Introduction
Bovine respiratory disease (BRD) is a multi-factorial disease complex involving an interaction of stressors (weaning, passage through auction markets, commingling, shipping, etc.), viral infections (bovine viral diarrhea virus, bovine respiratory syncytial virus, para-infulenza-3 virus, bovine herpes virus-1, etc.), and bacteria (Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, Mycoplasma bovis). Global economic impact of BRD is estimated to be >$3 billion year−1 (Watts and Sweeney, Reference Watts and Sweeney2010). Antimicrobial administration is a mainstay of both prevention and control of disease and treatment of clinical disease. Table 1 contains a non-exhaustive list of antimicrobial products currently licensed in the United States (USA) for the treatment of BRD. Widespread bacterial pathogen resistance to antimicrobials commonly used for BRD is a very real concern shared by producers, practitioners, and the animal health industry. In the USA, there are no routine surveillance programs to monitor antimicrobial resistance among BRD pathogens; however, independent and industry-sponsored research can be found reporting surveillance-type data from different areas of the world.
Table 1. Non-exhaustive list of antimicrobial products licensed in the USA for treatment and/or prevention/control of BRD
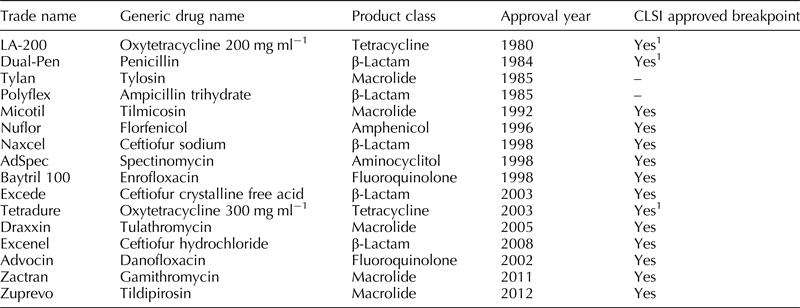
1 Generic label breakpoints established using field wild-type isolates, in vitro pharmacokinetic, and pharmacodynamic data in the absence of randomized clinical field trials with treatment outcomes.
Observations from the medical field on antimicrobial resistance were first reported by medical practitioners as early as the late 1940s to early 1950s (Glisan et al., Reference Glisan, Steele, Whitford, Christensen and Kapadia1982). The first documented case of multiple drug resistance transfer by conjugation was reported in 1959 by Akiba and Ochiai in Japan (Watanabe, Reference Watanabe1967). Specific to BRD, the first reports of multiple drug resistance in P. multocida and M. haemolytica (formerly Pasteurella haemolytica) were published by Chang and Carter in 1976 (Chang and Carter, Reference Chang and Carter1976).
The first publications of research into the mechanisms of antimicrobial resistance are from the early 1960s. Similar publications specific for pathogens of BRD did not begin appearing in the literature until the early 1980s. Resistance genes have been found and described for the common bacterial pathogens associated with BRD for the tetracyclines (Singer et al., Reference Singer, Case, Carpenter, Walker and Hirsh1998; Kehrenberg et al., Reference Kehrenberg, Catry, Haesebrouck, De Kruif and Schwarz2005; O'Connor et al., Reference O'Connor, Wellman, Rice and Funk2010; D'Amours et al., Reference D'Amours, Ward, Mulvey, Read and Morck2011; Klima et al., Reference Klima, Alexander, Read, Gow, Booker, Hannon, Sheedy, Mcallister and Selinger2011; Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b), fluoroquinolones (Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b; Pardon et al., Reference Pardon, Hostens, Duchateau, Dewulf, De Bleecker and Deprez2013), beta-lactams ( Klima et al., Reference Klima, Alexander, Read, Gow, Booker, Hannon, Sheedy, Mcallister and Selinger2011; Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b; Alexander et al., Reference Alexander, Cook, Klima, Topp and Mcallister2013a), macrolides (Desmolaize et al., Reference Desmolaize, Rose, Warrass and Douthwaite2011a, Reference Desmolaize, Rose, Wilhelm, Warrass and Douthwaiteb; Kadlec et al., Reference Kadlec, Brenner Michael, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Watts and Schwarz2011; Klima et al., Reference Klima, Alexander, Read, Gow, Booker, Hannon, Sheedy, Mcallister and Selinger2011; Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b), sulfonamides (Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b), lincosamides (Desmolaize et al., Reference Desmolaize, Rose, Wilhelm, Warrass and Douthwaite2011b; Kadlec et al., Reference Kadlec, Brenner Michael, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Watts and Schwarz2011; Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b), phenicols (Kehrenberg et al., Reference Kehrenberg, Wallmann and Schwarz2008; Katsuda et al., Reference Katsuda, Kohmoto, Mikami, Tamamura and Uchida2012; Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b), and aminoglycosides (Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012b; Alexander et al., Reference Alexander, Cook, Klima, Topp and Mcallister2013a).
