Introduction
Parasites are one of the most important factors exerting selection pressures in animal populations (Haldane, Reference Haldane1949). Generally, each species may act as a competent host for a very versatile and large community of parasites. However, only rarely do these communities show high similarity in terms of prevalence and diversity across the distribution range of the host (Freeman-Gallant et al. Reference Freeman-Gallant, O'Connor and Breuer2001; Poulin, Reference Poulin2003). Differences in parasite communities should translate into diverse selection pressures acting upon the host, and this in turn could result in variations of life-history traits among host populations. Consequently, in order to understand the role of parasites in shaping inter-population differences in fitness-associated traits, it is necessary to examine local parasite communities and investigate the patterns of variation in infection/infestation rates.
In birds, among the commonest parasites are vector-transmitted protozoans, which complete some stages of the life cycle in the blood of the vertebrate host (therefore, they are generally referred to as blood parasites or haemoparasites). This group includes, among others, parasites from the genera Plasmodium, Haemoproteus and Trypanosoma (Clark et al. Reference Clark, Boardman and Raidal2009). Avian haemosporidians and trypanosomes are transmitted by blood-sucking arthropods. Specifically, Plasmodium is transmitted by mosquitoes (Culicidae), Haemoproteus by hippoboscid flies (Hippoboscidae) and biting midges (Ceratopogonidae) and Trypanosoma by black flies (Simuliidae), hippoboscid flies (Hippoboscidae), biting midges (Ceratopogonidae), Culex spp. mosquitoes (Culicidae) and dermanyssid mites (Dermanyssidae) (Baker, Reference Baker, Lumsden and Evans1976; Molyneux, Reference Molyneux1977; Votýpka et al. Reference Votýpka, Szabová, Rádrová, Zídková and Svobodová2012). Despite being transmitted by the same type of vectors (blood-sucking arthropods) haemosporidians and trypanosomes differ in their transmission modes: haemosporidians enter the host organism through the bite of the vector, and trypansomes – through ingestion of the vector or contamination of abraded skin or conjunctiva with material containing infective parasite stages (Desser et al. Reference Desser, McIver and Jez1975; Molyneux, Reference Molyneux1977; Votýpka and Svobodová, Reference Votýpka and Svobodová2004; Votýpka et al. Reference Votýpka, Szabová, Rádrová, Zídková and Svobodová2012). Parasites from all three genera can negatively affect components of fitness in birds (Merino et al. Reference Merino, Moreno, Jose Sanz and Arriero2000; Knowles et al. Reference Knowles, Palinauskas and Sheldon2010; Martínez-de la Puente et al. Reference Martínez-de la Puente, Merino, Tomas, Moreno, Morales, Lobato, Garcia-Fraile and Belda2010), although a number of studies failed to find such associations (Dufva, Reference Dufva1996; Siikamäki et al. Reference Siikamäki, Rätti, Hovi and Bennett1997). The introduction at the beginning of the XXI century of molecular techniques for detection and identification of these parasites not only improved the detectability of infection (Hellgren et al. Reference Hellgren, Waldenström and Bensch2004; Waldenström et al. Reference Waldenström, Bensch, Hasselquist and Östman2004), but also uncovered their extreme genetic diversity (Bensch et al. Reference Bensch, Hellgren and Pérez-Tris2009). Importantly, by using a molecular approach, it was possible to show that genetic lineages identified as the same morphospecies may, in fact, represent separate biological entities (e.g. Bensch et al. Reference Bensch, Pérez-Tris, Waldenström and Hellgren2004; Nilsson et al. Reference Nilsson, Taubert, Hellgren, Huang, Palinauskas, Markovets, Valkiūnas and Bensch2016).
Examination of different populations of the same host species in the context of infection rates and diversity of blood parasites has shown that both parameters can vary geographically (Bensch and Åkesson, Reference Bensch and Åkesson2003; Pagenkopp et al. Reference Pagenkopp, Klicka, Durrant, Garvin and Fleischer2008; Szöllősi et al. Reference Szöllősi, Cichoń, Eens, Hasselquist, Kempenaers, Merino, Nilsson, Rosivall, Rytkönen, Török, Wood and Garemszegi2011; Spurgin et al. Reference Spurgin, Illera, Padilla and Richardson2012), even within a short distance (Sol et al. Reference Sol, Jovani and Torres2000; Ferrer et al. Reference Ferrer, García-Navas, Sanz and Ortego2012). However, the geographic variation of these parameters is not a universal phenomenon. In some host species, it has not been found or was restricted to a subgroup of individuals (Swanson et al. Reference Swanson, Lyons and Bouzat2014; Svoboda et al. Reference Svoboda, Marthinsen, Pavel, Chutný, Turčoková, Lifjeld and Johnsen2015). For example, in the breeding populations of the migratory lark sparrow (Chondestes grammacus) located across the USA, geographic differences in the overall prevalence were present only in juvenile birds, but not in the adults (Swanson et al. Reference Swanson, Lyons and Bouzat2014). Similarly, adult bluethroats (Luscinia svecica) sampled in seven populations ranging in location from central to northern Europe did not differ in overall infection rates (Svoboda et al. Reference Svoboda, Marthinsen, Pavel, Chutný, Turčoková, Lifjeld and Johnsen2015).
