1834; A. ijimai Kawamura, 1915; and A. chongqingensis Wei, Huang & Dai, 2001.
Materials and methods
Sample collection and identification
Aspidobothrean trematodes were obtained during parasitological field work in 2009–2011 from the European part of Russia (Rybinsk reservoir (58°5′N 38°17′E) and Tvertza River (56°56′N 35°41′E)) and from the Russian Far East (Khanka Lake, Primorskyi Region (44°31′N 132°22′E), and two locations of the stream canal of the Amur River – 140 km downstream from Khabarovsk city (49°13′N 136°14′E) and near Nikolaevsk–na–Amure city (53°6′N 140°41′E)).
Morphological data
Most trematode specimens were killed with hot tap water without crushing, and were flattened under slight pressure, fixed in 70% ethanol, stained with alum carmine and, after dehydrating and clearing, were mounted in Canada balsam. Species identification was performed according to different authors (Kawamura, Reference Kawamura1915; Timofeeva, Reference Timofeeva1973; Tang & Tang, Reference Tang and Tang1980; Bykhovskaya-Pavlovskaya, Reference Bykhovskaya–Pavlovskaya and Bauer1987; Pavljuchenko, Reference Pavljuchenko2007). Thus, we obtained aspidogastrean specimens that unambiguously belonged to A. conchicola, A. limacoides s. str. and A. ijimai (figs 1–3, table 1). Specimens of A. ijimai collected for the present study possessed a spined cirrus (fig. 3), which is in contrast to the original description of this species (Kawamura, Reference Kawamura1915). We have studied specimens of A. ijimai from the type host (Cyprinus carpio (L.) s. lato) and type location (Biwa Lake, Japan), which were provided by Dr T. Shimazu. These specimens are morphologically similar to those from Primorye, including details of the cirrus structure. Voucher specimens of the studied species were deposited in the Museum of Helminthological Collections at the Centre for Parasitology of the A.N. Severtsov Institute of