Introduction
Agricultural productivity is hampered by abiotic and biotic stresses throughout the world (Rao et al., Reference Rao, Nair and Nayyar2016; Gull et al., Reference Gull, Lone, Wani and Alexandre2019). However, development and dissemination of plant species across different environmental zones has been affected more by abiotic stresses compared to biotic ones (Chaves et al., Reference Chaves, Maroco and Pereira2003; Gong et al., Reference Gong, Rao, Yu, Stoytcheva and Zlatev2013). Among food crops, the role played by legumes in meeting the requirements of human nutrition and cropping systems is currently being considered more important (Arnoldi et al., Reference Arnoldi, Zanoni, Lammi and Boschin2014; Araujo et al., Reference Araujo, Beebe, Crespi, Delbreli, Gonzaliz, Gruber, Lejeune-Henaut, Link, Monteros, Prats, Rao, Vadez and Patto2015). Pulses are salt-sensitive crops because the high concentration of Na+ and Cl− ions in the root zone of water-scarce areas limits their geographical and agro-edaphic adaptations in arid and semiarid climates where evapo-transpiration exceeds the precipitation (Hopmans et al., Reference Hopmans, Qureshi, Kisekka, Munns, Grattan, Rengasamy, Ben-Gal, Assouline, Javaux, Minhas, Raats, Skaggs, Wang, De Jong van Lier, Jiao, Lavado, Lazarovitch, Lil and Taleisni2021). During irrigation, salts tend to accumulate in the soil surface of irrigated landscapes due to the rise in saline groundwater (Podmore, Reference Podmore2009). This may lead to high soil salinity levels which are negatively affecting the humans and the natural resources (Slinger and Tenison, Reference Slinger and Tenison2007). Inefficient irrigation and drainage systems are a potential source of excess leakage which increases the risk of salinity and water-logging stresses in irrigated areas (Podmore, Reference Podmore2009). In addition, pulses’ cultivation along the coastal areas is also adversely affected by their poor tolerance to salt stresses. Under salinity, plants are usually affected physiologically in terms of osmotic stress and ion toxicities, whereas legumes receive a third set back of reduced nodulation by Rhizobia as a result of salt stress (Rao et al., Reference Rao, Nair and Nayyar2016; Hopmans et al., Reference Hopmans, Qureshi, Kisekka, Munns, Grattan, Rengasamy, Ben-Gal, Assouline, Javaux, Minhas, Raats, Skaggs, Wang, De Jong van Lier, Jiao, Lavado, Lazarovitch, Lil and Taleisni2021). The crop response to salt stresses also varies with the plant species and their genotypes, stages of growth, cultural practices and soil fertility (Podmore, Reference Podmore2009).
Among food legumes, the genus Vigna encompassing cowpea, green gram, black gram, adzuki bean, etc. is an important taxonomic group for meeting the human nutritional needs (Arnoldi et al., Reference Arnoldi, Zanoni, Lammi and Boschin2014). This genus consists of more than 100 wild species found in tropical and subtropical areas of the world (Takahashi et al., Reference Takahashi, Somta, Muto, Iseki, Naito, Pandiyan, Senthil and Tomooka2016). The abiotic stress-affected habitats of some wild Vigna species such as saline lands on sandy beaches (V. marina), acidic lands (V. vexillata) and flood prone lands beside rivers (V. luteola) are home to important genetic resources required during breeding for abiotic stress tolerance in pulses (Tomooka et al., Reference Tomooka, Naito, Kaga, Sakai, Isemura, Ogiso-Tanaka, Iseki and Takahashi2014). Recently, a study was conducted to understand the comparative salt tolerance of seven cultivated Vigna species (moth bean, adzuki bean, black gram, mung bean, creole bean, rice bean and cowpea) along with 23 wild Vigna species (Yoshida et al., Reference Yoshida, Tomooka, Khaing, Shantha, Naito, Matsuda and Ehara2020). These findings revealed all six Asian Vigna crop species (moth bean, adzuki bean, black gram, mung bean, creole bean and rice bean) as susceptible except for African Vigna crop species, cowpea which was found moderately resistant under 200 mM NaCl stress spanning two weeks. On the contrary, among 23 wild Vigna species, five species viz. V. riukiuensis, V. trilobata, V. vexillata, V. luteola and V. marina showed considerably higher levels of tolerance.