In the present study, we molecularly examine the prevalence and community composition of parasites from the Haemoproteus, Plasmodium and Trypanosoma genera in a population of the pied flycatcher (Ficedula hypoleuca) located in central Poland. The pied flycatcher is a long-distance migrant which breeds in most of Europe and western Asia and winters in West Africa (Cramp and Perrins, Reference Cramp and Perrins1993; Ouwehand et al. Reference Ouwehand, Ahola, Ausems, Bridge, Burgess, Hahn, Hewson, Klaassen, Laaksonen, Lampe, Velmala and Both2016). It is a model species in the evolutionary ecology of cavity-nesting long-distance migrants with several closely monitored populations located across its breeding range (e.g. Laaksonen et al. Reference Laaksonen, Sirkiä, Calhim, Brommer, Leskinen, Primmer, Adamík, Artemyev, Belskii, Both, Bureš, Burgess, Doligez, Forsman, Grinkov, Hoffmann, Ivankina, Král, Krams, Lampe, Moreno, Mägi, Nord, Potti, Ravussin and Sokolov2015; Ouwehand et al. Reference Ouwehand, Ahola, Ausems, Bridge, Burgess, Hahn, Hewson, Klaassen, Laaksonen, Lampe, Velmala and Both2016). To date, several studies looked into the community of blood parasites in this species, but most of them were based on blood smear screening (Bennett et al. Reference Bennett, Siikamäki, Rätti, Allander, Gustafsson and Squires-Parsons1994; Dale et al. Reference Dale, Kruszewicz and Slagsvold1996; Siikamäki et al. Reference Siikamäki, Rätti, Hovi and Bennett1997; Sanz et al. Reference Sanz, Arriero, Moreno and Merino2001a) and consequently may have underestimated infection rates and the diversity of these parasites (Fallon et al. Reference Fallon, Ricklefs, Swanson and Bermingham2003; Garamszegi, Reference Garamszegi2010; Nilsson et al. Reference Nilsson, Taubert, Hellgren, Huang, Palinauskas, Markovets, Valkiūnas and Bensch2016). Importantly, this is one of the few studies molecularly examining avian trypanosomes. In contrast to haemosporidian parasites, infections with Trypanosoma have been largely understudied in birds, although such infections may have serious consequences for a host's fitness, including the study species (Rätti et al. Reference Rätti, Dufva and Alatalo1993; Sanz et al. Reference Sanz, Arriero, Moreno and Merino2001a; Dyrcz et al. Reference Dyrcz, Wink, Kruszewicz and Leisler2005, also see Siikamäki et al. Reference Siikamäki, Rätti, Hovi and Bennett1997; Sanz et al. Reference Sanz, Arriero, Moreno and Merino2001b), and are known to widely occur in birds (Sehgal et al. Reference Sehgal, Jones and Smith2001; Oakgrove et al. Reference Oakgrove, Harrigan, Loiseau, Guers, Seppi and Sehgal2014). Additionally, we: (i) studied the relationship between infection incidence and two host traits commonly considered in the analyses of the presence of infection, namely sex and age (McCurdy et al. Reference McCurdy, Shutler, Mullie and Forbes1998; Deviche et al. Reference Deviche, Greiner and Manteca2001; van Oers et al. Reference van Oers, Richardson, Sæther and Komdeur2010), and (ii) investigated geographic variation in the prevalence and diversity of haemosporidian parasites (Haemoproteus and Plasmodium) among four molecularly screened breeding populations of the pied flycatcher using data from the current and previously published studies (Wiersch et al. Reference Wiersch, Lubjuhn, Maier and Kampen2007; Kulma et al. Reference Kulma, Low, Bensch and Qvarnström2013).
Materials and methods
Data were collected from a small population (up to 17 pairs per year) of nest box-breeding pied flycatchers in 2011–2014. The study site is located in Sękocin Stary, 10 km south-west of Warsaw's city borders, Poland (52°05′N, 20°52′E). All study plots except for one are located in a pine (Pinus sylvestris) forest with an admixture of deciduous trees [birch (Betula spp.), sessile (Quercus petraea) and pedunculate (Quercus robur) oaks and common hornbeam (Carpinus betulus)] with a moderate to dense understory. Nest boxes (for their dimensions and distribution see Dubiec and Mazgajski, Reference Dubiec and Mazgajski2013) are primarily occupied by great tits (Parus major), blue tits (Cyanistes caeruleus) and pied flycatchers, and occasionally by coal tits (Periparus ater). The number of nest boxes varied among breeding seasons, from 156 in 2011 to 259 in 2014. Pied flycatchers bred only in pine-dominated plots. We additionally monitored over 35 nest boxes, differing in size and the height of attachment, located in close vicinity to the study plots. Each year at least one pied flycatcher breeding attempt was recorded in these boxes.
Before each breeding season nest boxes were cleaned of remaining nest material and feces of roosting birds. Regular controls of boxes, in order to record phenological data and breeding parameters, started in the beginning of April. Adult birds were caught when the nestlings were 13 days old (hatching day = day 1) or older, either by mist-netting or using traps installed inside the box. Birds were ringed, sexed, aged, measured (data not shown) and sampled for blood. Sexing and ageing (as yearlings or at least 2 years old) were based on plumage characteristics following Svensson (Reference Svensson1994). In the case of re-trapped birds, the assigned age was later verified based on ringing records from previous seasons. Blood, obtained by puncturing the brachial vein, was collected into non-heparinized capillaries placed in 96% ethanol. Until the molecular analyses were performed, blood samples were stored at ambient temperature. Because of the brood mortality at the early post-hatching stage or unsuccessful catching trials (mostly because the birds were not present in the vicinity of the box during a catching trial), a few individuals were not caught.
DNA was extracted using the standard ammonium acetate precipitation method (Bruford et al. Reference Bruford, Hanotte, Brookfield, Burke and Hoelzel1998). DNA isolates were checked for concentration and purity (absorbance ratios: A260/A280 and A260/A230) using a NanoDrop spectrophotometer. Samples were screened for the presence of blood parasites: (1) from the Haemoproteus and Plasmodium genera by amplifying a 478 bp fragment of the mitochondrial cyt b gene, using the nested polymerase chain reaction (Waldenström et al. Reference Waldenström, Bensch, Hasselquist and Östman2004; Cosgrove et al. Reference Cosgrove, Wood, Day and Sheldon2008); (2) from the Trypanosoma genus by amplifying a 326 bp fragment of the small subunit ribosomal RNA (SSU rRNA) gene as described in Sehgal et al. (Reference Sehgal, Jones and Smith2001). Four μL of final PCR products were run on 2% agarose gels stained with GelRed (Biotium, Hayward, CA, USA) and visualized under ultraviolet light. Each plate contained a positive (DNA from individuals with confirmed infection based on blood smear screening) and negative (ddH2O) control, to monitor for possible contamination or failures during PCRs. The quality of the DNA isolate of all samples yielding no PCR product was checked with sex-specific primers (Griffiths et al. Reference Griffiths, Double, Orr and Dawson1998). PCR products of positive samples were purified with FastAP (Fermentas) and then sequenced directly (mostly unidirectionally, except for a subset of trypanosome PCR products, which were sequenced bidirectionally) with an automated ABI 3130 DNA analyser (Applied Biosystems) using BigDye terminator ver. 3.1 (Applied Biosystems). The obtained sequences were aligned using BioEdit software (Hall, Reference Hall1999) and compared with the MalAvi database (Bensch et al. Reference Bensch, Hellgren and Pérez-Tris2009) in the case of Plasmodium and Haemoproteus and with the GenBank database in the case of Trypanosoma.
In total, we screened 97 samples from 68 individuals. Twenty-one individuals were sampled more than once. To avoid the issue of pseudoreplication, only one record per multiply sampled individuals was selected for further analyses. The record was selected either randomly (most cases) or non-randomly when information was lacking (age of the individual) in the datasets collected in different seasons. In such cases, the record with the full set of data was chosen for statistical analyses. The final data set included 43 females and 25 males.
