Introduction
Heat shock proteins (Hsps), known as stress proteins and molecular chaperones, are highly conserved and produced in all organisms from lower prokaryotic organisms to higher mammals (Bose et al., Reference Bose, Weikl, Bügl and Buchner1996; Shi et al., Reference Shi, Fu, Zhao, Zhou, Yang and Qiu2016). In insects, Hsps include ATP-independent small Hsps and the larger ATP-dependent proteins, Hsp60, Hsp70, and Hsp90 (King & MacRae, Reference King and MacRae2015). They perform diverse biological functions during insect normal growth phase or physiological state (Li et al., Reference Li, Li, Yu, Xiang, Kishino and Zhang2009; Sasibhushan et al., Reference Sasibhushan, Ponnuvel and Vijayaprakash2012), and play essential roles in transcription, transporting, protein synthesis, folding, assembling of degraded and misfolded proteins, cell signaling, and metabolism (Huang and Kang, Reference Huang and Kang2007). They are synthesized constitutively and induced significantly when insects are exposed to undesirable stressors, including extreme temperatures (Kayukawa et al., Reference Kayukawa, Chen, Miyazaki, Itoyama, Shinoda and Ishikawa2005; Huang and Kang, Reference Huang and Kang2007), crowding (Wang et al., Reference Wang, Wang, Zhou, Huang, Zhang, Guo and Kang2007), anoxia (Michaud et al., Reference Michaud, Teets, Peyton, Blobner and Denlinger2011), desiccation (Tammariello et al., Reference Tammariello, Rinehart and Denlinger1999), starvation (Wang et al., Reference Wang, Li, Zhu, Fang and Ye2012), heavy metals, other toxic particles and chemical substances (Ge et al., Reference Ge, Huang, Yang, Song, Stanley, Gurr and Wu2013; Zhang et al., Reference Zhang, Wang, Jing, Zhuang and Wu2015).
Hsp60 is a homolog of phage growth E large in bacteria (Gupta, Reference Gupta1995; Xu et al., Reference Xu, Sun, Liu, Zhang, Ru, Zhao and Yang2014), which is produced in the cytoplasm and mainly located in the mitochondria of eukaryotic cells (Neupert, Reference Neupert1997). In recent decades, extensive researches on multiple Hsp60 functions in diverse physiological processes have been conducted in various organisms, for example: involvement in apoptosis (Ghosh et al., Reference Ghosh, Dohi, Kang and Altieri2008), germ cell differentiation (Prinsloo et al., Reference Prinsloo, Setati, Longshaw and Blatch2009), tumorigenesis (Cappello et al., Reference Cappello, de Macario, Marasà, Zummo and Macario2008), and so on. However, researches on insect Hsp60 have focused on response under multiple environmental stress and involvement in certain important physiological processes. Hsp60 can enhance cold hardiness and promote membrane stability by stabilizing microfilaments on insects, such as: Delia antique (Kayukawa and Ishikawa, Reference Kayukawa and Ishikawa2009), Eurosta solidaginis (Zhang et al., Reference Zhang, Storey and Storey2011), and Chilo suppressalis (Cui et al., Reference Cui, Du, Lu and Qiang2010).
Hsp10, known as phage growth E small (in Escherichia coli), is a co-chaperone for Hsp60 and exerts its biological functions in diverse conditions (Boettinger et al., Reference Boettinger, Oeljeklaus, Guiard, Rospert, Warscheid and Becker2015). Hsp60 and Hsp10 form a folding cage through their rings and Hsp60–Hsp10 complex may accelerate polypeptide folding, denatured protein refolding, and misfolded protein correcting (Corrao et al., Reference Corrao, Campanella, Anzalone, Farina, Zummo, Conway, Macario, Cappello and Rocca2010; Clare & Saibil, Reference Clare and Saibil2013; Hayer-Hartl et al., Reference Hayer-Hartl, Bracher and Hartl2016). In insects, information about Hsp10 is far from sufficient. Establishing the molecular information of the Hsp10 and Hsp60 would be necessary for better understanding the tolerance mechanism associated with Hsp60–Hsp10 complex.
Galeruca daurica (Joannis) is a new pest occurring seriously in the Inner Mongolia grasslands in China in recent years, which has exclusively destructive effects on Alliums plants, the pest would lead to a devastating disaster in turf seeding growth on grassland, especially on barren pasture (Tan et al., Reference Tan, Zhou and Pang2016; Zhou et al., Reference Zhou, Gao and Pang2016). It was reported that extensive outbreaks of this pest beginning in 2009 have caused great losses to pasture in grasslands in northern China and levels of damage has increased year by year (Li et al., Reference Li, Zhou, Pang and Chang2014). G. daurica occurs one generation each year, and overwinters as eggs in its natural habitats. Their overwintering eggs have strong cold hardiness with the lower half lethal temperature (LT50) of −33.08°C after 12 h exposure (Gao et al., Reference Gao, Zhou, Pang, Bao, Luo and Erdengqimuge2015), and their 1st, 2nd, 3rd instars’ larvae have also high cold hardiness with the LT50 of −8.5 to −10.1°C after 2 h exposure (Li et al., Reference Li, Zhou, Pang and Chang2014). G. daurica might survive through undergoing huge temperature fluctuation during larvae development process in spring, but how they response to drastic temperature changes and possible mechanism is unknown.
Although Hsps have been proposed to involve in cold hardiness and heat shock responses, the mechanisms underlying temperature stress in G. daurica are hardly known. Hence it is important to understand temperature–stress response and molecular regulation in this outbreak insect. Hsps, as called heat stress proteins, naturally became our focus. At present, there were no Hsp genes in G. daurica that have been studied. Here, we reported the full length of Hsp10 and Hsp60 cDNA sequences of G. daurica, their deduced amino acid sequences and gene characterizations. Furthermore, we examined the mRNA expression pattern of the two Hsps of G. daurica in different developmental stages, five adult tissues, larvae exposed to different temperatures, and eggs under cold stress. Our research should be valuable and provide the basis for further studies on Hsp10 and Hsp60 functions, and also allowed us to explore the roles of the two Hsps associated with thermotolerance and cold hardiness on this seriously occurring pest species.
