Original:
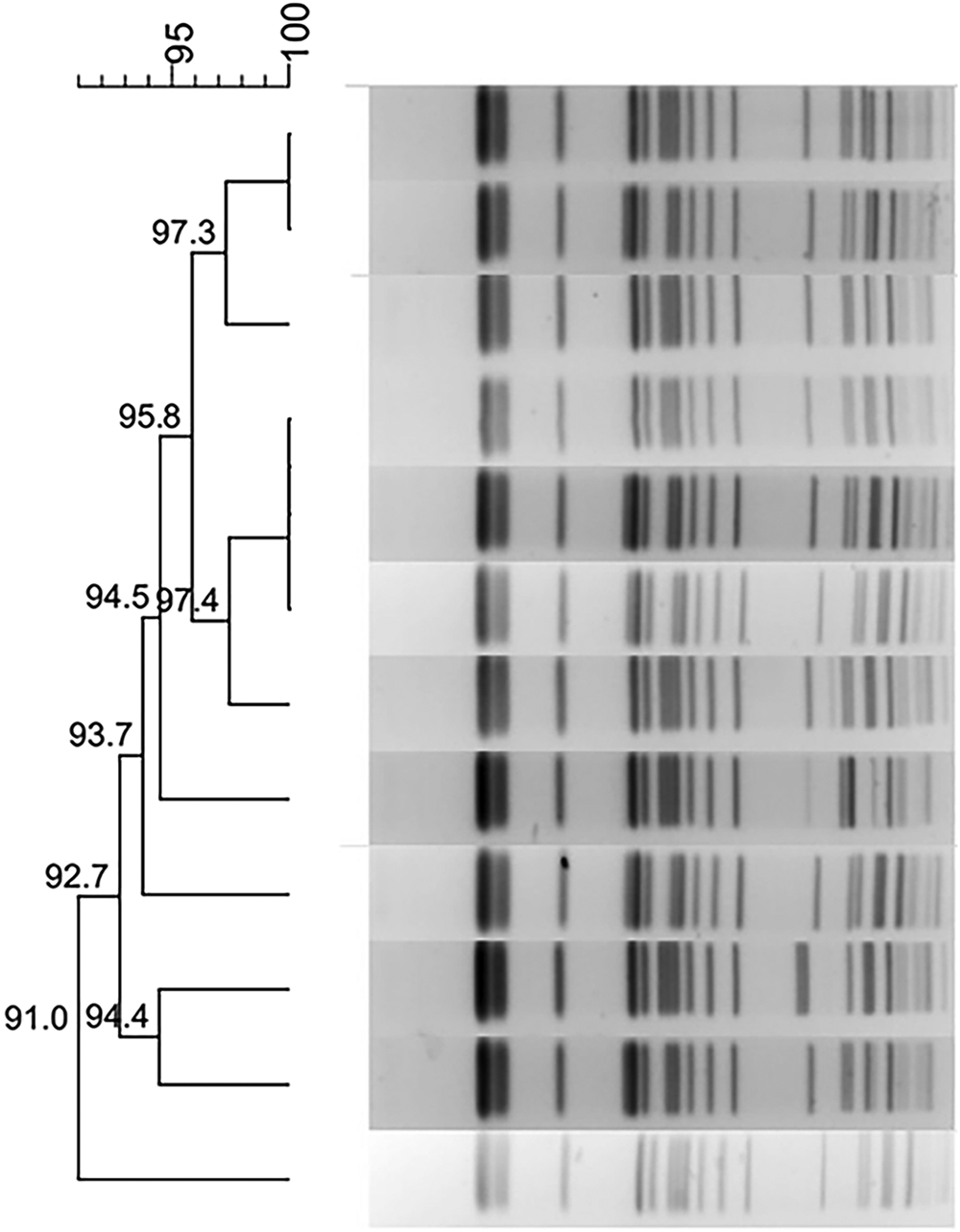
Fig. 1. Typing results of PFGE and MLST from 44 CRKP isolates. Two PFGE clones belong to ST337 and ST11, respectively.