Lubbers and Hanzlicek described the available literature on antimicrobial resistance by categorizing the information into two categories: (1) authors reporting the percentage of isolates that are susceptible or resistant, or (2) the minimal inhibitory concentration (MIC) distribution, i.e. the lowest concentration of an antimicrobial agent that prevents visible growth of a microorganism in an agar or broth dilution susceptibility test for either 50% (MIC50) or 90% (MIC90) of isolates tested (Lubbers and Hanzlicek, Reference Lubbers and Hanzlicek2013). Also present in the literature are molecular investigations into the mechanisms of resistance from small pools of field isolates or specific strains known to carry single or multidrug resistance.
Studies reporting either an MIC distribution or a percentage of susceptible/resistant isolates often, but not always, provide a description of criteria used to determine isolate classification as susceptible, intermediate, or resistant. Comparison of results between publications is difficult, if not impossible, when these criteria are not described adequately. Even when described appropriately, differences in methodology may make comparison between datasets inappropriate.
Standardized antimicrobial susceptibility testing methods and interpretive criteria are described by the Clinical and Laboratory Standards Institute (CLSI) in document VET01-A4 (CLSI VET01-A4, July 2013). The most recent listing of CLSI interpretive criteria are contained in the supplemental document VET01-S2 (CLSI VET01-S2, July 2013). The Veterinary Antimicrobial Susceptibility Testing subcommittee within the CLSI determines veterinary-specific interpretive criteria based on evaluating clinical isolates, wild-type isolates, and pharmacokinetic/pharmacodynamic cutoffs as described in CLSI document VET02-A3 (CLSI and VET02-A3, February 2008).
The CLSI, in VET01-A4, defines Susceptible, Intermediate, and Resistant as follows:
-
Susceptible – a category that implies that an infection due to the strain may be appropriately treated with the dosage regimen of an antimicrobial agent recommended for that type of infection and infection species, unless otherwise indicated;
-
Intermediate – a category that implies that an infection due to the isolate may be appropriately treated in body sites where the drugs are physiologically concentrated or when a high dosage of drug can be used; also indicates a ‘buffer zone’ that should prevent small, uncontrolled, technical factors from causing major discrepancies in interpretations;
-
Resistant – resistant strains are not inhibited by the usually achievable concentration of the agent with normal dosage schedules and/or fall in the range where specific resistance mechanisms are likely (e.g., β-lactamase), and clinical outcome has not been predictable in effectiveness studies.
As outlined in Table 1, 11 antimicrobials have (non-generic) veterinary breakpoints established for the treatment of BRD as described in VET01-A4 (CLSI VET01-A4, July 2013). A CLSI-approved veterinary breakpoint applies to a specific combination of disease, pathogen, animal species, and antimicrobial treatment regimen. When one of these parameters is altered, implied clinical outcome as related to the breakpoint is no longer valid, and therefore, the predictive value of the breakpoint is suspect. Unfortunately, the literature contains a plethora of studies containing information based on non-standardized testing methods or interpretive criteria not validated for the condition being treated (Apley, Reference Apley2003). Applying CLSI interpretive criteria to data generated from susceptibility testing which did not adhere to CLSI standards is an example of inappropriate reporting. As such, all studies described herein have either used the criteria described by CLSI or have adequate reasoning for using different criteria.
Literature review
The scientific literature contained in PubMed, AGRICOLA, and CAB were searched in February of 2014 using the following combinations of terms: (((((((bovine respiratory disease) AND antibiotic resistance) OR antimicrobial resistance) AND M. haemolytica) OR Pasteurella haemolytica) OR P. multocida) OR H. somni) OR Haemophilus somnus). Titles and abstracts were read and 105 articles that were relevant to the subject of BRD antibiotic resistance were attained for further review. Relevance, defined as an abstract stating an objective (or conclusion) related to an obvious attempt to characterize susceptibility or resistance of the three BRD pathogens within a population of cattle. Additionally, if the title was related to BRD and antimicrobial susceptibility or resistance and did not contain an abstract, the article was attained for review.
The first exclusion criteria involved removing publications not originating in North America or not in English. The next exclusion criterion applied was the removal of publications that did not utilize CLSI (or the former title NCCLS) criteria. The final exclusion criterion was the removal of articles not focused on BRD in beef cattle. Ten articles investigating phenotypic resistance and six articles on genetic components involved in macrolide resistance in BRD bacterial pathogens are included in this review.
Reports of phenotypic resistance in North America
Chang and Carter (Reference Chang and Carter1976) analyzed isolates of M. haemolytica (n = 262) and P. multocida (n = 141) from clinical cases of BRD sent to the Michigan State Laboratory during a 3-year period from 1971 to 1974 (Chang and Carter, Reference Chang and Carter1976). Careful interpretation of these data must be used as the publication predates the establishment of CLSI interpretive criteria. However, this report was the first report of multi-drug-resistant BRD bacterial pathogens in the scientific literature. They found that most of the isolates were resistant to at least one of the four antimicrobials tested (dihydrostreptomycin, tetracycline, penicillin, and chloramphenicol). Using susceptibility interpretive criteria described by Kirby and Bauer, they reported resistance to at least a single drug in 79.1% (148/187) and 96.1% (122/127) of P. multocida and M. haemolytica, respectively. This paper is included as a historical reference, but the interpretive criteria differ from current criteria, and the methods pre-date CLSI standards, so comparison to susceptibility distributions using current standards would be inappropriate.