Materials and methods
Experimental insects
G. daurica egg masses were collected from the Arxant village of Xilinhot City (44°62′N, 115°80′E), Inner Mongolia, China, in middle September in 2015, were brought back to the Research Center for Grassland Entomology (40°48′N, 111°42′E), Inner Mongolia Agricultural University (Hohhot, Inner Mongolia, China), and were maintained in an uninsulated container in outdoors. After 1 month, some of the egg masses were brought back and held in a growth chamber (20 ± 1°C, RH = 50 ± 10%, L16:D8). After 1 week, larvae hatched out and were fed on fresh garlic chives in a growth chamber under the same conditions; garlic chives were planted in our laboratory, which were without exposure to any chemicals and pesticides. The larval development durations were 8, 9, 15, and 7 days for 1st, 2nd, 3rd instars and pupal G. daurica, respectively, and then bugs enter the adult stage.
RNA isolation and cDNA cloning
Total RNA was isolated from test bugs (20–40 mg) using Takara Mini RNA Extraction Kit (Takara, Dalian, China) following the manufacturer's instructions. The RNA integrity was checked by analysis on a 1.0% (w/v) agarose gel; RNA concentrations were measured using a spectrophotometer. The first strand of cDNA was synthesized using PrimeScript First Strand cDNA Synthesis Kit (Takara, Dalian, China), following the manufacturer's protocol.
Based on the partial sequences annotated with Hsp10 and Hsp60 genes from our transcriptome analysis (unpublished), gene-specific primers were designed by Primer 5.0 (table 1). Cycling conditions for amplifying partial cDNA sequences of GdHsp10 and GdHsp60 were as follows: 5 min 94°C; 30 cycles of 20 s 94°C, 20 s 55°C, 30 s 72°C, and a final extension of 10 min at 72°C, using primers GdHsp10-f, GdHsp10-r, GdHsp60-f, and GdHsp60-r. Amplified fragments were eluted from agarose gels and cloned into the pMD™18-T vector (Takara) and then sequenced.
Table 1. PCR primers used in the experiment.

Rapid amplification of the GdHsp10 and GdHsp60 cDNA ends
The 5′ and 3′ ends of cDNA were synthesized employing SMARTer® rapid amplification of cDNA ends (RACE) 5′/3′ Kit as specified by the manufacture (Clontech). PCR amplification was performed using 5′ and 3′ anchor primers (enclosed in 5′/3′ RACE kit) and a specific primer designed from the obtained fragment sequence. Thermal cycling was done using the following program for semi-nested PCR, the 5′GSP1 and 3′GSP1 primers were used to amplify the sequence in the first PCR run; 5′GSP2 and 3′GSP2 primers were for the second round PCR. Before the second run, 5 µl of the primary PCR product was diluted into 245 µl of the EDTA buffer, which was as the cDNA template in the secondary PCR. The thermal cycling was performed as follows: 25 cycles of 30 s 94°C, 30 s 68°C, 2 min 72°C. The expected-size fragments were purified, tailed with A, ligated into a vector and sequenced.
Sequence and phylogenetic analysis
The full cDNA sequences of GdHsp10 and GdHsp60 were analyzed for homology with the BLAST programs at the National Center for Biotechnology Information (NCBI). DNAMAN V6.0 (Lynnon Biosoft, Canada) was used to assemble gene sequences and predict molecular mass and theoretical isoelectric point (pI). The functional sites or domains in the amino acid sequences were predicted by the SMART program (http://smart.embl-heidelberg.de/). Multiple sequence alignments and phylogenetic-tree construction applying the bootstrapped neighbor-joining method were created using ClustalX and MEGA 5.0 software.
Expression of GdHsp10 and GdHsp60 in different development stages and tissues
To investigate the mRNA expression level of GdHsp10 and GdHsp60 in different developmental phases, whole body samples were collected and pooled as follows: third day eggs, 1st, 2nd, 3rd instars’ larvae, pupae, female and male adults, respectively, then all of the samples were kept in −80°C for RNA extraction.
For examining the tissue distribution of GdHsp10 and GdHsp60 transcripts in G. daurica, antenna, head, leg, thorax, wing, and abdomen were cut off with dissecting scissors from third day mixed sex adults. About 100 mg five body tissue samples, including the head (without antenna), thorax (without legs), abdomen (without wings), antenna, and legs (including foreleg, midleg, hindleg) were obtained and frozen in liquid nitrogen immediately, and stored at −80°C until RNA isolation and quantitative PCR (qPCR) determination.
Expression of GdHsp10 and GdHsp60 in response to heat and cold stresses
Different temperature treatments
Ten constant temperatures (−14, −10, −5, 0, 5, 10, 15, 20, 25, and 30°C) were chosen to treat the 2nd instar larvae for 1 h without any recovery and determine mRNA expression level of GdHsp10 and GdHsp60, and the 20°C group was regarded as control. All of the samples were kept in −80°C until RNA extraction.
Low-temperature treatment for different times
Because expression variation of proteins to thermal stress not only influences the growth and development but also determine hatching of embryo (Punyavathi et al., Reference Punyavathi, Bhat and Manjunatha2017), the third day hatched eggs were set as sampling criteria. The tested eggs exposed to 0°C for 0 (control), 15, 30 min, 1, 1.5, 2, and 3 h without any recovery, were selected to test mRNA relative expression level. All samples were instantly frozen and stored at −80°C. Each of the processing was repeated at least four times.
Quantitative analysis of GdHsp10 and GdHsp60 mRNA expression
The relative transcription levels of GdHsp10 and GdHsp60 genes in different developmental periods, different adult tissues, larvae exposed to different temperatures, and eggs exposed to 0°C for a different time were examined using real-time qPCR. Total RNA isolation and first-strand cDNA synthesis were described above. The mRNA expression levels of GdHsp10 and GdHsp60 were examined using a SYBR Green® real-time PCR assay (SYBR PrimeScrip™ RT-PCR Kit II, Takara) with FTC-3000P Real-time PCR (Funglyn Biotech, Canada). The succinate dehydrogenase (SDHA) gene was confirmed stably expressed among different treatments in G. daurica (Tan et al., Reference Tan, Zhou and Pang2017), and was used as an internal control. Primers for qPCR were designed using online software Primer 3.0 (http://primer3.ut.ee/) and listed in table 1. The real-time PCR (final volume of 25 µl reactions) contained 1 µl cDNA template, 12 µl of SYBR Green Real-time PCR Master Mix (Takara, No. DRR420S), and 0.5 µl of each primer; sterile distilled water was added for a total of 25 µl. The experiment was performed as follows: 95°C for 5 min followed by 30 cycles of 95°C for 10 s, 52°C for 15 s, and 72°C for 20 s. The entire experiment was replicated four times with four determinations per sample. The amplification efficiency of each gene was estimated by using the equation: E = [10(−1/slope)−1] × 100 (Pfaffl, Reference Pfaffl2001). Relative expression analysis of GdHsp10 and GdHsp60 mRNA was conducted according to the 2−ΔΔCt value method (Livak & Schmittgen, Reference Livak and Schmittgen2001). All data were given as means ± SD; a statistical difference was determined by analysis of variance followed by the Tukey's multiple comparison test using SPSS 16.0 for Windows.