The association between occurrences of parasites representing different genera within a host was analysed with Fisher's exact test. To assess the association between the probability of infection (separately for each parasite genera) and sex and age of the host generalized linear mixed models (GLMM) with a binomial error distribution and a logit link function were used. The individual age and sex were defined as fixed categorical variables, a year as a random variable and infection status (0 for absence and 1 for presence) as a response variable. The interaction between sex and age was introduced into the model, but removed if not significant. The model was run using a dataset reduced by four females, because these individuals were not aged (final data set: females – 18 yearlings and 21 older birds, males – 12 yearlings and 13 older birds). Because Plasmodium infections occurred at a low rate (see ‘Results’ section), the analyses of haemoparasite co-occurrence and age- and sex-related variation were limited to Haemoproteus and Trypanosoma infections. GLMM models were run using the glmer function in the package ‘lme4’ (Bates et al. Reference Bates, Mächler, Bolker and Walker2015) in R (R Development Core Team, 2014) with the Laplace approximation of maximum likelihood (Bolker et al. Reference Bolker, Brooks, Clark, Geange, Poulsen, Stevens and White2009).
Geographic variation in the prevalence and ecological diversity of haemosporidian parasites among the breeding populations of the pied flycatcher was estimated for three previously molecularly screened populations – Lingen and Bahrdorf in Germany (sampling period: 1993 for Bahrdorf and 1996 for Lingen; Wiersch et al. Reference Wiersch, Lubjuhn, Maier and Kampen2007) and the island of Öland in Sweden (sampling period: 2004–2010, except 2008; Kulma et al. Reference Kulma, Low, Bensch and Qvarnström2013) – and the population examined in the present study (Fig. 1). In the case of four mitochondrial sequences from German populations (one out of six sequences in Lingen and three out of four sequences in Bahrdorf), it was not possible to assign them to specific lineages from the MalAvi database because only information about the GenBank entry most closely matching each of these sequences (the similarity was over 98% in all cases) was provided. These entries were used in the current study as a reference to assign the sequence to a parasite genus. Two ecological diversity indices were used: the species richness (S – the total number of detected lineages) and the Shannon index (H; combining information on species richness and relative abundance). The diversity indices were calculated in the ‘vegan’ package (Oksanen et al. Reference Oksanen, Blanchet, Kindt, Legendre, Minchin, O'Hara, Simpson, Solymos, Stevens and Wagner2016) in R, while the 95% confidence intervals (CIs) of the Shannon index and differences in those indexes between populations were calculated in the ‘boot’ package (Canty and Ripley, Reference Canty and Ripley2015) according to the recommendations of Gardener (Reference Gardener2014). CIs were estimated based on 1000 bootstrap resamples (Custom R Command: ‘H_boot’ from Gardener, Reference Gardener2014). The statistical significance of the difference in diversity index between populations was estimated using a randomization method based on 2000 bootstrap resamples (Custom R Command: ‘H_bss’ from Gardener, Reference Gardener2014).