Correction:
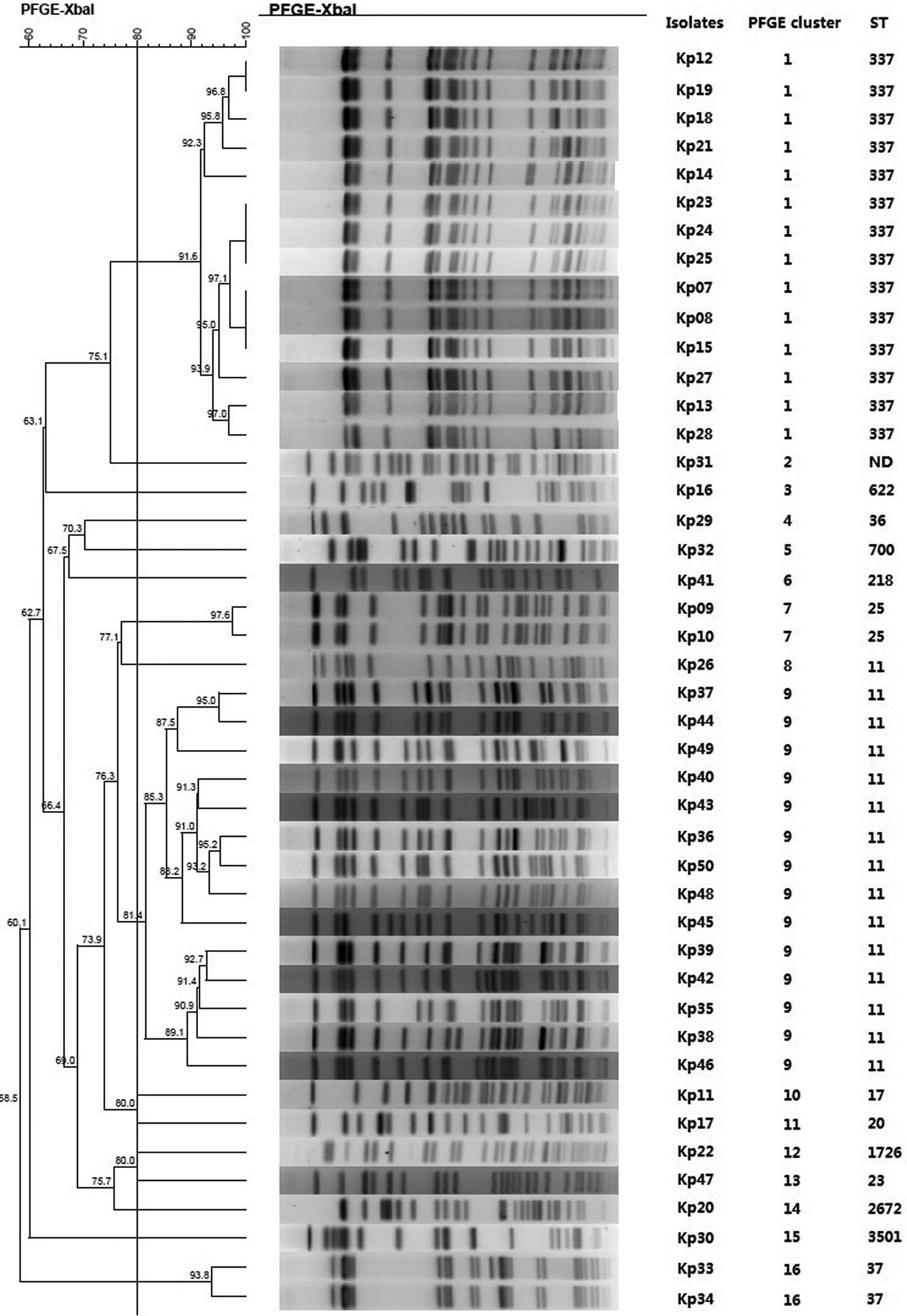
Fig. 1. Typing results of PFGE and MLST from 44 CRKP isolates. Two PFGE clones belong to ST337 and ST11, respectively.
Original:
Fig. 2. Three-dimensional plot generated by MALDI-TOF MS. Four PCA clusters were identified. Strains from distinct PFGE clusters (PC) were grouped into one MALDI-TOF cluster, and strains belonging to the same PFGE cluster were assigned to more than one MALDI-TOF clusters.
Correction:

Fig. 2. Three-dimensional plot generated by MALDI-TOF MS. Four PCA clusters were identified. Strains from distinct PFGE clusters (PC) were grouped into one MALDI-TOF cluster, and strains belonging to the same PFGE cluster were assigned to more than one MALDI-TOF clusters.