Ecology and Evolution, Moscow, Russia (IPEE RAS): A. conchicola, inventory number 14257; A. limacoides s. str., 14258; and A. ijimai, 14259 (table 1).
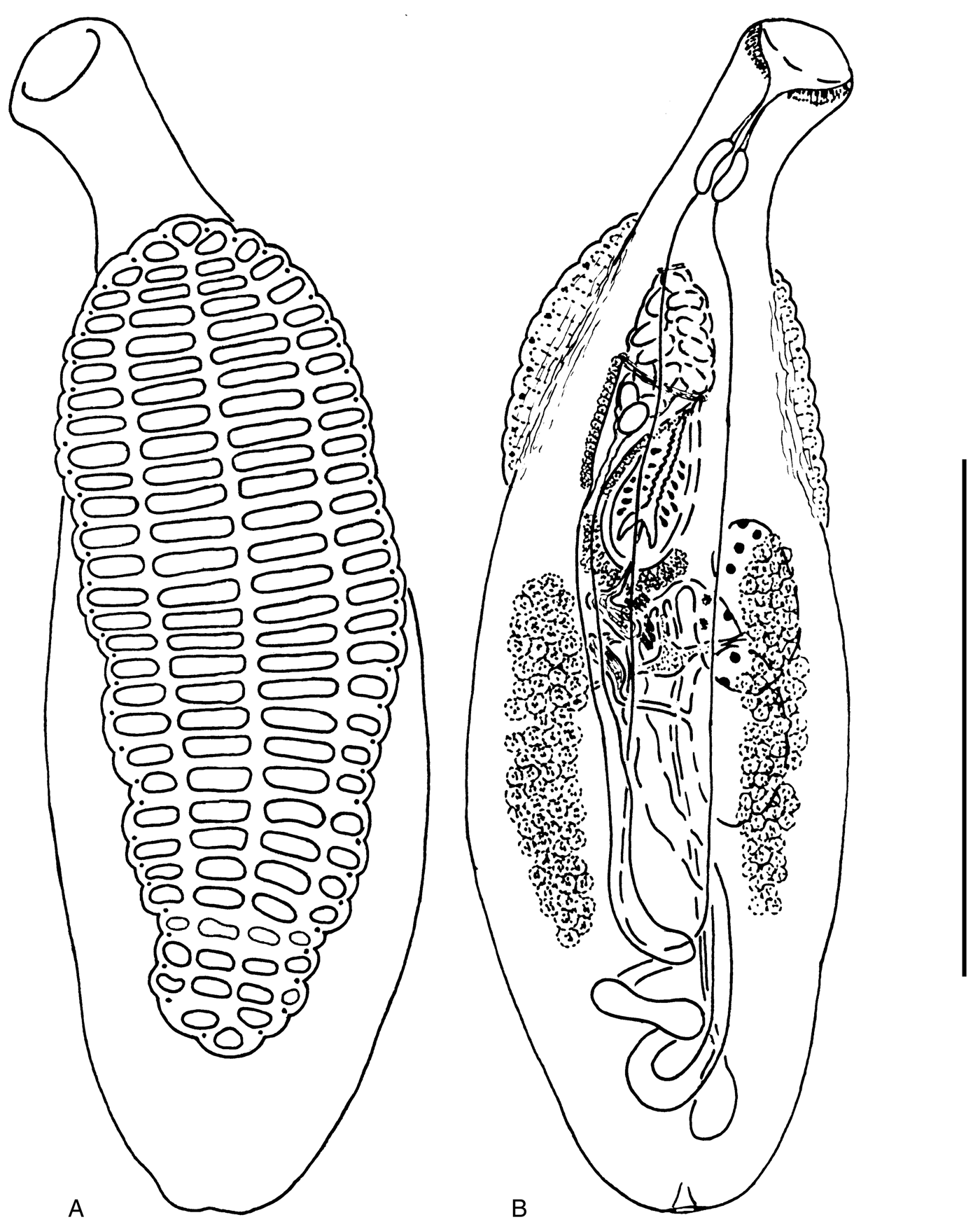
Fig. 1. Aspidogaster conchicola from Colletopterum anatinum, Tvertza River: (A) ventral view; (B) dorsal view. Scale bar: 1 mm.
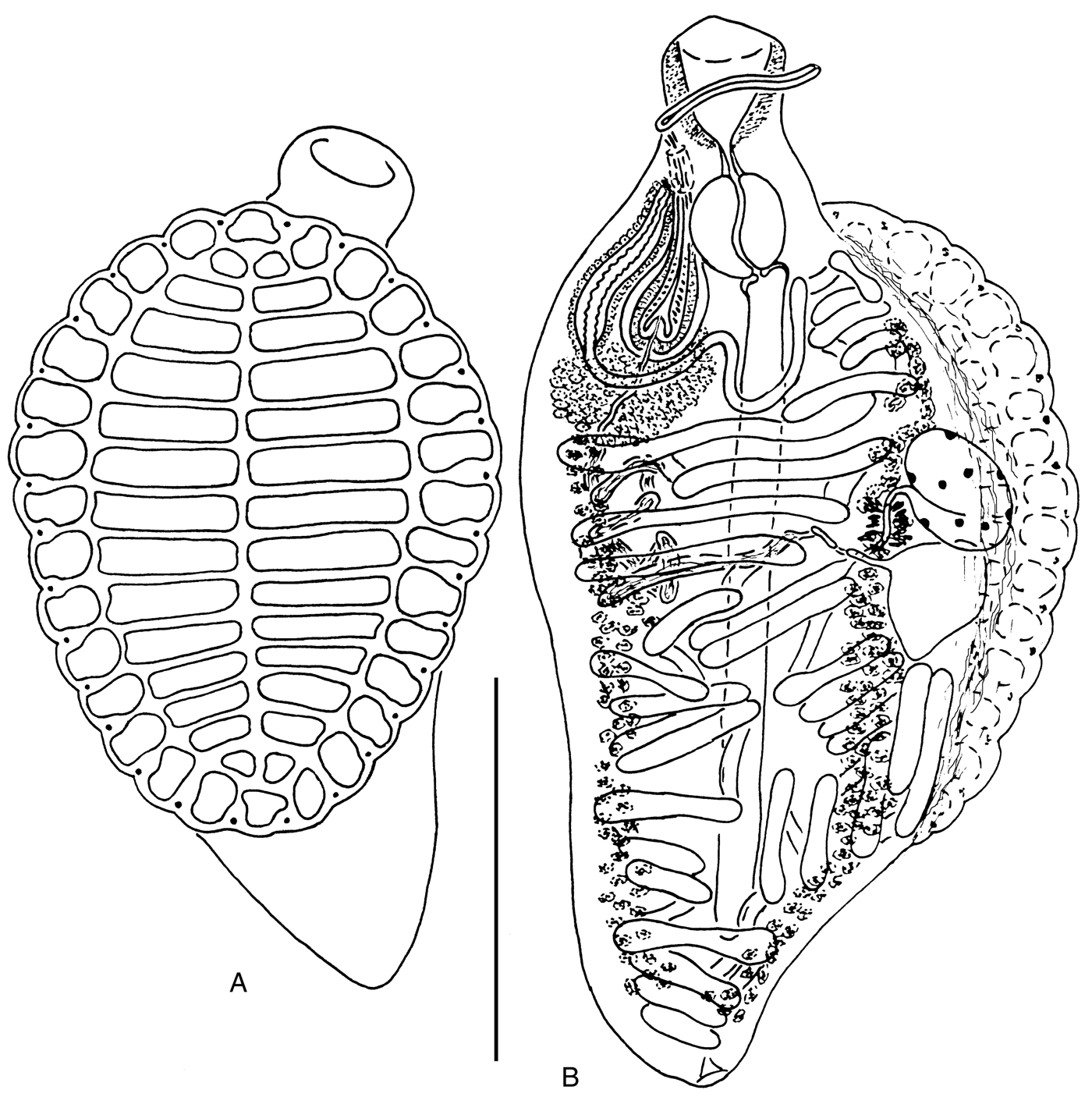
Fig. 2. Aspidogaster limacoides from Rutilus rutilus, Rybinsk Reservoir: (A) ventral view; (B) dorsal view. Scale bar: 1 mm.

Fig. 3. Aspidogaster ijimai from Cyprinus carpio s. lato: (A) ventral view; (B) dorsal view. Scale bars: (A) 0.4 mm; (B) 1 mm.
Table 1. List of the representatives of the genus Aspidogaster incorporated in sequence analysis (n, number of replicates; voucher accession numbers are in bold).
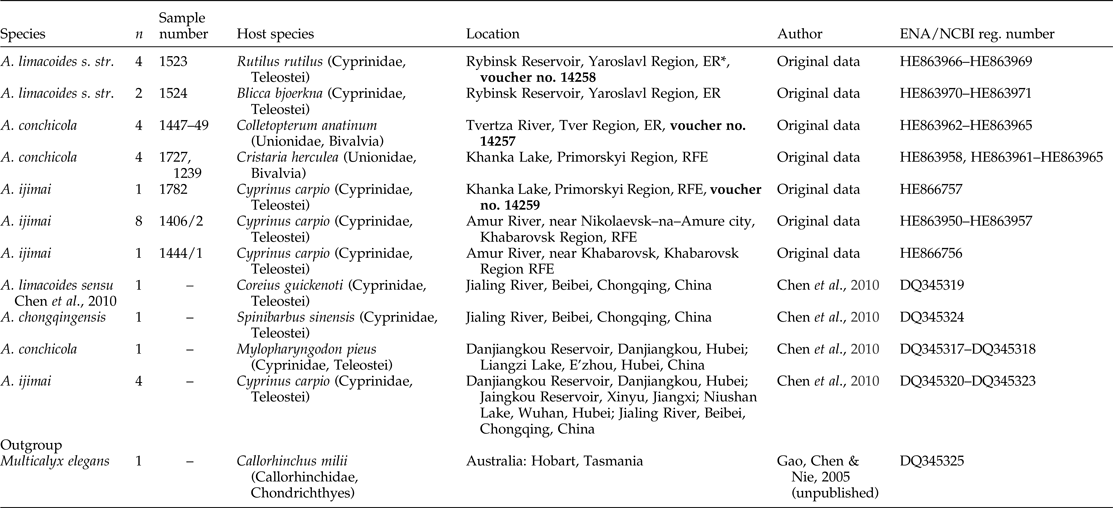
* ER, European part of Russia; RFE, Russian Far East.
ENA, European Nucleotide Archive; NCBI, National Center for Biotechnology Information.