Beachpea [Vigna marina (Burm.f.) Merr.], a wild leguminous plant which belongs to the family Fabaceae is generally found to occur on the tropical and subtropical beaches of the world (Verdcourt, Reference Verdcourt1970; USDA, 2011). Since V. marina thrives well along the sea-shore sandy beaches, it could be a potent donor of genes conferring tolerance to both drought and salinity stresses. Based on its habitats along sea coasts and higher salt tolerance, V. marina is considered as a halophytic (salt loving) species. Seeds of V. marina remained viable even after being submerged in seawater for many years (Lawn and Cottrell, Reference Lawn and Cottrell1988), whereas young plants could survive more than 1 month when grown in a NaCl solution of 400 mM concentration (Tomooka et al., Reference Tomooka, Kaga, Isemura, Vaughan and Kole2011).
Chankaew et al. (Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014) reported quantitative trait loci (QTLs) for salt tolerance in V. marina by studying F2 population developed from V. luteola × V. marina subsp. oblonga cross. In their study, the salt tolerance in V. marina subsp. oblonga was found to be controlled by a single major QTL or by a set of few genes. Therefore, this species could be utilized as a potential salt-tolerant donor in future pulses breeding programmes through inter-specific hybridization. Although India is the largest producer and consumer of pulses, yet there is urgent need to develop high yielding as well as potentially salt-tolerant varieties suitable to its inland saline areas as well as long sea-shore line to cater to the nutritional needs of growing population. It is also perceived that the effects of climate change and rising salinity are likely to affect coastal as well as island population more than inland eco-systems. Keeping these considerations in view, the current study was undertaken with the following objectives: (1) collection of V. marina germplasm from different locations in Andaman and Nicobar Islands and their morphological characterization and (2) molecular characterization of collected germplasm using simple sequence repeat (SSR) markers associated with salinity tolerance.
Materials and methods
Experimental material
The experimental material consisted of 20 Vigna genotypes including 15 accessions of V. marina collected from Andaman and Nicobar Islands during August to December 2018 (Table 1 and Fig. 1) and five check varieties of Vigna-cultivated species [three black gram (IPU-02-43, LBG 752 and VBN 7); two green gram (LGG 544 and TARM 1)]. Since the targeted plant species were at the flowering stage during collection time, both the leaf samples (for DNA isolation) and stem cuttings (for field establishment at ICAR-Central Island Agricultural Research Institute, Port Blair) of genotypes were collected from each site. Although flowering occurs throughout the year in this species, pod matures only in the spring/summer season. During field surveys, passport information with respect to sites including geographical coordinates and soil samples were collected. By subsequent exploration trips during March to May in the year 2019, the seeds of another two genotypes (one each from Car Nicobar and Campbell bay of Nicobar district) were also collected.

Fig. 1. (A) Location map of Andaman and Nicobar Islands along with collection sites of V. marina genotypes. (B) Individual collection sites: (a) South Andaman (Chidyatapu, Manjery and Wandoor in Port Blair), (b) North Andaman (Lamia bay in Diglipur), (c) Havelock Island (Kalapathar and Radhanagar), (d) Neil Island (Bharatpur, Lakshmanpur and Pearl park), (e) Baratang Island (Baludera at Nayagarh) and (f) Little Andaman (Harmindar bay beach at Hut bay) (Source: www.mapsofindia.com and website of APWD, Andaman and Nicobar Administration).
Table 1. Passport information of V. marina genotypes collected from Andaman and Nicobar Islands

SA, South Andaman; NMA, North and Middle Andaman; NA, not available.
Measurement of pH and electrical conductivity (EC) of soil samples
The pH of soil was measured using a Digital pH meter which consists of electrode and digital meter. Glass electrode had a silver-based electrical wire suspended in a solution of potassium chloride, contained inside a thin bulb (or membrane) made from a special glass containing metal salts (typically, compounds of sodium and calcium). The other reference electrode had a potassium chloride wire suspended in a solution of potassium chloride. The procedure for measuring pH included following steps: (1) 20 g of soil was taken in a 100 ml beaker; (2) added 40 ml of pure water into it using a pipette or suitable volumetric container and stirred with a glass rod and let the sample to stand for 30 min; (3) standardized the pH meter; (4) stirred the sample again immediately before measuring the soil pH and (5) the electrodes were positioned in the solution just above the sand layer and pH was recorded to the nearest 0.1 pH unit. Similarly, the procedure for measuring EC included the following steps: (1) 20 g of soil was taken in a 100 ml beaker; (2) added 40 ml of pure water into it using a pipette or suitable volumetric container and stirred with a glass rod and let the sample to stand for 30 min; (3) standardized the conductivity meter; (4) stirred the sample again immediately before measuring EC of the soil and (5) the electrodes were placed in the solution just above the sand layer and EC was recorded in dS/m unit.