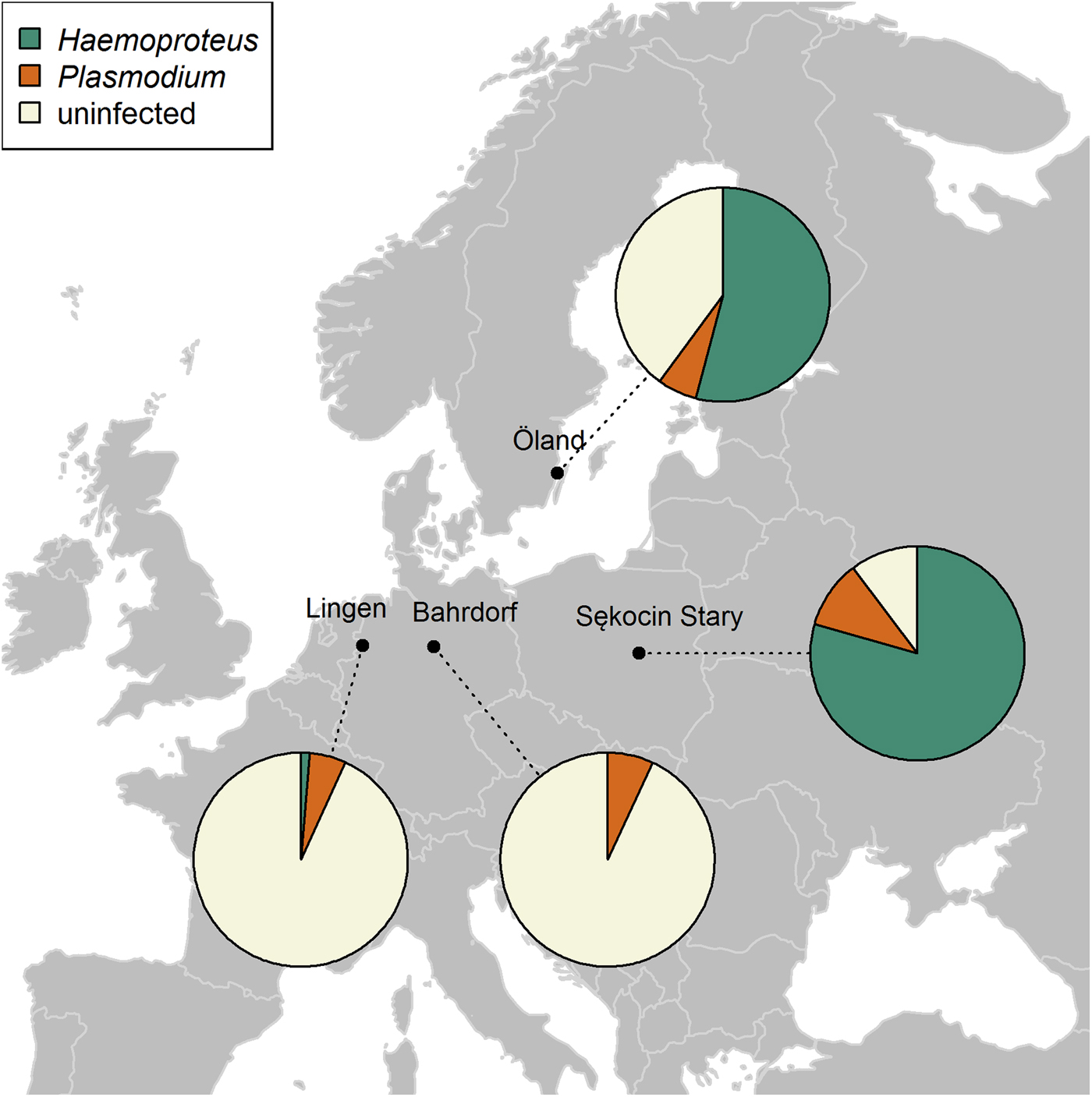
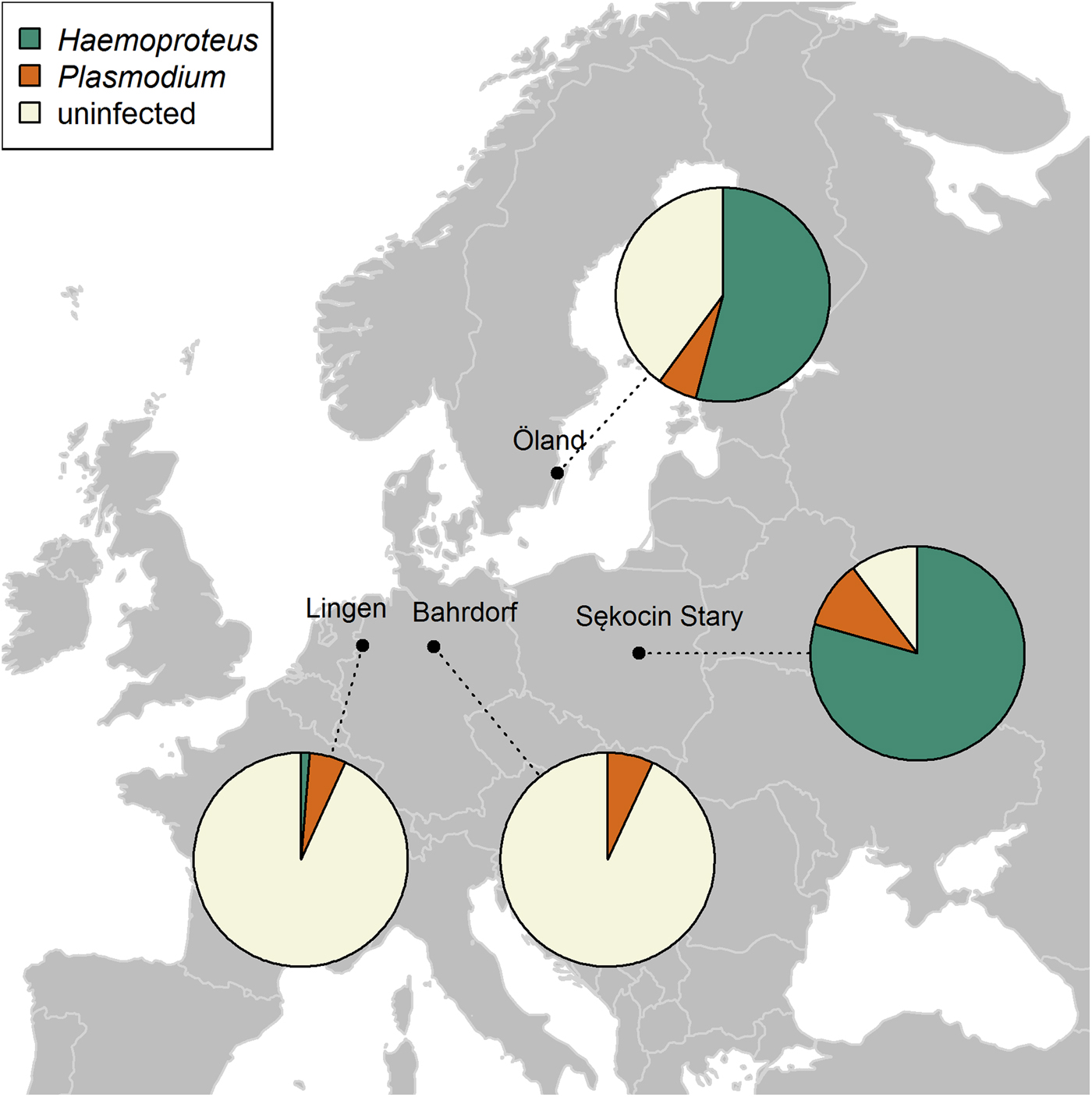
Fig. 1. Distribution and prevalence of Plasmodium and Haemoproteus parasites across four breeding populations of the pied flycatcher molecularly screened for the presence of parasites: Bahrdorf and Lingen in Germany (Wiersch et al. Reference Wiersch, Lubjuhn, Maier and Kampen2007), the island of Öland in Sweden (Kulma et al. Reference Kulma, Low, Bensch and Qvarnström2013) and Sękocin Stary in Poland (present study). Pie charts show the parasite genus frequencies of each population. Sample sizes: N Bahrdorf = 72; N Lingen = 147; N Öland = 353; N Sekocin Stary = 68. Note that Haemoproteus infection rates recorded in Bahrdorf and Lingen populations may be underestimated as a consequence of the applied protocol (see ‘Discussion’ for details).
The phylogenetic analysis includes sequences of Haemoproteus and Plasmodium lineages from the present study and two other European breeding populations of pied flycatchers [Lingen (five sequences that could be matched with specific lineages from the MalAvi database) and Öland] as well as additional lineages found previously in this species (birds caught on breeding grounds either during or outside of the breeding season), which we extracted from the MalAvi database (version: Malavi8120153_04102016; Bensch et al. Reference Bensch, Hellgren and Pérez-Tris2009). Bayesian reconstructions were carried out in MrBayes v. 3.1.2 (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003), following a General Time Reversible model including invariable sites and variation among sites (GTR + I + G) suggested by the software jModelTest 2.1.10 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012). The Leucocytozoon spp. lineage SISKIN2 was used as an outgroup and two separate runs of 10 million generations were conducted with a sampling frequency of every 100th generation. The burn-in fraction was set to 0·25.
Results
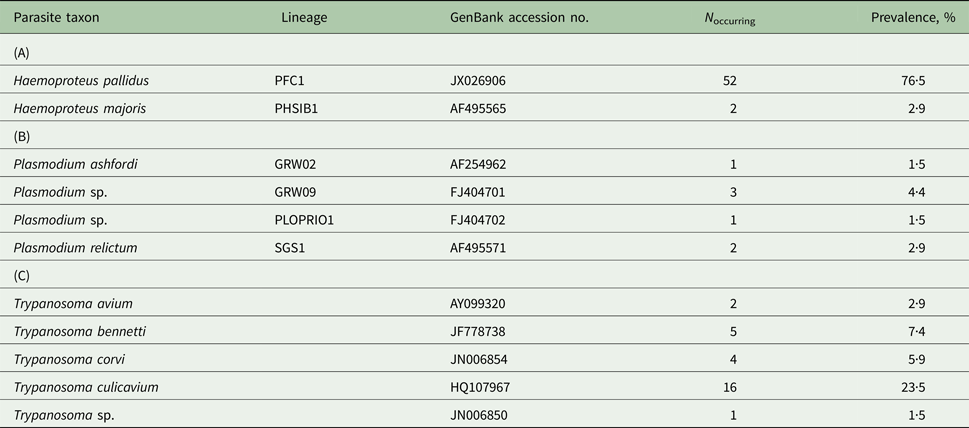
Infection rates of the study population of pied flycatchers varied from very frequent for Haemoproteus (79·4%), through moderate for Trypanosoma (39·7%) to rare for Plasmodium (8·8%). Parasites from the Haemoproteus and Plasmodium genera were represented by two and four cytochrome b lineages, respectively (Table 1). The most common lineage – PFC1, representing the morphospecies H. pallidus – was found in 96·3% of Haemoproteus-infected individuals. Other lineages occurred at a frequency of <5%, including two lineages detected only in a single individual each (Table 1B). Only two birds (2·9%) carried mixed haemosporidian infections: one individual with two Haemoproteus lineages and the other with Haemoproteus and Plasmodium lineages. In the case of Trypanosoma parasites, five types of parasite SSU rRNA sequences were detected: four assigned to a species level and one assigned to a genus level (Table 1C). The most common species – T. culicavium, occurred in 59·2% of Trypanosoma-infected individuals. Only one bird (1·5%) carried a mixed (double) infection.