DNA extraction, amplification and sequencing
Total DNA was extracted from separate mature worms fixed in 96% ethanol using a ‘hot shot’ technique, which has been described previously (Truett, Reference Truett and Kieleczawa2006). The nuclear ITS1–5.8S–ITS2 was amplified using the polymerase chain reaction (PCR) with the universal primers BD1 (5′-GTCGTAACAAGGTTTCCGTA-3′) and BD2 (5′-TATGCTTAA(G/A)TTCAGCGGGT-3′) (Luton et al., Reference Luton, Walker and Blair1992). The initial PCR reaction was carried out in a total volume of 20 μl containing 0.25 mm of each primer pair, 1 μl DNA in water, 1 × Taq buffer, 1.25 mm deoxynucleoside triphosphates (dNTP), 1.5 mm MgCl2 and 1 unit of Taq polymerase. The amplification of a 1200-bp fragment of ITS1–5.8S–ITS2 was performed in a GeneAmp 9700 (Applied Biosystems, Foster City, California, USA) with a 3-min denaturation hold at 94°C; 40 cycles of 30 s at 94°C, 30 s at 54°C and 2 min at 72°C; and a 7-min extension hold at 72°C. Negative and positive controls were amplified using both primers. The PCR products were directly sequenced using an ABI Big Dye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems), as recommended by the manufacturer, with the internal sequencing primer 3S (5′-GGTACCGGTGGATCACGTGGCTAGTG-3′) (Luton et al., Reference Luton, Walker and Blair1992). The PCR products were analysed using an ABI 3130 genetic analyser at the Institute of Biology and Soil Sciences, Far Eastern Branch of Russian Academy of Sciences. The sequences have been submitted to the European Nucleotide Archive (ENA)/GenBank with the following accession numbers: HE863950–HE863971, HE866756–HE866757.
Alignment and phylogenetic analysis
The ribosomal DNA sequences were assembled with SeqScape v.2.6 software (Applied Biosystems) and aligned with sequences of aspidogastrids from China, retrieved from the GenBank database using ClustalW DNA weight matrix within MEGA 5.0 software alignment explorer (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011).
Regions that could not be unambiguously aligned were excluded from the analyses. A number of variable, parsimony-informative sites, nucleotide compositions and substitution ratio analyses were performed using MEGA 5.0. Genetic divergence was estimated by calculating genetic p-distance (d) values. Phylogenetic analysis of the nucleotide sequences was undertaken, using maximum likelihood (ML) and Bayesian (BI) methods. Prior to analysis, the nucleotide substitution model was estimated using Akaike's information criterion (AIC) for ML (Akaike, Reference Akaike1974) and Bayesian information criterion (BIC) for BI (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001) using the jModeltest v.3.07 software (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). The models TIM1 + G (Posada, Reference Posada2003) and HKY + G (Hasegawa et al., Reference Hasegawa, Kishino and Yano1985) were estimated as those fitting the data best for ML and BI analyses, respectively. Phylogenetic trees were reconstructed with PhyML 3.1 (Guindon & Gascuel, Reference Guindon and Gascuel2003) and MrBayes v.3.1.2 software (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001). A Bayesian algorithm was performed using the MCMC option with ngen = 1,000,000, nruns = 2, nchains = 4 and samplefreq = 100. Burn-in values were 250,000 for ‘sump’ and ‘sumt’ options. Optimization of the Bayesian inference algorithm was performed by setting up priors using the Tracer v.1.5.0 software (Rambaunt & Drummond, Reference Rambaut and Drummond2009). Phylogenetic relationship significance was estimated using a bootstrap analysis (Felsenstein, Reference Felsenstein1985) with 100 replications and posterior probabilities (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001) for ML and BI analyses, respectively. Nucleotide sequences of the ITS1–5.8S–ITS2 fragment of the ribosomal cluster from the GenBank database were used in our study to evaluate the phylogenetic relationships of European and Far Eastern representatives of the genus Aspidogaster Baer, 1827: A. conchicola Baer, 1827; A. limacoides Diesing, 1834; A. ijimai Kawamura, 1915; and A. chongqingensis Wei, Huang & Dai, Reference Wei, Huang and Dai2001 (table 1).
Results
The amplification procedure produced a 1500-bp fragment of the ITS1–5.8S–ITS2 rDNA for all Aspidogaster specimens. After assembly and alignment procedures, the resulting ITS rDNA sequences were 1437–1517 bp in length for different species. The fragment contained 442 variable and 414 parsimony-informative sites.
Genetic divergence between the ITS sequences from Aspidogaster species and sequences from Multicalyx elegans was estimated by the calculation of p-distances (table 2). The genetic p-distance between A. limacoides s. str. from the European part of Russia and A. limacoides sensu Chen et al., 2010 from China was 13.7%, whereas between A. limacoides sensu Chen et al., 2010 and A. chongqingensis from China it was only 0.09%. The genetic divergence between A. ijimai and A. limacoides s. str. from the European part of Russia was two times higher (d = 13.4%) than between A. ijimai and A. limacoides sensu Chen et al., 2010 from China (d = 6.6%). Genetic differentiation between A. ijimai specimens from the Russian Far East and China was 0.28%. The highest values of p-distances were obtained between A. conchicola and other Aspidogaster species: d = 16.9–18.5%.