In 1991, Post et al. published a report regarding the antimicrobial susceptibility patterns and MICs of P. multocida (n = 158) and M. haemolytica (n = 421) isolated from nasal swabs and tissue specimens submitted from cattle with BRD to the Texas Veterinary Medical Diagnostic Laboratory between February and June 1989 (Post et al., Reference Post, Cole and Raleigh1991). The susceptible or resistant findings from this study are only marginally useful for comparison to current findings because of changes in CLSI interpretive criteria and uncertainty in comparing the plates used in this study and the commercial plates currently used in microwell dilution testing. The M. haemolytica MIC50 and MIC90 values for ceftiofur were at or below 0.125 µg ml−1, the lowest dilution tested. For tetracycline, the corresponding values for MIC50 and MIC90 were 2 and 16 µg ml−1, respectively.
Watts et al. published a survey of antimicrobial susceptibility findings involving a total of 888 isolates recovered from BRD cases over a 4-year period from 1988 to 1992 (Watts et al., Reference Watts, Yancey, Salmon and Case1994). The isolates (461 M. haemolytica, 318 P. multocida, and 109 H. somni) were sent from veterinary diagnostic laboratories to an Upjohn (Zoetis) laboratory for MIC determinations. Only isolates of M. haemolytica, P. multocida, and H. somni obtained from lungs of animals that had died from acute BRD were requested. No more than two isolates of each species from each herd (or feedlot) were received from the following states: Pennsylvania, Wyoming, Iowa, Washington, California, Missouri, Nebraska, Oregon, Kansas, Arizona, Texas, South Dakota, Montana, Minnesota, Oklahoma, Colorado, and Utah. The Canadian provinces of Saskatchewan, Alberta, and Quebec were included in year four. Minimum inhibitory concentrations using CLSI approved interpretive criteria were determined for the following antimicrobials: ceftiofur, tilmicosin (breakpoint not yet established at publication), and spectinomycin. The other antimicrobials were evaluated using CLSI interpretive criteria adapted from human medicine, not approved in relation to bovine respiratory disease (BRD).
The cumulative results of this 4-year study are reported in Table 2. These MIC distributions represent the earliest published documentation of MIC distributions for major BRD pathogens using CLSI methods and microdilution methods consistent with current methods in diagnostic laboratories. The last 2 years of the studies utilized commercially available serial dilution microwell dilution plates.
Table 2. Cumulative results of MIC data for M. haemolytica, P. multocida, and H. somni from a 4-year study of isolates collected from cattle with BRD (Watts et al.)

1 These values for tilmicosin were determined using a breakpoint of ≤4 for susceptible. The interpretive criteria subsequently approved for tilmicosin by the CLSI are ≤8, 16, and ≥32 µg ml−1 for susceptible, intermediate, and resistant, respectively.
The finding of significant resistance to tilmicosin in M. haemolytica and P. multocida using the breakpoints in the study led to Watts et al. speculating on reasons for resistance to a new antimicrobial. Although the findings of tilmicosin resistance would have been dramatically decreased with the use of the subsequently approved breakpoints for M. haemolytica, their speculation of cross-resistance within the macrolide class of antimicrobials has since been substantiated (Van Donkersgoed et al., Reference Van Donkersgoed, Merrill and Hendrick2008; Kadlec et al., Reference Kadlec, Brenner Michael, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Watts and Schwarz2011; Desmolaize et al., Reference Desmolaize, Rose, Wilhelm, Warrass and Douthwaite2011b).
Welsh et al. monitored the trends in antimicrobial susceptibility on a total of 842 isolates over the span of 1994–2002 (Welsh et al., Reference Welsh, Dye, Payton and Confer2004). In total, they collected 390 M. haemolytica, 292 P. multocida, and 160 H. somni were isolated at the Oklahoma Animal Disease Diagnostic Laboratory from lungs from 6- to 18-month-old beef cattle succumbed to pneumonia. They reported variable susceptibility to tetracycline (range 23–74%) and relatively stable susceptibility to both ceftiofur (96–100%) and enrofloxacin (89–98%) for M. haemolytica isolates over that time period. Pasteurella multocida susceptibility profiles remained constant for both ceftiofur (96–100%) and enrofloxacin (96–100%) but a decline in florfenicol susceptibility from 100 to 86% was observed from when florfenicol first came to use in 1996 through 2002. As for H. somni, they found that susceptibility to commonly used antimicrobials remained consistently high (87–100%). A summary of their observed data is reported for each pathogen across all years in Table 3.
Table 3. Summary of susceptibly data of M. haemolytica, P. multocida, and H. somni isolated from lungs of BRD cases submitted to the Oklahoma Animal and Disease Diagnostic Laboratory between 1994 and 2002 (Welsh et al.)

* Indicates significant decline (P < 0.05).
CLSI approved interpretive criteria related to BRD and this pathogen for these antimicrobials.
Note: Tilmicosin has CLSI approved interpretive criteria only for M. haemolytica in BRD.