Results
cDNA cloning, sequencing, and characterization of the GdHsp10 and GdHsp60 genes
After amplifying genes using two pairs of specific primers of GdHsp10-f, -r, and GdHsp60-f, -r, two fragment sequences with 206 and 215 bp were obtained and sequenced, respectively, which were identified as section fragments of Hsp10 and Hsp60 genes by Blastn. For cloning the 5′ and 3′ ends of cDNA of Hsp10, RACE and semi-nested PCR were explored; two fragments of 525 and 402 bp were amplified by 3′ and 5′ RACE techniques, respectively. By sequence assembly of the above sequences using DNAMAN V6.0 (Lynnon Biosoft, Canada), a full length of GdHsp10 cDNA from G. daurica was obtained, which is 822 bp, containing open reading frame (ORF) of 315 bp encoding a protein of 104 amino acid, a 129 bp 5′-UTR, and a 378 bp 3′-UTR (fig. 1). The predicted molecular mass of GdHsp10 was 11.34 kD and the theoretical pI was 9.58. Chaperonin's hsp10/cpn10 signature (LVPLFDRILIKKFEATTKTKGGIVI) of the deduced amino acid sequence of GdHsp10 cDNA is located at 7–31 (shown in gray). Furthermore, a mobile loop, which functioned as an interaction surface with the large subunit of the chaperonin complex, was found in the GdHsp10 sequence (underlined). The complete cDNA sequence of GdHsp10 gene was deposited in GenBank under accession no. KY460464.

Fig. 1. Nucleotide sequence and deduced the amino acid sequence of GdHsp10 cDNA clone. The deduced amino acid sequence is shown below the nucleotide sequence. The initiation and termination codons are marked boxes. The chaperonin hsp10/cpn10 signature is shown in gray and the predicted mobile loop is underlined.
For cloning the 5′ and 3′ ends of cDNA of Hsp60, two fragments of 2017 and 658 bp were amplified by 3′ and 5′ RACE techniques, respectively. After assembling, the full length of GdHsp60 cDNA with 2365 bp was obtained, including a 5′-UTR of 159 bp, a 3′-UTR of 484 bp, and an ORF of 1722 bp encoding 573 amino acids (fig. 2). The calculated molecular mass of GdHsp60 was 61.15 kD and the pI was 5.17. A classical mitochondrial Hsp60 signature shown in gray is shown at amino acid positions of 430–441, and the ATP-binding motif is underlined. In addition, the typical GGM repeat motif at the C-terminus is shown as waved lines. With the 3′-UTR, one putative polyadenylation signal was observed at positions 1919–1923. The nucleotide sequence of GdHsp60 gene was deposited in GenBank under accession no. KY460463.
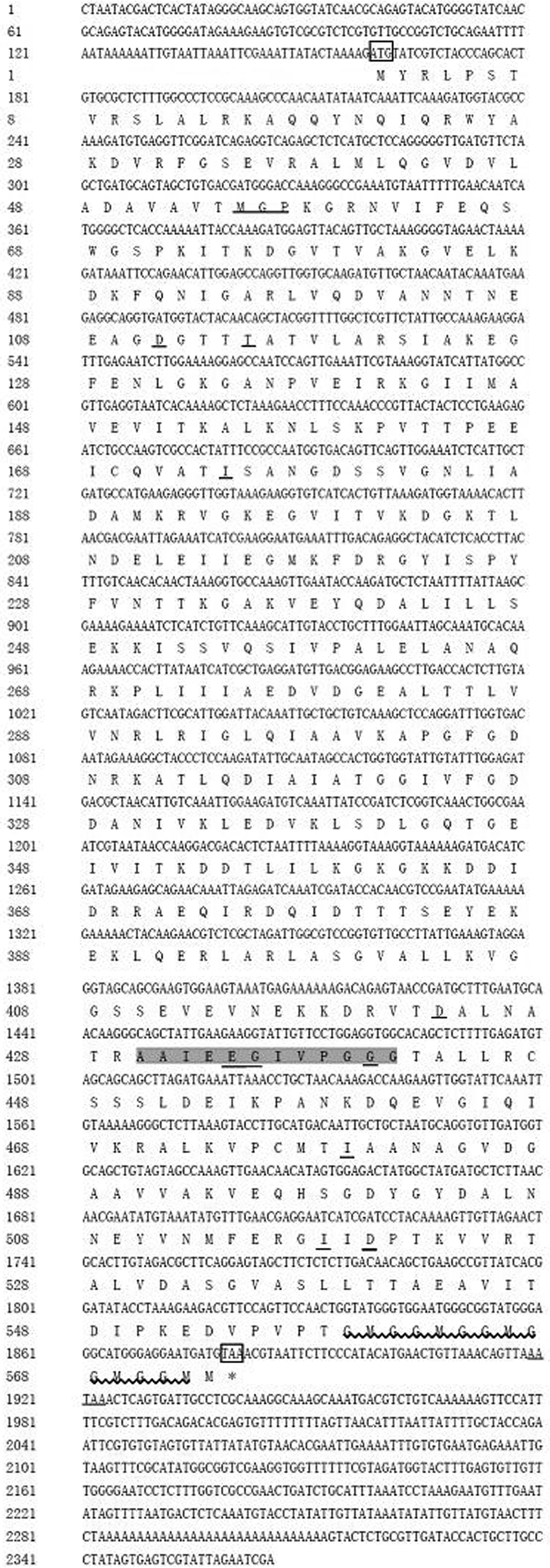
Fig. 2. Nucleotide sequence and deduced the amino acid sequence of GdHsp60 cDNA clone. The deduced amino acid sequence is shown below the nucleotide sequence. The initiation and termination codons are marked boxes. The classical mitochondrial Hsp60 signature is shown in gray and the ATP-binding motif is underlined. The typical GGM repeat motif at the C-terminus is shown as waved lines.