Table 1. The community of blood parasites [genera Haemoproteus (A), Plasmodium (B) and Trypanosoma (C)] in the pied flycatcher population from central Poland (Sękocin Stary) in 2011–2014 (n = 68)
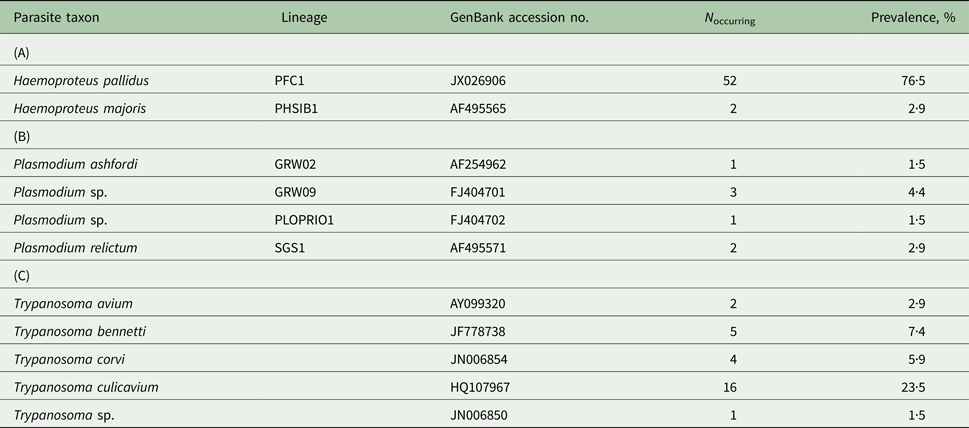
The dataset includes only one record per individual for birds sampled more than once. The prevalence of each parasite taxon is presented as the percentage of individuals harbouring infection (either as a single or as a multiple infection) with a given taxon.
Among all screened individuals, 35·3% carried simultaneous infections of Haemoproteus and Trypanosoma parasites. Because of differences between males and females in Trypanosoma infection rates (see below), the analyses of the association between the occurrences of these two parasite genera within an individual host were performed separately for each sex. The occurrence of Haemoproteus was not associated with the occurrence of Trypanosoma either in males or females (Fisher's exact test, males: P = 0·565, females: P = 0·290).
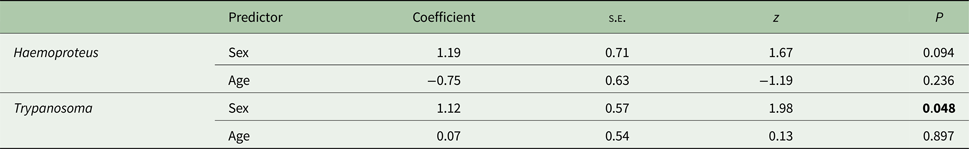
The probability of carrying a Haemoproteus infection was not associated with either the sex or age of the host (sex: males – 88·0%, females – 69·2%, age: yearlings – 83·3%, older birds – 70·6%; Table 2). In the case of Trypanosoma, the probability of infection varied in relation to the host's sex, with males being more frequently infected than females (sex: males – 56·0%, females – 30·8%, age: yearlings – 40·0%, older birds – 41·2%; Table 2).
Table 2. The association between host's sex and age and the probability of infection with parasites from the Haemoproteus and Trypanosoma genera
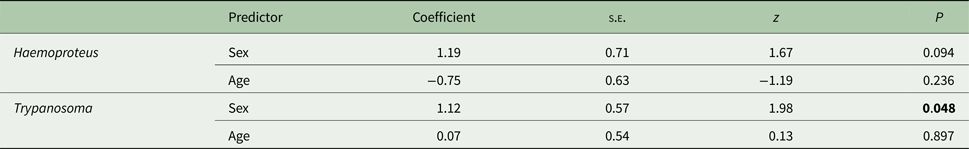
Data were analysed with generalized linear mixed models (GLMM) with sex and age as categorical explanatory variables and study year as a random variable (effect not shown). Non-significant interactions were removed from the model. Significant terms (P < 0·05) are shown in bold.