In a study on healthy feedlot cattle, Klima et al. isolated M. haemolytica from deep nasal swabs from a random selection of approximately 10% of animals from 30% of feedlot pens within two feedlots in Southern Alberta, Canada during 2007 and 2008 (Klima et al., Reference Klima, Alexander, Hendrick and Mcallister2014a). Swabs were taken from the same cattle on arrival and within 30 days of feedlot exit. Cattle were subjected to normal commercial practices; BRD cases were treated with either tulathromycin or ceftiofur. During the feeding period, in-feed ionophores (lasalocid or monensin) were administered, chlortetracycline was fed at levels for liver abscess control, cattle deemed to be at high risk of BRD were administered pulse (1–6 g hd−1 day−1) doses of chlortetracycline in the feed, and tylosin was fed in the diet for liver abscess control.
Susceptibility testing by disk diffusion was performed on 409 M. haemolytica isolates collected during the Klima et al. study. Antimicrobial resistance was found to consistently be low. All isolates were found to be susceptible to ceftiofur, enrofloxacin, and florfenicol. Resistance was most common for oxytetracycline (n = 16) and one isolate was found intermediate to tilmicosin. They reported no obvious trend in resistance among these isolates at entry or exit from either feedlot. Additionally, they found no trend correlating isolate expression of resistance and the antimicrobial that was administered to their host. The authors used CLSI Interpretive breakpoints for applications other than BRD for amoxicillin/clavulanic acid, sulfamethoxazole/trimethoprim, ampicillin, gentamicin, and oxytetracylcine.
McClary et al. identified cattle that had received tilmicosin for treatment of BRD based on records of 16 randomized clinical trials conducted in confined cattle feeding facilities in seven different states from 1996 to 2004 (McClary et al., Reference Mcclary, Loneragan, Shryock, Carter, Guthrie, Corbin and Mechor2011). The cattle in these studies had no previous history of antimicrobial usage (including metaphylaxis) and met the individual study definition for BRD. Isolates of M. haemolytica (n = 878) and P. multocida (n = 359) were collected via deep nasal swab prior to treatment with tilmicosin; calves yielding >1 pathogen were not included in the analysis. The minimum inhibitory concentration to tilmicosin was determined using CLSI standard methods. Only 0.8% of M. haemolytica and 6.9% of P. multocida pre-treatment isolates were phenotypically resistant to tilmicosin and most (73.7%) of the M. haemolytica isolates categorized as either not susceptible or resistant were obtained during two of the 16 clinical trials. Additionally, they found no associations between case outcome and the classification (susceptible or not susceptible) of the isolate for either pathogen. The authors reported the case outcome data in figures only, with approximate clinical success rates for M. haemolytica of 62% for susceptible isolates (n = 688), 47% for intermediate susceptibility isolates (n = 57), and 38% for resistant isolates (n = 6). The P value for a difference in clinical success rate between susceptible isolates as compared to intermediate or resistant isolates was 0.08. The authors acknowledge the lack of power in the study due to the low number of resistant isolates. While the paper did not find a significant difference in clinical outcome between susceptible and resistant isolates, the low power of the paper does not allow the conclusion that there is no difference.
In an attempt to find correlations of ante-mortem treatment regimens with post-mortem susceptibility patterns, Lamm et al. searched the Oklahoma Animal Disease Diagnostic Laboratory database for animals that died at the Oklahoma State University Willard Sparks Beef Research Center in 2007 (Lamm et al., Reference Lamm, Love, Krehbiel, Johnson and Step2012). Cattle were included in the analysis if they had died due to BRD, had chronicity of the lesions recorded, bacterial organisms were isolated, and susceptibility patterns of the bacterial organisms determined.
The authors (Lamm et al.) reported highly variable susceptibility patterns to tilmicosin between M. haemolytica, P. multocida, and H. somni with 88% (15/17), 42% (6/14), and 0% (0/12) of those isolates, respectively, either being intermediate or resistant. When grouping all three pathogens together, only a small portion of the isolates were susceptible to tetracycline (17/42, 40%) but a large number of isolates were susceptible to enrofloxacin (42/43, 98%), ceftiofur (38/42, 90%), and florfenicol (31/43, 72%); the authors indicated that ceftiofur and enrofloxacin were administered to these cattle prior to death. Overall, they reported that their study showed susceptibility patterns of the bacterial organisms isolated from the lungs of feedlot cattle that died with bronchopneumonia may not always relate to the antimicrobial treatments administered prior to death.
A large investigation was undertaken by Portis et al. using isolates from pre- and post-mortem bovine specimens to detect changes of in vitro susceptibility of BRD clinical isolates from 2000 to 2009 (Portis et al., Reference Portis, Lindeman, Johansen and Stoltman2012). Isolates of M. haemolytica (n = 2977), P. multocida (n = 3291), and H. somni (n = 1844) from 24 diagnostic labs across the USA (isolates from 47 states) and Canada (isolates from six provinces) were included in the study. All isolates were from diseased or deceased animals without any knowledge of either the age or the previous antimicrobial treatments administered to the animals. All susceptibility testing was carried out in two Pfizer (Zoetis) laboratories using CLSI standardized testing methods and approved interpretive criteria. The testing panel consisted of ceftiofur, enrofloxacin, florfenicol, penicillin, tetracycline, and tilmicosin. Danofloxacin and tulathromycin were added to the panel in 2004. Individual MIC distributions are reported for each drug and year for M. haemolytica (n = 2977), P. multocida (n = 3291) and H. somni (n = 1844) in this report. The reader is referred to the original publication for review of the extensive tables.