Homology and phylogenetic analysis of GdHsp10 and GdHsp60 genes
From the Blastn program, the nucleotide sequences of the GdHsp10 and GdHsp60 genes shared high homology with other known Hsp10 and Hsp60 genes, which indicated that the two cloned genes encode Hsp10 and Hsp60 proteins. The deduced amino acid sequences of GdHsp10 revealed high similarity with Hsp10 from three insect species: Apis florae (67.3%), Bombyx mori (71.3%), and Musca domestica (59.4%), and three vertebrate species: Penaeus monodon (57.0%), Danio rerio (59.0%), and Homo sapiens (59.0%). Multiple sequence alignment of GdHsp60 with other known Hsp60 amino acid sequences of seven insects revealed that it was highly conserved. GdHsp60 showed the highest identity with Tribolium castaneum (90.4%), as well as Nasonia vitripennis (81.3%), Apis mellifera (80.2%), B. mori (79.2%), Plutella xylostella (79.2%), M. domestica (78.7%), and Myzus persicae (73.8%).
Twenty-two different Hsp10 sequences and 23 Hsp60 sequences of invertebrate and vertebrate species were downloaded from NCBI to conduct phylogenetic analyses. For Hsp10, G. daurica was clustered together with four Lepidoptera insects, and this clade was closed to the other clade with several Diptera and Hymenoptera species. Besides, other invertebrate and vertebrate species were well grouped based on their taxonomic status (fig. 3). Gd Hsp60 was most closely related to sequences from Coleoptera species with high bootstrap support, and they fell into distinct clades with sequences from mammals and Osteichthyes (fig. 4). The phylogenetic trees indicated that the evolution of Hsp60 was almost in accord with the evolution of species.
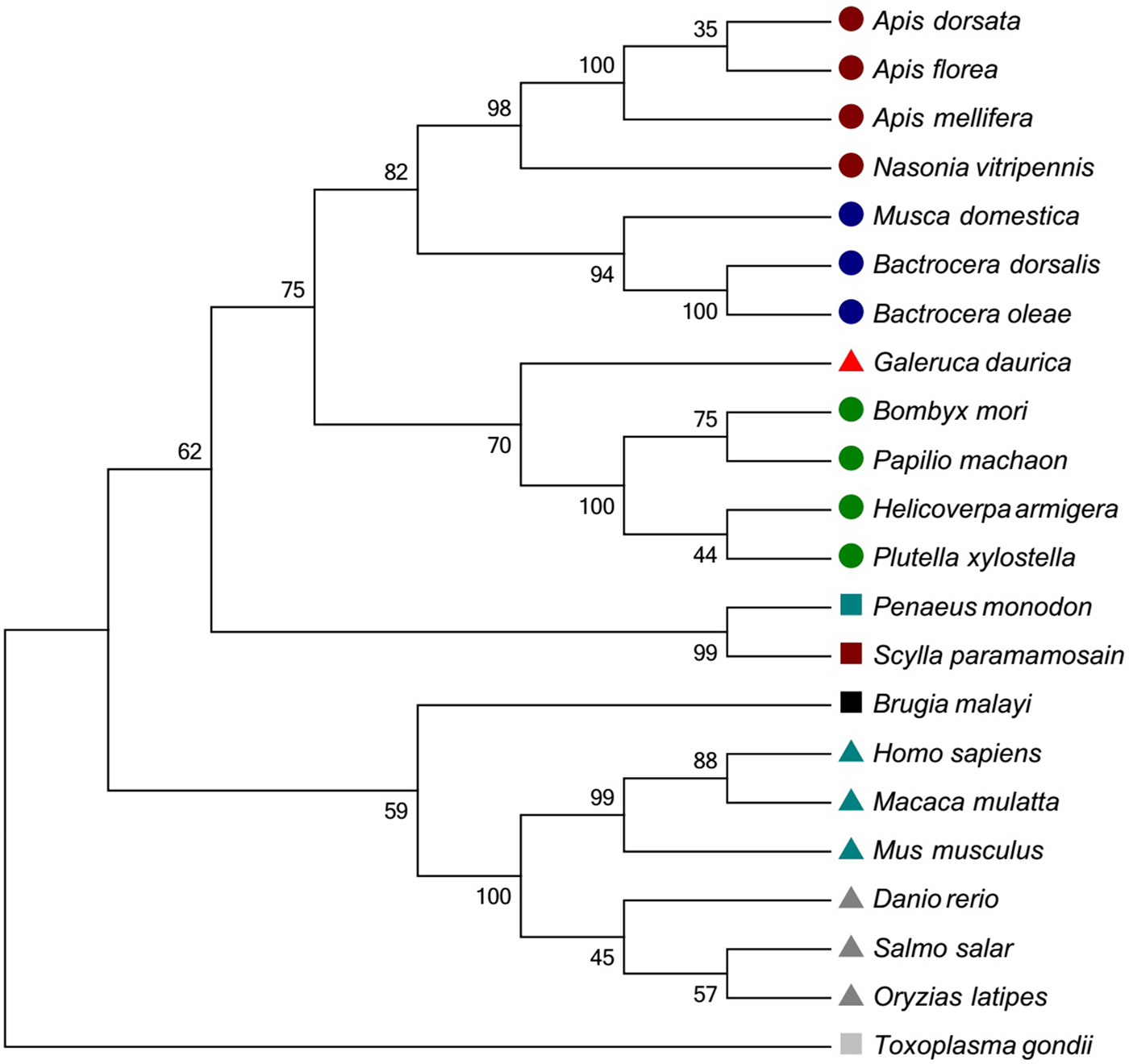
Fig. 3. Phylogenetic trees of GdHsp10. The deduced amino acid sequences were aligned with other known species Hsp10 by the ClustalW program. The tree was constructed using MEGA 5.0 software with the neighbor-joining method. GenBank accession numbers encoding Hsp60 are as follows: Apis dorsata (XM_006622720.1); Apis florae (XM_01245356.1); Apis mellifera (XM_624907.4); Nasonia vitripennis (XM_00159992.4); Bombyx mori (XM_004932889.2); Papilio machaon (XM_014506630.1); Helicoverpa armigera (KC689791.1); Plutella xylostella (XM_011556418.1); Bactrocera dorsalis (XM_011201618.2); Musca domestica (XM_005175857.3); Bactrocera oleae (XM_014241027.1); Homo sapiens (NM_002157.2); Macaca mulatta (XM_001100531); Mus musculus (NM_008303.4); Salmo salar (NM_001139672.1); Danio rerio (NM_131526.1); Oryzias latipes (NM_001104762.1); Penaeus monodon (KT001212.1); Scylla paramamosain (AGI74966.1); Toxoplasma gondii (AY644772.1); Brugia malayi (XM_001902716.1); Galeruca daurica (KY460464). ![]() : Hymenoptera;
: Hymenoptera; ![]() : Diptera;
: Diptera; ![]() : Lepidoptera;
: Lepidoptera; ![]() : Coleoptera (G. daurica);
: Coleoptera (G. daurica); ![]() : Primates;
: Primates; ![]() : Osteichthyes;
: Osteichthyes; ![]() : Coccidae;
: Coccidae; ![]() : Malacostraca;
: Malacostraca; ![]() : Crustacea;
: Crustacea; ![]() : Nematoda.