Table 2. Genetic divergence of Aspidogaster species, estimated with p-distance calculations by means of ITS1–5.8S–ITS2 rDNA nucleotide sequences.

*AN, Amur River, Nikolaevsk-na-Amure; AK, Amur River, Khabarovsk; Kh, Khanka Lake; Chi, China; ER, European part of Russia; Aus, Australia.
Intraspecific molecular differentiation was revealed between A. conchicola and A. ijimai from different localities. Genetic p-distance values within A. conchicola ranged from 0.45% (between specimens from China and European part of Russia) to 0.94–0.96% (between specimens from the Russian Far East and the European part of Russia or China). The variation within A. ijimai ranged from 0 (Amur River, Khabarovsk/Khanka Lake) to 0.3% (Amur River, Nikolaevsk–na–Amure/China). Transition/transversion bias (R) between different Aspidogaster species ranged from 1.14 to 2.97 (table 3). A minimal range of R values was observed by pairwise comparison of A. conchicola with other Aspidogaster species (R = 1.14–1.25).
Table 3. Transition/transversion ratio bias (R*), obtained by pairwise comparision of ITS1–5.8S–ITS2 rDNA sequences of different Aspidogaster species.

*R = [A*G*k1 + T*C*k2]/[(A + G)*(T + C)], where k1 and k2 are frequencies of transitions between purines and pyrimidines, respectively. R becomes 0.5 when there is no bias towards either transitional or transversional substitution, because when the two kinds of substitution are equally probable, there are twice as many possible transversions as transitions.
The phylogenetic relationships of Aspidogaster species were reconstructed using ML and BI methods (fig. 4). Both phylogenetic tree topologies showed the differentiation of Aspidogaster species into four clades, corresponding to different species, and were highly statistically supported. The first contained specimens of A. conchicola, which were subdivided according to geographical origin with high statistical support. Specimens of A. ijimai formed the second distinct clade with high support. Aspidogaster ijimai was subdivided into three groups, corresponding to ‘Russian’ and ‘Chinese’ samples. It is notable that Chinese samples of A. ijimai formed a distinct compact group within the Russian A. ijimai cluster. The third clade included A. limacoides sensu Chen et al., 2010 and A. chongquingensis from China, and the fourth clade contained A. limacoides s. str. from the European part of Russia.

Fig. 4. Phylogenetic tree based on analysis of ITS1–5.8S–ITS2 rDNA sequences of species of the genus Aspidogaster using the Bayesian method of phylogenetic reconstructions. Nodal numbers give bootstrap statistical support for ML/BI analyses. AN, Amur River, Nikolaevsk-na-Amure; AK, Amur River, Khabarovsk; Kh, Khanka Lake; Chi, China; ER, European part of Russia.
Discussion
Our results showed considerable molecular differentiation between A. limacoides s. str. from the European part of Russia and A. limacoides sensu Chen et al., 2010 from China. Aspidogaster limacoides s. str. is reliably known only from European, and Central and Western Asian territories (reviewed by Alves et al., Reference Alves, Viera, Santos, Scholz and Luque2015). Records of A. limacoides in China were presented by several authors (Jin et al., Reference Jin, Dai, Liu, Zeng, He and Xiang1993; Wang et al., Reference Wang, Li, Yu, Feng, Xiao, Wang, Yao, Feng and Song1997; Zhang et al., Reference Zhang, Qiu and Ding1999; Chen et al., Reference Chen, Zhang, Wen, Sun and Gao2010), but these data are not supported by the morphological description of the parasite (Kawamura, Reference Kawamura1915; Timofeeva, Reference Timofeeva1973; Tang & Tang, Reference Tang and Tang1980; Bykhovskaya-Pavlovskaya, Reference Bykhovskaya–Pavlovskaya and Bauer1987; Pavljuchenko, Reference Pavljuchenko2007). These circumstances raised some doubts about the presence of A. limacoides s. str. in China. Our molecular data also indicate that the reports of A. limacoides sensu Chen et al., 2010 from China are not reliable. These worms are conspecific with A. chongqingensis from Spinibarbus sinensis (Bleeker, 1871) caught from the Jialing River of Chongqing, China, as evidenced by the extremely low genetic p-distance values, corresponding to intraspecific genetic variation for trematodes (Jousson et al., Reference Jousson, Bartoli and Pawlowski2000; D. Chen et al., Reference Chen, Gao and Nie2007). Based on ITS1–5.8S–ITS2 rDNA sequence data, A. chongqingensis is phylogenetically closer to A. ijimai than to A. limacoides s. str. (fig. 4). These results are unexpected because A. chongqingensis is morphologically closer to A. limacoides s. str. in a number of significant features: size ratio of the buccal funnel and pharynx, degree of development of the external field of prostatic cells and presence of an unarmed cirrus (fig. 2) (Wei et al., Reference Wei, Huang and Dai2001).