Among M. haemolytica isolates Portis et al. observed no apparent changes in MIC distributions for penicillin and ceftiofur. There was a decline in the percentage of isolates susceptible to danofloxacin over this study period. Declines in susceptibility, upward shifts in MIC distributions, and increases in MIC90 were reported for enrofloxacin, tilmicosin, and tulathromycin. A marked decrease in isolate susceptibility to tilmicosin from 89.4% in 2000 to 59.5% in 2009 was observed. Additionally, a decrease in florfenicol susceptibility by 10% over the 10-year period was observed. Approximately 50% of all M. haemolytica isolates demonstrated in vitro susceptibility to tetracycline over the 10-year study period.
In this same study, isolates of P. multocida showed little or no change in MIC distributions, MIC50 or MIC90 for both penicillin and ceftiofur. Approximately 88% of isolates were susceptible to danofloxacin in 2004 and little change was observed over the 5 years, and it was included in the panel. There was an emergence of a few resistant isolates and a small increase in MIC90 for enrofloxacin. Tilmicosin displayed a shift toward a higher MIC distribution and the percentage of susceptible isolates decreased to 59.7% by the end of the study. No discernable trends up or down were observed for florfenicol or tetracycline. While a susceptibility of at least 90% was observed to tulathromycin, the MIC90 increased 3-fold over the 6-year period in which it was included in the panel.
Trends in isolates of H. somni included more than 90% susceptibility to penicillin, a steady increase of MIC90 for danofloxacin, and a drop in susceptibility for enrofloxacin from 100% in 2000 to 86% in 2009. Additionally, while the MIC50 remained constant, a decrease in florfenicol susceptibility and a shift toward higher MIC distributions was reported. The MIC50 of tetracycline increased multiple dilutions and therefore a decrease in tetracycline susceptibility was observed. A shift in MIC distributions for both tilmicosin and tulathromycin was reported.
While commenting on trends across all three bacteria, Portis et al. reported that a majority of BRD isolates remained susceptible to danofloxacin, enrofloxacin, and florfenicol, but they did observe a slow increase in resistance to these drugs. All three pathogens remained 100% susceptible to ceftiofur throughout the 10-year study period. Although there was substantial variation in levels of susceptibility to tetracycline, MIC distributions did not appear to change over the study period in this report. This report once again demonstrated a cross-resistance effect in the macrolide class of antimicrobials. In 2004, one year prior to the marketing of tulathromycin, a 2–6% resistance rate was noted across the three major BRD bacteria pathogens.
Using isolates from two previous studies (Klima et al., Reference Klima, Alexander, Read, Gow, Booker, Hannon, Sheedy, Mcallister and Selinger2011; Alexander et al., Reference Alexander, Jin, Li, Cook and Mcallister2013b), Alexander et al., evaluated tulathromycin resistance in M. haemolytica isolated from cattle with a known history of antimicrobial use over a 3-year period from four commercial feedlots in Southern Alberta, Canada (Alexander et al., Reference Alexander, Cook, Klima, Topp and Mcallister2013a). This period started one year after approval of tulathromycin for treatment and prevention of BRD in Canada. Tulathromycin was approved in 2005 in the USA and in 2006 in Canada. As described above, deep nasal swabs were obtained from a random selection of 10% of animals from 30% of feedlot pens of healthy cattle upon arrival and again at ≥60 days on feed (DOF). All M. haemolytica isolates (n = 4548 isolates from 796 of the 5814 cattle sampled) were initially screened for tulathromycin susceptibility by plating onto brain heart infusion plates supplemented with 2 µg ml−1 tulathromycin. Isolates that grew on these plates (n = 5) were isolated from three calves and were subjected to PCR analysis for resistance genes. All five isolates contained aphA-1 and tet(H), conferring resistance to neomycin and oxytetracycline. Two isolates contained bla ROB−1 encoding resistance to ampicillin and penicillin. However, none of the isolates contained the macrolide resistance genes screened for in this study, namely, erm(A), erm(B), erm(F), erm(X), erm(42), msr(E)-mph(E). The three animals that harbored the resistant isolates received metaphylactic doses of tulathromycin and were isolated only at ≥60 DOF. However, in the opinion of those authors, the remarkably low rates of resistance in that study did not support any association between macrolide use and tulathromycin resistance. The current study showed that tulathromycin resistance from this population of feedlot cattle in Western Canada was exceptionally low even after this antimicrobial had been used in Canada for a period of 4 years. In their opinion (Klima et al. and Alexander et al.), there was no evidence that the commercial practices used by the feedlots in this study selected for tulathromycin-resistant M. haemolytica.