: Nematoda.
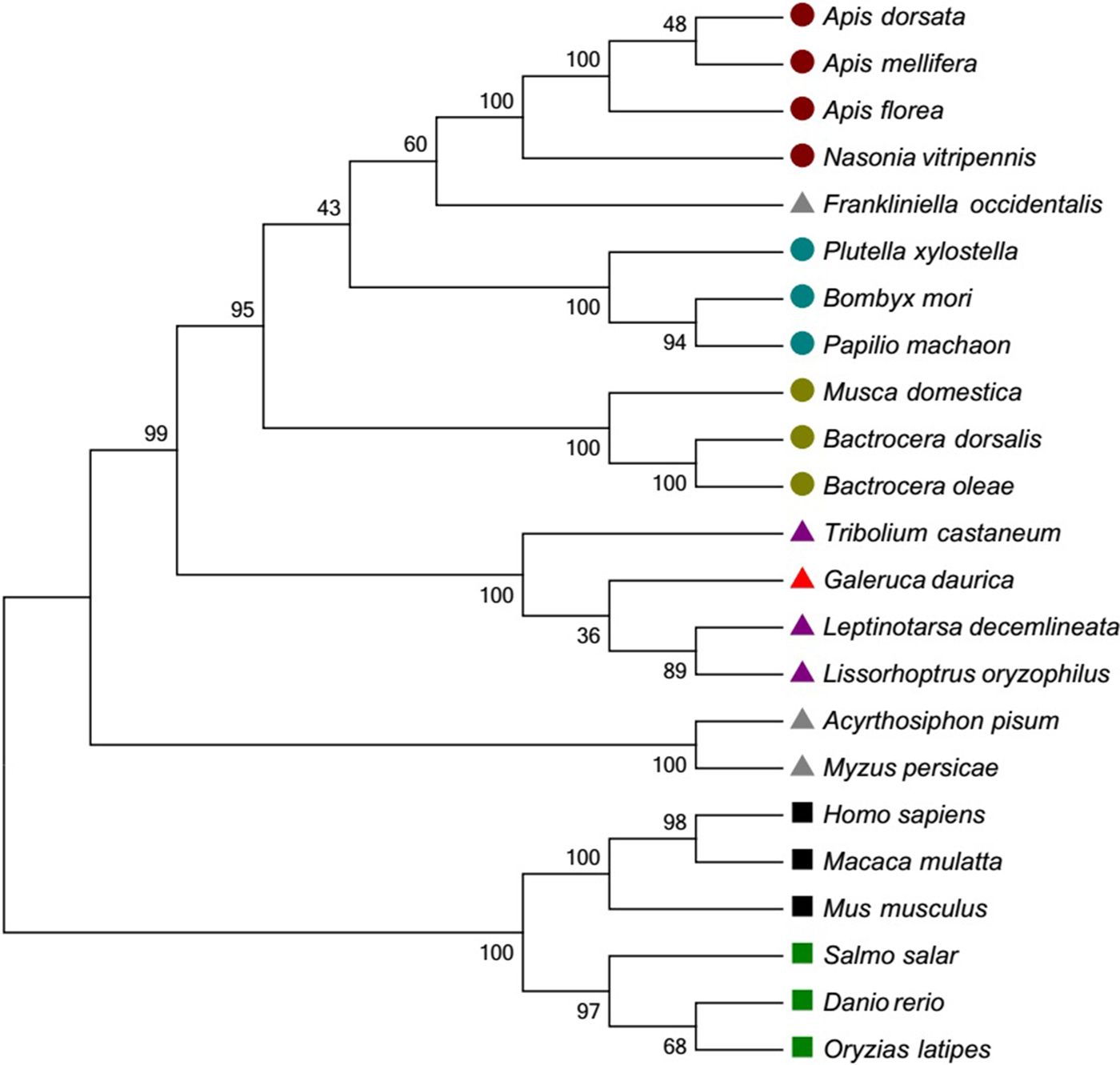
Fig. 4. Phylogenetic trees of GdHsp60. The deduced amino acid sequences were aligned with other known species Hsp60 by the ClustalW program. The tree was generated by MEGA 6.0 using the neighbor-joining (NJ) method. GenBank accession numbers encoding Hsp60 are as follows: Apis dorsata (XM_006622721.1); Apis florae (XM_003691238.2); Apis mellifera (XM_3392899.6); Nasonia vitripennis (XM_003427885.3); Bombyx mori (XM_004923900.2); Papilio machaon (XM_014505853.1); Plutella xylostella (KM215269.1); Bactrocera dorsalis (NM_001317412.1); Musca domestica (XM_005179343.3); Bactrocera oleae (XM_014238382.1); Tribolium castaneum (XM_966537.4); Leptinotarsa decemlineata (KC556801.1); Galeruca daurica (KY460463); Lissorhoptrus oryzophilus (KC620435.1); Frankliniella occidentalis (JX967580.1); Myzus persicae (AJ250348.1); Acyrthosiphon pisum (XM_008180647.2); Homo sapiens (ABB01006.1); Macaca mulatta (NM_034607.3); Mus musculus (AF137008); Salmo salar (ACN1137.01); Danio rerio (BC068415.1); Oryzias latipes (XM_004086882.2). ![]() : Hymenoptera;
: Hymenoptera; ![]() : Lepidoptera;
: Lepidoptera; ![]() : Diptera;
: Diptera; ![]() : Coleoptera;
: Coleoptera; ![]() : Hemiptera;
: Hemiptera; ![]() : Primates;
: Primates; ![]() : Osteichthyes.