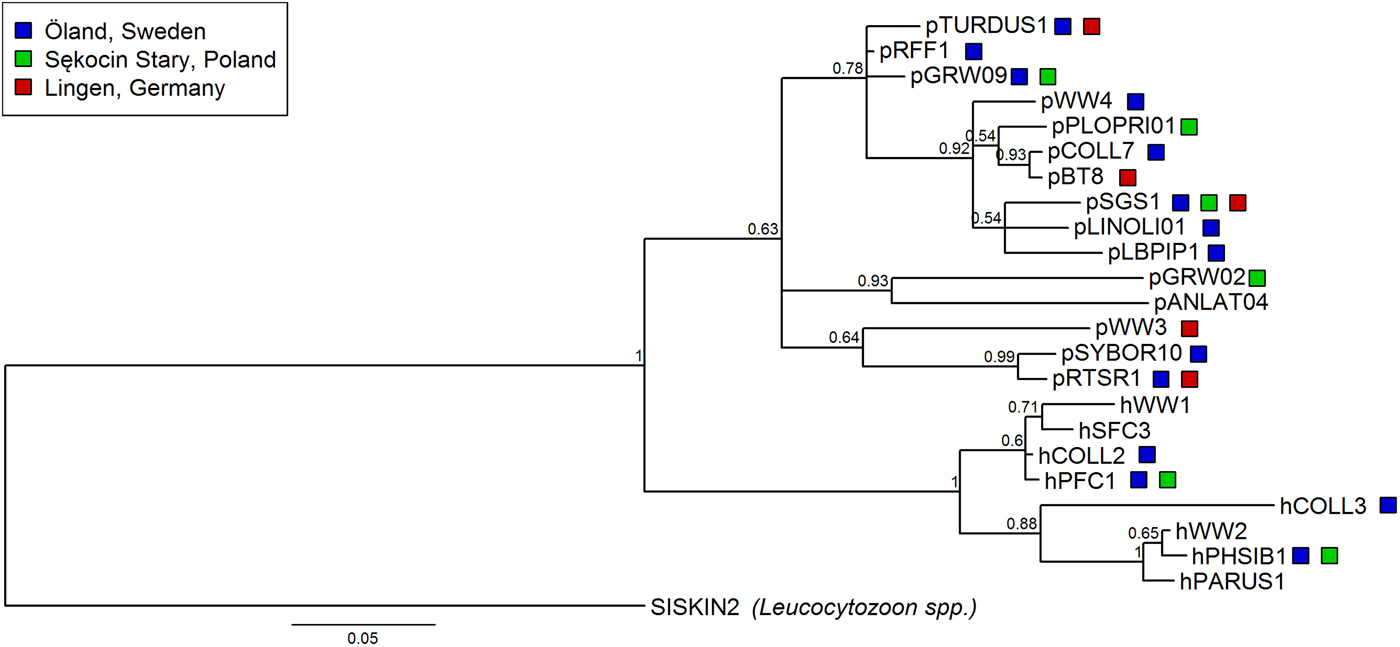
The prevalence of haemosporidian infections varied among four breeding populations of the pied flycatcher, with the lowest incidence of infection in the two German populations, intermediate in Öland and the highest in the population from central Poland (χ 2 = 213·81, df = 3, P < 0·001, Table 3). Also, the proportion of infections caused by each parasite genus differed among populations (Fisher's exact test, P < 0·001). In general, infections in the two German populations were dominated by Plasmodium, while in the two other populations, by Haemoproteus (Fig. 1, Table 3). Lineage richness was the highest in the Öland population with over twice as many lineages as in each of the three other populations (Table 3), while the Shannon index was over two times lower in Sękocin Stary than in the three other populations (Table 3). However, only the difference between the Sękocin Stary and Öland populations was statistically significant (Öland vs Sękocin Stary: z = 5·79, P < 0·001; Lingen vs Sękocin Stary: z = 1·92, P = 0·08; Lingen vs Öland: z = 0·82, P = 0·42; Lingen vs Bahrdorf: z = 0·66, P = 0·53; Öland vs Bahrdorf: z = 1·53, P = 0·14; Sękocin Stary vs Bahrdorf: z = 0·51, P = 0·62). Among 18 lineages detected in Lingen, Öland and Sękocin Stary, only one (pSGS1) was common for all populations, while 66·7% of the lineages occurred only in a single population (Fig. 2).
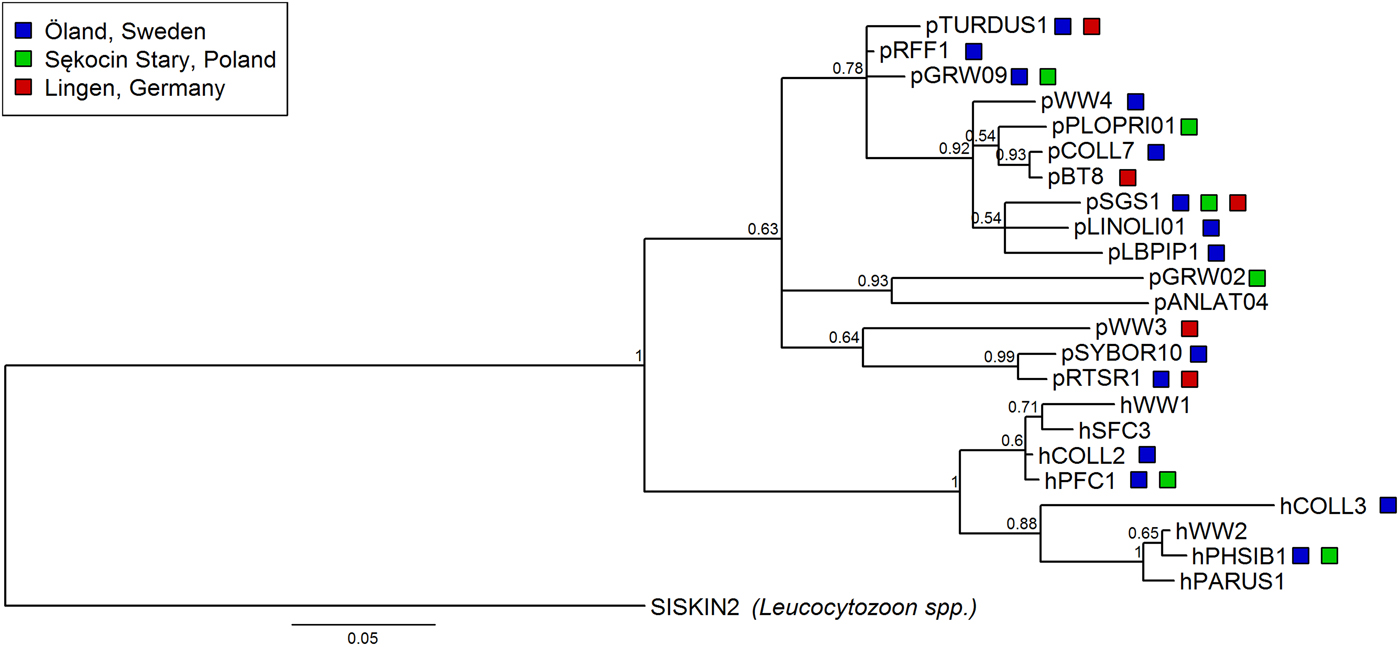
Fig. 2. Phylogenetic relationships of 23 parasite lineages detected in pied flycatchers (8 of Haemoproteus spp. and 15 of Plasmodium spp.) from the three analysed breeding populations [Lingen (Germany), Öland (Sweden) and Sękocin Stary (Poland); indicated with colour boxes] and previously found in this species (birds caught on breeding grounds either during or outside of the breeding season) extracted from the MalAvi database. In the case of the Lingen population one lineage was excluded from the analysis because, based on available information, it was not possible to match it with the lineage in the MalAvi database. The phylogenetic tree was constructed from partial cytochrome b sequences (479 bp) and rooted with Leucocytozoon spp. lineage SISKIN2. Nodal support values near branches indicate Bayesian posterior probabilities.