Isolates from the Kansas State Veterinary Diagnostic Laboratory were used by Lubbers and Hanzlicek in determining the prevalence of multidrug-resistant M. haemolytica from BRD cases over the time period of 2009–2011 (Lubbers and Hanzlicek, Reference Lubbers and Hanzlicek2013). Isolates (n = 389) included in the analysis had to be bovine lung specimen culture positive for M. haemolytica from clinical cases (research cases excluded) that had susceptibility test results available. Citing a low rate of resistance to ceftiofur (n = 2) the authors reported that no single isolate was resistant to all six drugs (ceftiofur, enrofloxacin, florfenicol, oxytetracycline, spectinomycin, and tilmicosin) analyzed in this study. Enrofloxacin and danofloxacin yielded equivalent results, so only enrofloxacin was included in the analysis. Tilmicosin and tulathromycin yielded the same susceptibility interpretation in 85.5% (153/179) of the isolates; of the remaining 26 isolates, 14 were interpretation discrepancies of intermediate and susceptible, which had no effect on the resistant finding. Seven isolates were resistant to tulathromycin and intermediate or susceptible to tilmicosin; five isolates had the opposite relationship, displaying resistance to tilmicosin but susceptible or intermediate to tulathromycin.
The prevalence of multidrug-resistance was alarming and increased over time in the Lubbers and Hanzlicek study. Using resistance to three or more antimicrobials as the definition for multidrug resistance, 42% (23/55), 46% (71/155), and 63% (113/179) of the isolates were classified as multidrug resistant in 2009, 2010, and 2011, respectively. By 2011, 25% of the isolates were resistant to four of six key antimicrobials (typically all but florfenicol and ceftiofur), and 35% were resistant to five of six key antimicrobials (all but ceftiofur). In this study, isolates found to be resistant to oxytetracycline were 3.52 times more likely (P = 0.04) to be resistant to one or more additional antimicrobials compared to non-oxytetracycline-resistant isolates. Isolates resistant to tilmicosin were 2.64 times more likely (P = 0.06) to be resistant to at least one other antimicrobial. There were no statistically significant coresistance patterns for enrofloxacin, florfenicol, or spectinomycin over the 3-year period.
Investigating the effects of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in M. haemolytica, Zaheer et al. conducted a study on 40 11-month-old beef steers in Alberta, Canada (Zaheer et al., Reference Zaheer, Cook, Klima, Stanford, Alexander, Topp, Read and Mcallister2013). All steers originated from the same ranch and had not received antimicrobials during their lifetime prior to inclusion in this study. Steers were housed in individual pens with ten replicates per treatment (controls no antimicrobials, tilmicosin single subcutaneous injection, tulathromycin single subcutaneous injection, and tylosin phosphate at 11 ppm in the feed). Deep nasopharyngeal swabs were collected on arrival, prior to administration of antimicrobials and then weekly thereafter for the 28-day study period. Their data demonstrated that 7 days post-injection, M. haemolytica was detected in only one steer treated from the tulathromycin treatment group and none of the steers treated with tilmicosin, whereas 60% of all steers were positive for this bacterium upon arrival (day 0). This is suggestive that M. haemolytica were not macrolide resistant upon arrival and likely a reflection that these calves had no previous exposure to antimicrobials. In contrast to injectable macrolides, tylosin, in the feed, had no effect on the number of M. haemolytica in steers receiving this antimicrobial as compared to cattle that received no antimicrobials. M. haemolytica isolated from animals belonging to control and all three macrolide treatment groups throughout the study were found to be susceptible to all tested macrolides, an indication that both therapeutic and subtherapeutic administration did not contribute to macrolide resistance in M. haemolytica during the study. Isolates in the present study were also susceptible to all other antimicrobials.
Genetic components of BRD resistance
While this review focuses on phenotypic characterization of antimicrobial resistance in BRD isolates, a selected review of genetic components helps in understanding the epidemiology of resistance spread. The understanding of macrolide resistance genetics in M. haemolytica and P. multocida were greatly advanced in a flurry of publications starting in 2011.
In 2011, Desmolaize et al. reported on a novel rRNA methylase gene, erm(42), which had diverged from all previously characterized erm genes and therefore was previously undetected with PCR assays. Genetic analysis suggested acquisition from other members of the Pasteurellaceae and recent gene transfer among M. haemolytica and P. multocida.
Soon thereafter, Kadlec et al. published a whole-genome sequencing evaluation of a 2005 Nebraska P. multocida isolate which was resistant to tulathromycin (Kadlec et al., Reference Kadlec, Brenner Michael, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Watts and Schwarz2011). Prior to this analysis, multiple techniques had failed to determine the mechanism of resistance. Three new resistance genes were identified in this study: the rRNA methylase gene erm(42), the macrolide transporter gene msr(E), and the macrolide phosphotransferase gene mph(E).
Desmolaize et al. demonstrated that these three genes are arranged in three distinct classes of resistance in M. haemolytica and P. multocida (Desmolaize et al., Reference Desmolaize, Rose, Wilhelm, Warrass and Douthwaite2011b). The erm(42) gene alone confers resistance to macrolides, lincosamides, and streptogramin group B antimicrobials. The second class consists of a tandem arrangement of msr(E) and mph(E). The third class contains all three resistance genes and displayed high resistance to all macrolides tested in this study (tulathromycin, gamithromycin, and tilmicosin).