: Osteichthyes.
Expression of GdHsp10 and GdHsp60 in different developmental stages and tissues
The qPCR was used to determine mRNA expression levels of GdHsp10 and GdHsp60 in the whole life cycle of G. daurica; GdHsp10 and GdHsp60 were expressed at all developmental stages tested. For GdHsp10, it was revealed that the mRNA expression levels were significantly high in the eggs and 1st instar larvae with 4.43 and 4.19 times, respectively. Furthermore, the expression levels of G. daurica in 2nd, 3rd instars’ larvae, pupae, and female adults were 2.82, 2.02, 2.77, and 1.25 times of that in the male adults, in which the relative expression level was set as fiducial value (1.00) (F 6,27 = 2.66, P = 0.06) (fig. 5a). The GdHsp60 was highly expressed in the eggs, and the relative expression level was 2.68 times of that in the male adults. Besides, the expression levels of G. daurica in the 1st and 3rd instar larvae were 1.60 and 2.10 times of that in the male adults. However, the GdHsp60 was lowly expressed in the 2nd instar larvae, pupae, and female adults, and their relative expression levels were 0.80, 0.82, and 0.53 times of that in the male adults (F 6,27 = 27.42, P < 0.05) (fig. 5b).
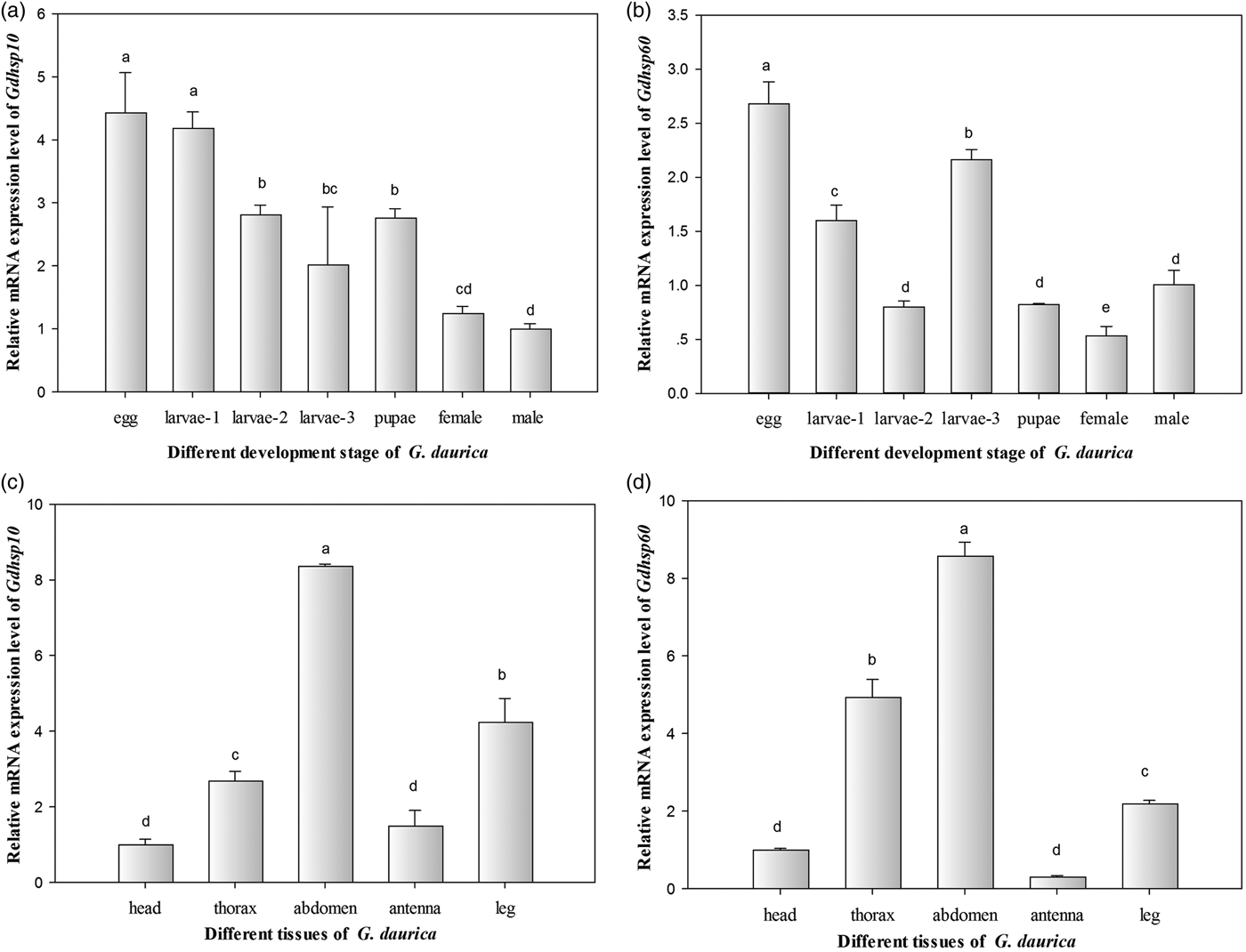
Fig. 5. Relative mRNA expression levels of GdHsp10 and GdHsp60 gene in different development stages and tissues. (a) Gdhsp10, (b) GdHsp60 in different development stages; (c) Gdhsp10, (d) Gdhsp60 in different tissues of adults. The bar colored light brown in each figure was considered as control. Different lowercase letters indicate significant differences (P<0.05). Each bar represents the mean ± SD (N = 4).
In order to investigate the tissue distribution of GdHsp10 and GdHsp60, the relative expression levels of their mRNA in five adult body tissues (head, thorax, abdomen, antenna, and leg) were examined by qPCR in the third day adults. Both of the two Hsp genes were expressed omnipresently in five tissues examined in G. daurica adults, but were abundantly expressed in the abdomen, while the head and antenna showed the lowest expression (fig. 5c, d). The relative expression levels of GdHsp10 and GdHsp60 showed a significant difference (P < 0.05) among different tissues (F 4,19 = 209.09, P < 0.05; F 4,19 = 518.81, P < 0.05).