Table 3. Prevalence and diversity of haemosporidian parasites in four molecularly screened breeding populations of the pied flycatcher: Bahrdorf and Lingen in Germany (Wiersch et al. Reference Wiersch, Lubjuhn, Maier and Kampen2007), the island of Öland in Sweden (Kulma et al. Reference Kulma, Low, Bensch and Qvarnström2013) and Sękocin Stary in Poland (present study)
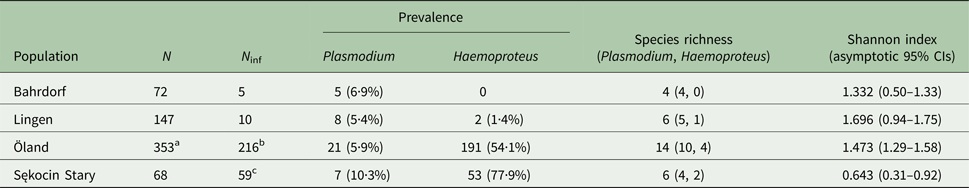
N and N inf refer to the number of screened samples/individuals and the number of samples/individuals positive for the presence of a parasite.
a Includes multiple samples from the same individuals, the total number of screened individuals: 319.
b Includes four individuals with unidentified infection.
c Includes two individuals with mixed infections: hPFC1/pSGS1 and hPFC1/hPHSIB1.
Lineages included in the phylogenetic analysis (18 lineages from Lingen, Öland, Sękocin Stary and five additional lineages detected in pied flycatchers on breeding grounds, Fig. 2) form two major clades of haemosporidian parasites: Haemoproteus and Plasmodium. The lineages found in all three breeding populations are dispersed in the clades and do not form monophyletic groups.
Discussion
The incidence of infection with haemosporidians and trypanosomes in a population of the pied flycatcher located in central Poland is one of the highest reported so far for this species. Specifically, we show that the prevalence of infection with Haemoproteus and Plasmodium parasites reaches 86·8%, while infection with Trypanosoma – 39·7%. Previous studies in populations of this species screened either molecularly or using blood smears showed the prevalence range between 6.9 and 61·2% in the case of haemospiridians (Bennett et al. Reference Bennett, Siikamäki, Rätti, Allander, Gustafsson and Squires-Parsons1994; Dale et al. Reference Dale, Kruszewicz and Slagsvold1996; Siikamäki et al. Reference Siikamäki, Rätti, Hovi and Bennett1997; Sanz et al. Reference Sanz, Arriero, Moreno and Merino2001a; Wiersch et al. Reference Wiersch, Lubjuhn, Maier and Kampen2007; Kulma et al. Reference Kulma, Low, Bensch and Qvarnström2013) and between 1·1 and 40·6% in the case of trypanosomes (Bennett et al. Reference Bennett, Siikamäki, Rätti, Allander, Gustafsson and Squires-Parsons1994; Dale et al. Reference Dale, Kruszewicz and Slagsvold1996; Siikamäki et al. Reference Siikamäki, Rätti, Hovi and Bennett1997; Sanz et al. Reference Sanz, Arriero, Moreno and Merino2001a). The detected haemosporidian prevalence is also much higher than the prevalence (estimated based on molecular screening) reported in two closely related species – the collared (Ficedula albicollis) and the semi-collared flycatchers (Ficedula semitorquata) (Szöllősi et al. Reference Szöllősi, Hellgren and Hasselquist2008; Kulma et al. Reference Kulma, Low, Bensch and Qvarnström2013; Peev et al. Reference Peev, Zehtindjiev, Ilieva, Träff, Briedis and Adamík2016).
The comparison of four pied flycatcher populations, which were screened molecularly for the presence of haemosporidian infections, showed some geographic differences both in the infection rate and the diversity of the haemosporidian community. Based on the overall prevalence and frequency of infection with Plasmodium and Haemoproteus parasites, four populations can be clustered into two groups: one containing both German populations (Wiersch et al. Reference Wiersch, Lubjuhn, Maier and Kampen2007), and the other containing the populations from Öland, Sweden (Kulma et al. Reference Kulma, Low, Bensch and Qvarnström2013) and Sękocin Stary, Poland (present study). Populations from Bahrdorf and Lingen were characterized by a low prevalence (6·9%) and the dominance of Plasmodium infections, while populations from Öland and central Poland by a high prevalence (61.2–86·8%) and the dominance of Haemoproteus lineages. It has to be noted, however, that Haemoproteus infection rates recorded in two German populations, may be underestimated as a consequence of the applied protocol. In contrast to Kulma et al. (Reference Kulma, Low, Bensch and Qvarnström2013) and our study, which assessed infection status based solely on Waldenström et al. (Reference Waldenström, Bensch, Hasselquist and Östman2004) protocol, Wiersch et al. (Reference Wiersch, Lubjuhn, Maier and Kampen2007) additionally carried out an initial screening and chose for the subsequent analysis only positive samples. Since the primer set used in this step, targeting 18 SSU rRNA gene fragment, presumably detects mostly Plasmodium specimens (Hulier et al. Reference Hulier, Pétour, Snounou, Nivez, Miltgen, Mazier and Rénia1996), it may, in general, fail at amplification of the majority of Haemoproteus parasites. Because this specific primer set was only very occasionally used in avian haemosporidian studies (Ribeiro et al. Reference Ribeiro, Sebaio, Branquinho, Marini, Vago and Braga2005; Wiersch et al. Reference Wiersch, Lubjuhn, Maier and Kampen2007), it is very difficult to reliably assess the success rate of Haemoproteus detection with its use. Although Öland and Sękocin Stary populations are similar in some aspects, they differ in some indices of parasite diversity. For example, the Öland population is characterized by higher species richness (the total number of detected lineages) and ecological diversity (as described by the Shannon index). In the case of the Haemoproteus community a noticeable difference between these two populations is a fairly common occurrence of two lineages on Öland (PFC1 – 42·0%, PHSIB1 – 34·4%) and an almost complete dominance of one lineage in Sękocin Stary (PFC1 – 96·3%). The higher lineage richness reported for Öland may be at least partly associated with the much larger sample size, since the number of detected lineages increases with the number of screened individuals (Neto et al. Reference Neto, Pérez-Rodríguez, Haase, Flade and Bensch2015). In general, the geographic variation of parasite infection rates and diversity indicates that pied flycatchers from different breeding populations may be exposed to different parasite-mediated selection pressures.
Observed differences in the composition of haemosporidian communities as well as in the incidence of infection in geographically distant populations of the same host species might result from various ecological, climatic and population factors, which affect haemosporidian distribution directly or indirectly by shaping the abundance and diversity of vectors (e.g. Lachish et al. Reference Lachish, Knowles, Alves, Wood and Sheldon2011; Krama et al. Reference Krama, Krams, Cīrule, Moore, Rantala and Krams2015). It is currently not possible to assess to what extent vectors drive the observed patterns because the distribution of vectors, both on wintering and breeding grounds, is poorly understood. That is especially true for Haemoproteus vectors.