Michael et al. characterized these genes within an integrative conjugative element (ICE) in an isolate of P. multocida that contained 12 antimicrobial resistance genes (Michael et al., Reference Michael, Kadlec, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Murray, Watts and Schwarz2012c). This ICE, designated ICEPmu1, contains the resistance genes aadA25 (streptomycin/spectinomycin), strA and strB (streptomycin), aadB (gentamicin), aphA1 (kanamycin/neomycin), tetR-tet(H) (tetracycline), floR (chloramphenicol/florfenicol), sul2 (sulfonamides), erm(42) (tilmicosin/clindamycin), and msr(E)-mph(E) (tilmicosin/tulathromycin). A complete bla OXA-2 (penicillins, first- and second-generation cephalosporins) was also identified but appeared to be non-functional in this isolate. The authors pointed to the sequences obtained as evidence suggesting that plasmids, gene cassettes, and insertion sequences have contributed to the development of the two resistance gene regions in this ICE. The presence of these 12 resistance genes in a single ICE demonstrates the potential for transfer of multiple antimicrobial resistance genes in one horizontal gene transfer event.
Michael et al. (Reference Michael, Eidam, Kadlec, Meyer, Sweeney, Murray, Watts and Schwarz2012a) evaluated the same three genes in a recently published investigation of MICs of gamithromycin and tildipirosin using field isolates of M. haemolytica (n = 29) and P. multocida (n = 40) collected between 1999 and 2007 (Van Donkersgoed et al., Reference Van Donkersgoed, Merrill and Hendrick2008). These isolates had previously been shown to carry the genes erm(42) and/or msr(E)-mph(E) (Kadlec et al., Reference Kadlec, Brenner Michael, Sweeney, Brzuszkiewicz, Liesegang, Daniel, Watts and Schwarz2011). If all three genes were present, the P. multocida isolates showed MICs of 16–64 µg ml−1 for gamithromycin and 16–32 µg ml−1 for tildipirosin, whereas similar MICs of 32–64 µg ml−1 for both macrolides were seen for the corresponding M. haemolytica isolates.
Ten P. multocida isolates that carried only erm(42) exhibited low MICs of 2–4 µg ml−1 for gamithromycin, but had higher MICs of 16–32 µg ml−1 for tildipirosin (Van Donkersgoed et al., Reference Van Donkersgoed, Merrill and Hendrick2008). The single M. haemolytica that harbored only erm(42) showed MICs of 4 and 32 µg ml−1 for gamithromycin and tildipirosin, respectively. The two P. multocida isolates that carried only the msr(E)-mph(E) operon exhibited a high MIC of 32 µg ml−1 for gamithromycin and a low MIC of 2 µg ml−1 for tildipirosin. The genes erm(42) and msr(E)-mph(E) are part of the resistance gene regions of the recently identified integrative and conjugative element ICEPmu1, which has been shown to move across genus boundaries and express its resistance genes in different hosts, such as P. multocida and M. haemolytica. Pronounced increases in the gamithromycin MICs were seen in the presence of msr(E)-mph(E), whereas distinct increases in the tildipirosin MICs were detected in the presence of erm(42). This report, yet again, demonstrates cross-resistance exists in the macrolide class of antimicrobials and is further evidence of BRD bacterial pathogens, due to cross-resistance, exhibiting resistance to antimicrobials prior to them being available for use in the industry.
The most recent evaluation of ICE-mediated antimicrobial resistance in BRD pathogens was published by Klima et al., in Reference Klima, Zaheer, Cook, Booker, Hendrick, Alexander and Mcallister2014b, and documents the presence of ICE in M. haemolytica and H. somni isolated from US feedlots (Klima et al., Reference Klima, Zaheer, Cook, Booker, Hendrick, Alexander and Mcallister2014b). Their investigation centered on 42 BRD mortalities in Alberta, Canada, 6 mortalities in Texas, and 20 mortalities in Nebraska. Isolates of M. haemolytica (55), P. multocida (8), and H. somni (10) were collected from lungs and nasal swabs. Forty-five percent (33/73) of all bacterial isolates displayed resistance to three or more antimicrobials. Thirty-three percent (18/55) of M. haemolytica isolates, 37.5% (3/8) of P. multocida isolates, and 30% (3/10) of H. somni were resistant to more than seven antimicrobial classes, including aminoglycosides, penicillins, fluoroquinolones, lincosamides, macrolides, pleuromutilins, and tetracyclines. All of the multidrug-resistant isolates originated from the Texas and Nebraska feedlots. These isolates varied between 60 and 100% similarity based on PFGE analysis, which the authors point out indicates that resistance was not spread strictly by clonal dissemination.
Eighteen of the M. haemolytica isolates, three of the P. multocida isolates, and three of the H. somni isolates contained ICE that conferred resistance for up to seven antimicrobial classes. These ICEs were demonstrated to be transferred by conjugation from P. multocida to Escherichia coli and from M. haemolytica and H. somni to P. multocida.