Expression of GdHsp10 and GdHsp60 in response to heat and cold stresses
The expression patterns of GdHsp10 and GdHsp60 were tested using qPCR after different temperature treatments for 1 h on the 2nd instar larvae. Both of the GdHsp10 and GdHsp60 genes had a significant response to heat and cold stresses. For cold treating, the GdHsp10 expression was significantly induced to increase more than triple times in the −5 and 0°C treatments, and relative expression levels in the −14, −10, 5, 10, and 15°C treatments were 1.81, 2.75, 1.93, 2.10, and 2.11 times of that in the 20°C treatment, respectively. For heating, the 25 and 30°C heat treating larvae showed overexpressed levels (2.46 and 1.96 times than in the 20°C group) (F 9,39 = 8.33, P < 0.05) (fig. 6a). A similar pattern appeared in the expression of GdHsp60 gene under different temperature exposures. As fig. 6b showed, the expression of GdHsp60 gene under cold temperature gradient was obviously higher than in the 20°C treatment, their expression levels were 3.66, 5.81, 4.38, 3.19, 4.13, 4.78, and 3.29 times for 15, 10, 5, 0, −5, −10, and −14°C. Moreover, the 25 and 30°C heat treating larvae showed overproduced GdHsp60 transcripts (3.38 and 3.36 times than in the 20°C group) (F 9,39 = 65.10, P < 0.05) (fig. 6b). The mRNA expression levels of GdHsp10 and GdHsp60 at 0°C for different times in the third day eggs were examined by qPCR. It was clearly exhibited that GdHsp10 and GdHsp60 genes were upregulated when G. daurica eggs were exposed at 0°C, and a clear time-dependent expression pattern of the two genes was observed. The GdHsp10 mRNA expression reached peak (5.19-fold higher than that of the control) after 1.5 h and then decreased (F 6,27 = 33.21, P < 0.05) (fig. 6c), and the GdHsp60 transcript increased to a maximum level (2.82-fold higher than that of the control) after 30 min and declined gradually (F 6,27 = 14.21, P < 0.05) (fig. 6d).

Fig. 6. Relative mRNA expression levels of GdHsp10 and GdHsp60 genes in response to thermal stress. (a) Gdhsp10 and (b) GdHsp60 in different temperature treatments of the 2nd instar larvae; (c) Gdhsp10 and (d) GdHsp60 in eggs under 0°C low-temperature exposure for different times. The bar colored light brown in each figure was considered as control. Different lowercase letters indicate significant differences (P < 0.05). Each bar represents the mean ± SD (N = 4).
Discussion
As a new and severely outbreaking insect species in grasslands in northern China, G. daurica has an adaptive strategy to escape extremely low-temperature condition in eggs and tolerate great daily temperature fluctuation in larvae to guarantee adequate population density, they mainly reply on their physiological stress responses. This survival strategy represents the first layer of self-defense in decreasing potentially negative effects of adverse conditions (Rosic et al., Reference Rosic, Kaniewska, Chan, Ling, Edwards, Dove and HoeghGuldberg2014). Gao et al. (Reference Gao, Zhou, Pang, Bao, Luo and Erdengqimuge2015) reported that the half-lethal time (LT50) at −30°C was 33.33 days in overwintering eggs of G. daurica, and Zhou et al. (Reference Zhou, Gao and Pang2016) discovered that exposure to low temperatures was required to terminate egg diapause; furthermore, the longer chilling would shorten post-diapause time and accelerate egg development. So far, more efforts have been concentrated on studying the effects of temperature stress and treating time on the physiology of G. daurica, while little is known about the mechanism of responses and adaptability to temperature variation at molecular levels. Hsp is associated with cryoprotection of cells and tissues from thermal stress (Denlinger, Reference Denlinger2002), and has been studied in some organisms subjected to thermal stress from ecological and physiological aspects (Feder & Hofmann, Reference Feder and Hofmann1999).
In the present study, the two genes encoding GdHsp10 and GdHsp60 were cloned and characterized from G. daurica. The full-length cDNAs are 822 and 2365 bp with ORF of 315 and 1722 bp for GdHsp10 and GdHsp60, respectively, encoding 104 and 573 amino acids with predicted molecular weights of 11.34 and 61.15 kD. A mobile loop structure and hsp10/cpn10 signature was detected in GdHsp10 cDNA, a highly conserved mitochondrial Hsp60 signature, the ATP-binding sites, and four Gly-Gly-Met repeat motifs at the C-terminus were found in the GdHsp60 sequence, in accordance with the structures of Hsp10/Hsp60 in marine organisms’ studies (Xu et al., Reference Xu, Horwich and Sigler1997; Reference Xu, Sun, Liu, Zhang, Ru, Zhao and Yang2014). A dome-shaped heptameric ring of Hsp10 subunit and Hsp60 cylinder with ATPase form special structure called chaperon folding cage (Saibil & Ranson, Reference Saibil and Ranson2002); the ATP hydrolysis triggers conformational changes of cage structure, producing a protected environment favoring protein folding (Clare & Saibil, Reference Clare and Saibil2013); meanwhile, the mobile loop in Hsp10 and an apical domain in Hsp60 cylinder bind and function as folding protein (Hayer et al., Reference Hayer-Hartl, Bracher and Hartl2016). However, how Hsp60/Hsp10 complex co-works in cells, what proteins they can protect, and how temperature variation regulates Hsp60/Hsp10 expression and triggers protein protection mechanism are hardly known in insects; therefore, these researches are our main focus and interest on G. daurica in the future.
Homologous alignment analysis demonstrated that both GdHsp10 and GdHsp60 had high similarity with other Hsp10 and Hsp60 from other insects which have the closest relationship with G. daurica based on morphologic taxonomy. Phylogenetic trees showed that the evolution of Hsp10 and Hsp60 was almost in accord with the evolution of species. These analyses further suggested that GdHsp10 and GdHsp60 belonged to Hsp10 and Hsp60 families.