The species A. conchicola possesses maximal values of intraspecific molecular differentiation among the investigated species of aspidogastreans (table 2, fig. 4). Specimens from the Russian Far East were collected from the Amur River basin (Khanka Lake), whereas samples from China (Hubei province) and the European part of Russia were from different river systems, which have no connections with the Amur River. Genetic p-distance values between different geographical samples indicate that A. conchicola from the Russian Far East has existed separately from European and Chinese specimens for twice as long as the two latter groups have been separated from each other. Despite the wide distribution of A. conchicola in molluscs, this species has also been found in freshwater fishes and even aquatic reptiles (reviewed by Alves et al., Reference Alves, Viera, Santos, Scholz and Luque2015; see also Vosnesenskaya, Reference Vosnesenskaya1968; Dvodryadkin, Reference Dvodryadkin1976). For this reason, there may have been a possible exchange of A. conchicola specimens between different territories in the past by host-switching processes, which have been described for some digenetic trematodes (Attwood et al., Reference Attwood, Upatham, Meng, Qiu and Southgate2002, Reference Attwood, Upatham, Zhang, Yang and Southgate2004, Reference Attwood, Faith, Mondal, Alim, Fadjar, Rajapakse and Rollinson2007). Moreover, aspidogastrids are well-known for their low host specificity, which can favour their dispersion (Alves et al., Reference Alves, Viera, Santos, Scholz and Luque2015). However, this assumption needs to be confirmed by detailed studies of the phylogeography of Aspidogaster species and their host fish species; facultative definitive hosts are potential distributors of these flatworms.
Molecular differentiation between A. conchicola and other Aspidogaster species studied here were also characterized by maximal values (table 2). Transition/transversion bias values (R) varied with the same pattern, suggesting a higher amount of transversion substitution type between ITS1–5.8S–ITS2 sequences of A. conchicola and other Aspidogaster species. Molluscs are obligate hosts for Aspidogaster species. However, the final stages of the life cycles of these worms electively occur in fish species (Timofeeva, Reference Timofeeva1975, Reference Timofeeva2005). Freshwater fish species are important for circulation of the three species examined in the present study – A. chongqingensis, A. ijimai and A. limacoides s. str. – and this is evidenced by the occurrence of these species in these vertebrates (Achmerov, Reference Achmerov1956; Gao et al., Reference Gao, Nie and Yao2003; Alves et al., Reference Alves, Viera, Santos, Scholz and Luque2015). Aspidogaster conchicola is generally known to infect freshwater bivalves and gastropods (Michelson, Reference Michelson1970; Dvodryadkin, Reference Dvodryadkin1976; Dugarov, Reference Dugarov2010), but there are few reports about this worm within fish species (Alves et al., Reference Alves, Viera, Santos, Scholz and Luque2015). The phylogenetic relationships of species obtained in the present study showed earlier divergence of the ancestral form of the parasite species group, namely A. chongqingensis, A. ijimai and A. limacoides s. str. in comparison with A. conchicola (fig. 4). This may be evidence that a wide inclusion of fish species into the aspidogastrean life cycle is plesiomorphic. This assumption corresponds with the hypothesis of aspidogastrean evolution, based on morphological and ecological data. These consider A. conchicola maturing within bivalve molluscs as a progenetic phenomenon, which appeared after fish-specific adaptation (Timofeeva, Reference Timofeeva2005).
Acknowledgements
The authors are deeply grateful to Dr T. Shimazu for providing preparations of aspidogastrids from Biwa Lake, Japan, to V.V. Bogatov for help with mollusc species identification, and to Dr S.V. Shedko for helpful comments.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflict of interest
None.