Agro-morphological characterization
Observations on 10 agro-morphological traits mainly seed characters were recorded on five V. marina genotypes collected from Andaman and Nicobar Islands. These traits included six quantitative traits viz. pod length (cm), number of seeds per cluster, seed length (mm), seed width (mm), length:breadth ratio (L:B ratio) and 100-seed weight (g) along with four qualitative traits viz. seed colour, lustre on seed, seed shape and hilum.
DNA extraction and selection of primers
The total genomic DNA of V. marina genotype was isolated from young leaf samples collected from each collection site (~15 mg each) using the cetyltrimethylammonium bromide (CTAB) method as described by Saghai-Maroof et al. (Reference Saghai-Maroof, Soliman, Jorgensen and Allard1984) with minor modifications. Three grams of freshly weighed leaf sample were taken in mortar and pestle and crushed into fine powder using liquid nitrogen. Ten millilitres of CTAB buffer was added to the fine powder which was already pre-heated in a water bath at 60°C for 10 min. The samples were then subjected to incubation at 65–70°C in the water bath and mixed by inversion every 10 min. After incubation, the samples were centrifuged at 10,000 rpm for 15 min. The supernatants were transferred into new centrifuge tubes. Equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added and mixed by inversion for 2 min. Then the samples were centrifuged for 10 min at 10,000 rpm. The upper aqueous phase was transferred into new tubes. Equal volume of chilled 70% ethanol was added and incubated at −20°C for 1 h. The samples were then centrifuged at 10,000 rpm for 10 min. The pellet obtained was washed with 70% ethanol, air dried and then dissolved in nuclease-free water. Seven SSR primers associated with QTLs related to salt tolerance as identified by Chankaew et al. (Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014) were used for the molecular diversity analysis of 20 Vigna genotypes/varieties. The details of these seven primers such as forward and reverse primer sequences, expected product size, name of QTL where primer is located and the trait governed by the QTL are listed in Table 2.
Table 2. Details of the SSR markers used for molecular characterization of 20 Vigna genotypes/varieties

PRS, plant recovery score; PSS, percentage of surviving seedlings; LWS, leaf wilt score at vegetative stage; NAm, not available due to monomorphic nature of marker; NA, not available due to no amplification of marker.
Forward and reverse primer sequences, expected product size (EPS), name of QTL where primer located and the trait governed by the QTL, annealing temperature (AT), number alleles detected (NAD), allele size (AS), PIC value and expected H e.
Polymerase chain reaction (PCR) amplification and gel electrophoresis
For optimization of component concentrations, PCR amplification was carried out with 30 ng of genomic DNA, 2.5 mM MgCl2, 1 U Taq DNA polymerase, 1× PCR buffer without MgCl2, 0.6 μM of each of primers and 200 mM of dNTP mix. The volume was made up to 25 μl with sterile distilled water. PCR tubes containing above components were capped and centrifuged at 10,000 rpm for 2 min to allow proper settling of reaction mixture. Thermo-cycling was carried out in a C1000 Thermal cycler (Bio-Rad). First, the gel casting tray was cleaned and sealed with cello-tape. Then agarose was weighed for 0.8% (0.4 g in 150 ml according to the sample) and dissolved in 1× TAE buffer. Agarose solution was heated until it would become transparent and the agarose solution was allowed to cool. Three microlitres of ethidium bromide was added and mixed properly. The agarose solution was poured into a gel plate with gel comb placed in it and allowed to solidify. After solidification, the gel comb was removed and the solidified gel placed in gel electrophoresis unit with 1× TAE running buffer. Four microlitres of DNA and 2 μl of loading dye were mixed and added to every well. The samples were run at 70 V for 1 h and the gel was visualized under gel doc machine, Gel Doc XR+ Imager (Bio-Rad).
Data analysis
For marker data analysis, the amplification products were scored as present (1) or absent (0). The polymorphism information content (PIC) and heterozygosity (H e) value of SSR markers were calculated using online software available at: http://www.liv.ac.uk/~kempsj/pic.html. The un-weighted paired group method with arithmetic means (UPGMA)-based Jaccard's similarity coefficient was used for cluster analysis and dendrogram was generated. The principal co-ordinate analysis (PCoA) was also carried out by using molecular data. Two-dimensional scatter plot was generated to represent the accessions with their genetic variability. PAST3 software (Hammer et al., Reference Hammer, Harper and Ryan2001) was used for the multivariate analysis of molecular data viz., cluster analysis and PCoA.