Some Hsps played essential roles in regulating growth, development, and cellular differentiation in insects, with tissue and time specificity (Michaud et al., Reference Michaud, Marin and Tanguay1997; Sharmas et al., Reference Sharmas, Rohilla and Tiwari2007). GdHsp10 and GdHsp60 were proven to be expressed during the whole life cycle, but expressed highest in embryonic development stage (fig. 5a, b). As tissue distribution of mRNA expression analysis showed, GdHsp10 and GdHsp60 were expressed in five tissues tested in adults, and to be abundant in the abdomen of G. daurica (fig. 5c, d). It was reported that Hsps exist in high concentration in cells and fulfill a crucial role as a defense mechanism by sustaining protein homeostasis during cellular stress through overproduction (Kastle & Grune, Reference Kastle and Grune2012; Bozaykut et al., Reference Bozaykut, Kartal and Karademir2014). G. daurica overwinters as eggs in bitter cold northern China; it was suggested that high expression of Hsp10 and Hsp60 in eggs might be helpful for tolerating extreme low temperatures, transmitting signals for cell proliferation, protein denaturation, programed death to protect them from severe cold injury, which might be one important cold resistance mechanism. More experiments on function and structure of GdHsp10/GdHsp60 are required to prove our speculation in further research.
From mid-April to early May, G. daurica in larval stage can emerge overnight and rapidly cause destructive losses on young grasses in the vast pasture. If the population density can be reduced below the economic threshold before 3rd instar larvae outbreak, the losses would be relieved. For low age larvae, G. daurica experienced daily temperature oscillation even more than 20°C in the field; the potential mechanism of tolerating severe temperature variation is our main concern. Hsps are commonly used as gene markers to study temperature-induced cell stress and explore potential protective mechanism when the organism is exposed to thermal stress (Chow et al., Reference Chow, Ferriere-Pages, Khalouei, Reynaud and Brown2009; Olsen et al., Reference Olsen, Ritson, Ochrietor, Paul and Ross2013). From our results, both heat and cold stresses can induce mRNA expression of GdHsp10 and GdHsp60 after 1 h exposure (fig. 6a, b). The similar results were reported in other studies (Fang et al., Reference Fang, Huang and Lin1997; Chow et al., Reference Chow, Ferriere-Pages, Khalouei, Reynaud and Brown2009; Meng et al., Reference Meng, Liu, Li, Gao and Chen2014; Xu et al., Reference Xu, Sun, Liu, Zhang, Ru, Zhao and Yang2014; Rinehart et al., Reference Rinehart, Li, Yocum, Robich, Hayward and Denlinger2007). Hsp10 and Hsp60 in the sea cucumber were slightly upregulated when temperature increased to 2–4°C and sharply overexpressed after exposed to 6°C rising (Xu et al., Reference Xu, Sun, Liu, Zhang, Ru, Zhao and Yang2014); HSP60 expression in the reef-building coral increased after exposed to moderate, severe cold and heat in short time of 6 h (Seveso et al., Reference Seveso, Montano, Strona, Orlandi, Galli and Vai2016). These results suggested that Hsp10 and Hsp60 were rapidly induced to protect the threatened proteins in organisms under elevated and depressed temperatures, in line with the role of Hsps as rapid and temporally dynamic cellular machinery associated with cellular defense (Georgopoulos and Welch Reference Georgopoulos and Welch1993; Feder & Hofmann, Reference Feder and Hofmann1999). However, some researchers considered that Hsp10 and Hsp60 expression trends and modulation magnitudes have close relationship with the intensity of the thermal stress and exposure time, either hot or cold (Rodriguez-Lanetty et al., Reference Rodriguez-Lanetty, Harii and Hoegh-Guldberg2009; Meyer et al., Reference Meyer, Aglyamova and Matz2011; Seveso et al., Reference Seveso, Montano, Strona, Orlandi, Galli and Vai2016). Further studies are required to verify the conclusion on G. daurica.
For 0°C exposure of G. daurica eggs, we also tracked the change of expression of GdHsp10 and GdHsp60 over time; it was indicated that the transcriptional expression levels of the two Hsp genes have a peculiar time-dependent response and both of them showed similar patterns of mRNA expression (fig. 6c, d). The expression of GdHsp10 and GdHsp60 reached a peak at 1.5 h and 30 min, respectively, then decreased slowly. The expression of Hsp10 and Hsp60 in other organisms was strongly induced under thermal stress and decreased gradually (Georgopoulos and Welch, Reference Georgopoulos and Welch1993; Xu et al., Reference Xu, Sun, Liu, Zhang, Ru, Zhao and Yang2014). Researchers explained that upregulation of Hsp10 or Hsp60 might be due to cellar defenses triggered to counteract stresses in short time (Georgopoulos and Welch, Reference Georgopoulos and Welch1993; King & MacRae Reference King and MacRae2015); but for subsequent stabilization or gradual downexpression of the two genes, more explanations mainly focus on energy balance strategy and metabolic disorder. The former considers that organisms cannot afford the excess energy consumption required for the synthesis of Hsps after long exposure to threatened stressors (Krebs & Feder, Reference Krebs and Feder1997, Reference Krebs and Feder1998). The later reflects strong repression of the metabolic activities, such as restricted enzymatic activities, cell division, development, growth (Sokolova & Portner, Reference Sokolova and Portner2001; Roth et al., Reference Roth, Goericke and Deheyn2012). However, some researchers put forward a viewpoint that the phenomena were associated with host mitochondria injury, causing electron transport and ATP produce degradation, because Hsp60s are ATP-dependent (Dunn et al., Reference Dunn, Pernice, Green, Hoegh-Guldberg and Dove2012). In other studies, it was found that Hsp60 expression returned to the basal value, and they speculated that it might be due to a rapid physiological acclimation of the organism's ability (Seveso et al., Reference Seveso, Montano, Strona, Orlandi, Galli and Vai2016). At the cellular level, the cellular stress responses could transitorily increase the stress tolerance threshold to remain homeostatic (Kultz, Reference Kultz2005).
In summary, the GdHsp10 and GdHsp60 cDNA sequences were cloned and characterized from G. daurica for the first time. We found that both the Hsp genes were induced by heat and cold stresses in the 2nd instar larvae, and their transcriptional levels promptly increased with an apparent time-dependent response after eggs were exposed to freezing point. This information provides a foundation to research the biological function of the Hsp10/Hsp60 complex, tolerating temperature upheaval mechanism and cold resistance mechanism on G. daurica, meanwhile, confirms the practicability of Hsps as molecular markers to test biological responses to heat and cold stresses.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31360441).