Discussion
Antimicrobial resistance among the bacterial pathogens commonly associated with BRD is well documented in the scientific literature and the occurrence of resistant isolates appears to be steadily increasing in reports with the ability to analyze temporal trends. Foreign researchers, although utilizing different industry wide production practices and drug prescription practices, cite the same concerns over increases and patterns in resistance in BRD pathogens, especially in concern to the fluoroquinolone class of antimicrobials (Shin et al., Reference Shin, Kang, Nabin, Kang and Yoo2005; Kaspar, Reference Kaspar2006; Katsuda et al., Reference Katsuda, Kohmoto and Mikami2013; Pardon et al., Reference Pardon, Hostens, Duchateau, Dewulf, De Bleecker and Deprez2013). Japanese researchers have observed resistance rates in fluoroquinolone antimicrobials to have increased up to 4-fold from the period of 2006–2009 (Katsuda et al., Reference Katsuda, Kohmoto and Mikami2013).
In the USA, publications from diagnostic laboratory submissions (Watts et al., Reference Watts, Yancey, Salmon and Case1994; Chang and Carter, Reference Chang and Carter1976; Welsh et al., Reference Welsh, Dye, Payton and Confer2004; Lamm et al., Reference Lamm, Love, Krehbiel, Johnson and Step2012; Portis et al., Reference Portis, Lindeman, Johansen and Stoltman2012; Lubbers and Hanzlicek, Reference Lubbers and Hanzlicek2013), utilizing specimens from animals that likely died from un-responsive BRD, report consistently higher rates of resistance as compared to those that report resistance rates of isolates from pre-treatment (McClary et al., Reference Mcclary, Loneragan, Shryock, Carter, Guthrie, Corbin and Mechor2011) or healthy cattle (Klima et al., Reference Klima, Alexander, Read, Gow, Booker, Hannon, Sheedy, Mcallister and Selinger2011; Alexander et al., Reference Alexander, Cook, Klima, Topp and Mcallister2013a; Zaheer et al., Reference Zaheer, Cook, Klima, Stanford, Alexander, Topp, Read and Mcallister2013). These contrasts fuel the controversy over whether diagnostic laboratory data are a valid means of monitoring antimicrobial resistance trends due to the biased nature of diagnostic laboratory submissions. Unless the bias toward challenging cases with high morbidity and/or case fatality has dramatically changed, then trends in diagnostic laboratory data suggest that the pathogens involved in non-responsive and high-morbidity BRD challenges are displaying an increasing incidence of resistance to many of the antimicrobials used in control and therapy of this disease.
Studies which point to limited or non-existent resistance prevalence in small, confined populations exposed to antimicrobials do little to inform the discussion concerning the potential for antimicrobial use to enable spread of resistance through plasmids, ICEs, or clonal spread. Exposure of a population to antimicrobials in the absence of an existing resistant pathogen population relies on de-novo mutations for resistance and then selection for the isolates with these genetic characteristics or phenotypic expression of quiescent genes already present.
The fact that many of the isolates discussed in this review contain multiple resistance genes grouped together on some type of transferrable element argues against the concept of de-novo mutation in individual animals or small populations, and instead supports the hypothesis that much of our challenge in resistant BRD pathogens stems from selection for existing multi-drug resistant mobile genetic elements or the resistant pathogens which bear them.
The characterization of these pathogen isolates related to the combination of antimicrobial resistance, virulence, and dominance in colonization remains to be established. It is reasonable to hypothesize that pathogens which contain this ‘trifecta’ would be an enormous challenge within existing beef production systems. The data reviewed in this paper suggest that extensive investigations are needed on where selection pressure is being applied for organisms with resistance phenotypes encoded for by mechanisms such as an ICE. The design of these investigations will be informed by findings of recent and ongoing studies evaluating the epidemiology of BRD pathogens, including investigations of M. haemolytica population distributions within individual animals and across cattle populations.
Conclusion
Due to the disparate data from the few studies that investigate susceptibility testing of BRD pathogens, it is difficult, if not impossible to perform a quantitative assessment or meta-analysis of the studies presented in this review. Even though diagnostic lab data is heavily criticized by some, it is difficult to argue against the appearance of a trend in these data. Following these reports chronologically shows an apparent trend of a decrease in susceptibility of the three major BRD pathogens to most of the antimicrobials commonly used for treatment and control of BRD. It is possible that the cattle in diagnostic studies represent a specific niche in the population that has a preponderance for harboring resistant pathogens and this needs further investigation. However, the fact that there are isolates within the population that phenotypically express high levels of pan-resistance to the antimicrobials used both in treatment and control might suggest that it is unwise to use the same antimicrobials for both treatment and control.
An additional point to consider is the previous lack in technology allowing for the detection of the presence of genetic resistance. Most studies in this review focus on culture and sensitivity which are essentially testing phenotypic expression without any knowledge of the genotype. Perhaps, the ‘healthy cattle’ that display lower levels of resistance have not been exposed to the selection pressure through disease and subsequent treatment, to necessitate the phenotypic expression of resistance genes currently present in their genome as ICE elements or other stand-alone resistance mechanisms? Owing to the relatively recent advances in technology allowing rapid and economically efficient genetic sequencing methods, our research group is currently investigating this in further detail.
This review highlights the continued importance of judicious use of antimicrobials in all sectors veterinary medicine to ensure antimicrobials will remain effective into the future.