Results
Field survey and collection
Field surveys were conducted across the sea-shore beaches of Andaman and Nicobar Islands for the occurrence of V. marina during August to December 2018. Our collection drive covered many sea-shore beaches located in South Andaman district such as Baratang (Baludera beach), Chidya Tapu, Wandoor, Manjery, Havelock Island (Kalapather and Radhanagar), Neil Island (Bharatpur, Lakshmanpur and Pearl Park) and Little Andaman (Harminder bay). However, we could not find the occurrence of V. marina in few areas of South Andaman [Corbyn's cove, Little Andaman (Hut bay, Ramakrishnapur and Netaji nagar)], North and Middle Andaman [Betapur, Diglipur (Arial Bay and Ramnagar) and Mayabunder (Karmatang)].
Habitat
Most of the germplasm collection sites of the surveyed sea shore areas were characterized by sandy and saline soils. The occurrence of V. marina under natural conditions across the Andaman and Nicobar Islands is presented in online Supplementary Fig. S1. Generally, it was found growing well as creeper vine in our survey.
Soil analysis
The pH and EC of 10 collected soil samples from different locations of Andaman and Nicobar Islands are presented in Table 1. The pH values ranged from 7.65 in Wandoor beach (South Andaman) to 9.21 in Lamia bay of Diglipur (North and Middle Andaman), whereas, the EC of soil ranged from 9.92 dS/m in Kalapather beach-2 of Havelock Island (South Andaman) to 13.30 dS/m in Lakshmanpur beach of Neil Island (South Andaman).
Agro-morphological characterization
Five V. marina genotypes collected from Andaman and Nicobar Islands were subjected to morphological/seed characterization (online Supplementary Table S1). Field view of different stages of V. marina under natural conditions in Andaman and Nicobar Islands is presented in online Supplementary Fig. S2. Co-efficient of variation among genotypes varied from 8.82 (number of seeds per pod) to 3.94 (seed width). The values of traits such as number of seeds per pod, pod length (cm), seed length (mm), length:breadth ratio (L:B ratio) and 100-seed weight (g) ranged from 4.0 to 5.0, 4.8 to 5.7, 5.8 to 7.0, 1.2 to 1.4 and 6.9 to 7.9, respectively, with the corresponding mean values of 4.36, 5.26, 6.42, 1.32 and 7.48. Variability for seed traits of five V. marina accessions under study is shown in online Supplementary Fig. S3. All qualitative traits showed no variation except for seed shape. Beachpea genotypes under study showed mixed seed colour, dull luster on seed with non-concave hilum. In case of seed shape, all genotypes were oval shaped except for Car Nicobar genotype (drum shaped).
Molecular characterization
Molecular characterization of 20 genotypes of Vigna species (V. marina – 15; V. mungo – 3 and V. radiata – 2) was performed by using seven SSR markers associated with salt tolerance as identified by Chankaew et al. (Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014). However, only four SSR primers were found to exhibit amplification in the germplasm studied. Among these four SSR primers, three showed higher polymorphism.
The details of SSR primers, number of alleles deducted, PIC values and expected H e are presented in Table 2. The SSR marker profiles of Beachpea genotypes are presented in Fig. 2. The number of alleles detected per primer ranged from 1 to 3 whereas, the size of alleles ranged from 100 to 325 bp. The PIC values ranged from 0.305 to 0.537 with an average value of 0.422. The H e values ranged from 0.375 to 0.612. Two SSR loci revealed PIC and H e values higher than 0.40 and 0.50, respectively and the pertinent SSR markers are CEDG087 and CEDG007.

Fig. 2. SSR profiles of 20 Vigna genotypes/varieties generated by selected polymorphic primers: (a) CEDG007 and (b) CEDG087 using individual DNA in which M denotes marker lane and 1–20 denotes genotypes lane.