The current study is one of the few molecularly characterizing the incidence and richness of trypanosomes in populations of a specific avian host, as opposed to avian guilds (e.g. Soares et al. Reference Soares, Ellis and Ricklefs2016; Valkiūnas et al. Reference Valkiūnas, Iezhova and Sehgal2016). The infection rate with trypanosomes in the population from central Poland is over three times higher than the estimates in most pied flycatcher populations screened up to date. Specifically, the incidence of infection varied between 1·1% in south-eastern Sweden, 12–16·2% in central Finland, 15% in south-eastern Norway, 16% in central Spain and 40·6% in northern Finland (Bennett et al. Reference Bennett, Siikamäki, Rätti, Allander, Gustafsson and Squires-Parsons1994; Dale et al. Reference Dale, Kruszewicz and Slagsvold1996; Siikamäki et al. Reference Siikamäki, Rätti, Hovi and Bennett1997; Sanz et al. Reference Sanz, Arriero, Moreno and Merino2001a). Because the aforementioned prevalences were derived from blood smear screening, they may be underestimated in comparison with PCR-based diagnostics, which have been shown to be generally more sensitive in detecting Trypanosoma (Sehgal et al. Reference Sehgal, Jones and Smith2001; Nagata, Reference Nagata2006, but see Valkiūnas et al. Reference Valkiūnas, Iezhova and Sehgal2016). Moreover, actual prevalences, regardless of the technique used (smear- or PCR-based diagnostics using blood), may be underestimated because in infected birds Trypanosoma parasites are abundant in the bone marrow, while they usually occur at very low densities or may be absent in the peripheral blood (Apanius, Reference Apanius1991).
Trypanosome infections in the study population were dominated by T. culicavium – a recently described novel species of avian trypanosomes, which is transmitted by mosquitoes (subgenera Culex and Barraudius) (Votýpka et al. Reference Votýpka, Szabová, Rádrová, Zídková and Svobodová2012). Votýpka et al. (Reference Votýpka, Szabová, Rádrová, Zídková and Svobodová2012), based on experimentally identified transmission mode (ingestion of infected vectors) and detection of the parasite in two individuals of the collared flycatcher, suggested that insectivorous songbirds are natural vertebrate hosts of this species. By demonstrating the detection of this trypanosome in another insectivorous bird species, our study confirms this statement. Information about the diet of adult pied flycatchers is very limited, but available data indicate that they ingest Diptera, although at a much lower frequency than other prey (mainly Hymenoptera and Coleoptera) (Bibby and Green, Reference Bibby and Green1980; Silverin and Andersson, Reference Silverin and Andersson1984). So far, T. culicavium has only been detected in European mosquitoes (Votýpka et al. Reference Votýpka, Szabová, Rádrová, Zídková and Svobodová2012; Svobodová et al. Reference Svobodová, Weidinger, Peške, Volf, Votýpka and Voříšek2015a). Among the three other detected trypanosome species, T. corvi [vector: hippoboscid flies (Mungomba et al. Reference Mungomba, Molyneux and Wallbanks1989; Votýpka et al. Reference Votýpka, Lukeš and Oborník2004) has been shown to be transmitted in Europe (Zídková et al. Reference Zídková, Cepicka, Szabová and Svobodová2012)], T. avium [vector: blackflies (Bennett, Reference Bennett1961; Votýpka and Svobodová, Reference Votýpka and Svobodová2004)] – both in Europe and Africa (Svobodová et al. Reference Svobodová, Volf and Votýpka2015b; Valkiūnas et al. Reference Valkiūnas, Iezhova and Sehgal2016), while T. bennetti (vectors currently unknown) – in Europe (Zídková et al. Reference Zídková, Cepicka, Szabová and Svobodová2012). However, since information about the vectors and occurrence of trypanosome species in birds is currently very limited, the areas of transmission of these parasites are not definite.
The incidence of infection with Haemoproteus and Trypanosoma in the study population was unrelated. In birds, the correlated occurrence of these parasites has been found in some [a positive association in white-winged crossbills (Loxia leucoptera)] (Deviche et al. Reference Deviche, Fokidis, Lerbour and Greiner2010), but not in other studies (Oakgrove et al. Reference Oakgrove, Harrigan, Loiseau, Guers, Seppi and Sehgal2014; Svobodová et al. Reference Svobodová, Weidinger, Peške, Volf, Votýpka and Voříšek2015a). In general, a non-independent incidence of infection may occur if, e.g. infection with one parasite causes immunosuppression allowing the other parasite to successfully invade and develop in the host (positive association) or if infection with one parasite prevents the development of the other parasite via cross-immunity (negative association) (Pedersen and Fenton, Reference Pedersen and Fenton2007).
We also looked at each parasite genus separately to search for any relationship between infection incidence and two host traits: age and sex. Despite the fact that susceptibility to parasitic infection is known to increase with age as new infections accumulate during life (Deviche et al. Reference Deviche, Greiner and Manteca2001; Podmokła et al. Reference Podmokła, Dubiec, Drobniak, Arct, Gustafsson and Cichoń2014), we did not find such associations. In the case of the host sex, we found that males were more prone to be infected with Trypanosoma than females, while there was no difference between sexes in the probability of infection with Haemoproteus. Sex differences in parasite prevalence are generally being attributed to an unequal exposure of the sexes to vectors (van Oers et al. Reference van Oers, Richardson, Sæther and Komdeur2010; Egizi et al. Reference Egizi, Farajollahi and Fonseca2014) and differences between males and females in the immune-endocrine system (Schuurs and Verheul, Reference Schuurs and Verheul1990).
In conclusion, by using the molecular approach we showed that the pied flycatcher population from central Poland exhibits one of the highest infection rates with haemoparasites across the breeding range of this species. The comparison of molecularly screened breeding populations revealed some geographic differences in haemosporidian communities, which indicates that pied flycatchers from different populations may experience diverse parasite-mediated selection pressures. However, to be able to confidently make such a conclusion, more thorough study is needed, including of populations with balanced and synchronized sampling, conditions which were not met by the current study. The latter condition means that samples should be collected in the same years as well as at the same point in a breeding period, since both factors are known to be associated with changes in the prevalence and/or composition of the parasite community (annual variation: Bensch and Åkesson, Reference Bensch and Åkesson2003; Bensch et al. Reference Bensch, Waldenström, Jonzén, Westerdahl, Hansson, Sejberg and Hasselquist2007; Knowles et al. Reference Knowles, Wood, Alves, Wilkin, Bensch and Sheldon2011, but see Pagenkopp et al. Reference Pagenkopp, Klicka, Durrant, Garvin and Fleischer2008; variation within a season: Szöllősi et al. Reference Szöllősi, Garamszegi, Hegyi, Laczi, Rosivall and Török2016, Dubiec et al. Reference Dubiec, Podmokła and Gustafsson2017).
Acknowledgments
We thank Bartosz Matuszczak for help in the field, Przemysław Chylarecki for statistical advice and Barbara Przybylska for improving the manuscript's English. The study conforms to the legal requirements of Poland.
Financial support
This work was supported by the Polish Ministry of Science and Higher Education/National Science Centre to TDM [grant number N304 345139].