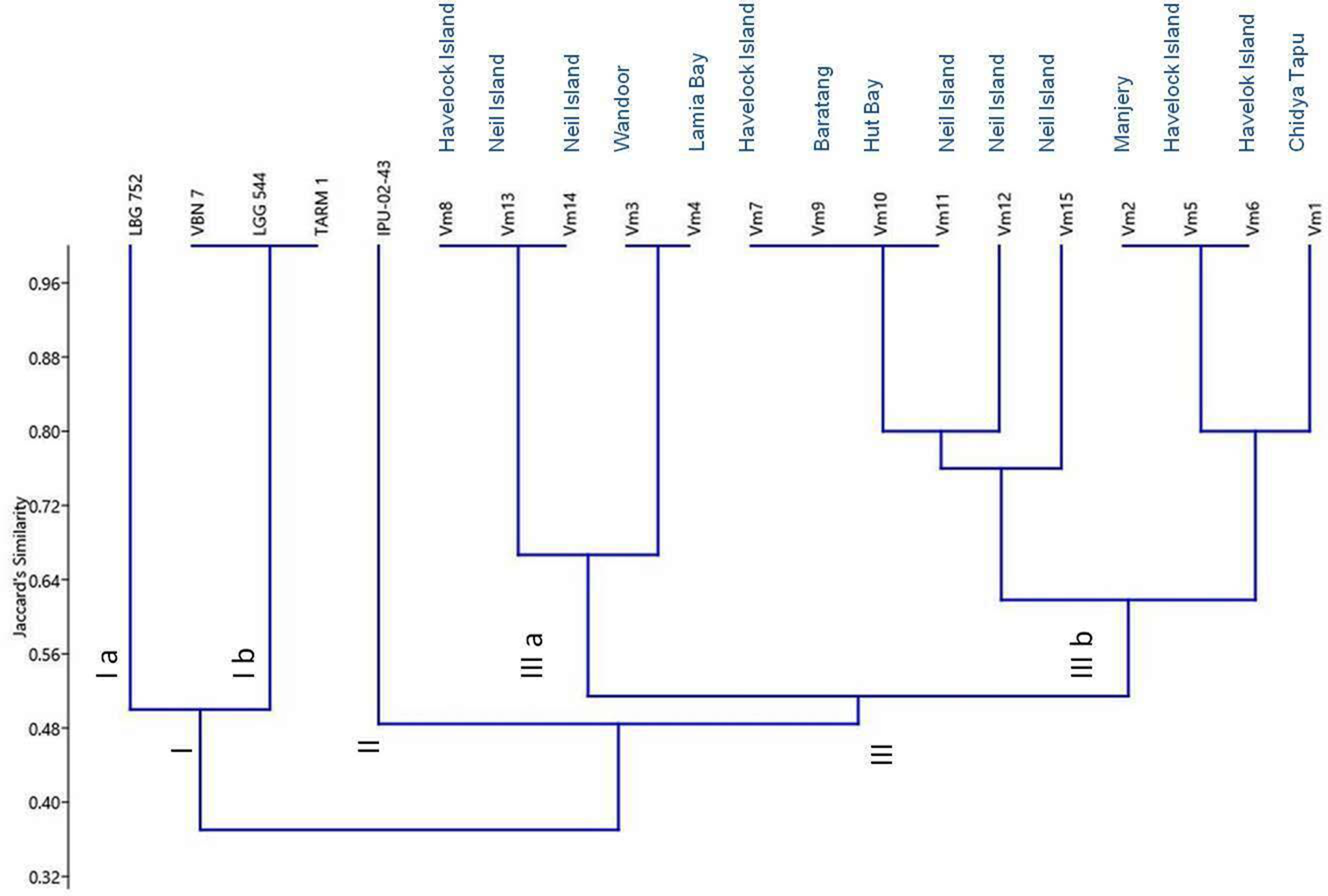
Cluster analysis based on Jaccard's similarity coefficient using the UPGMA method clearly distinguished 20 Vigna genotypes into three major clusters viz. I, II and III at a similarity coefficient value of 0.48 (Fig. 3). This cluster analysis grouped the check varieties (green gram and black gram lines) in clusters I and II. On the contrary, all V. marina genotypes collected from Andaman and Nicobar Islands were grouped in cluster III. Cluster I was formed with four check varieties whereas, only one variety (IPU-02-43) was included in cluster II. Cluster I was divided at a similarity coefficient value of 0.50 into two sub-clusters viz. sub-cluster Ia consisting of one genotype LBG752 and sub-cluster Ib having three varieties (VBN 7, LGG 544 and TARM 1). Similarly, cluster III of V. marina was also divided at a similarity coefficient value of 0.53 into two sub-clusters viz. IIIa consisting of five accessions (Vm8, Vm13, Vm14, Vm3 and Vm4) and IIIb having 10 accessions (Vm7, Vm9, Vm10, Vm11, Vm12, Vm15, Vm2, Vm5, Vm6 and Vm1). Overall, cluster III showed 100% similarity among genotypes of V. marina collected from Andaman and Nicobar Islands at various similarity coefficient values [within sub-cluster IIIa as three (Vm8, Vm13 and Vm14) and two (Vm3 and Vm4) at 0.66; within sub-cluster IIIb as four (Vm7, Vm9, Vm10 and Vm11) and one (Vm12) at 0.80; three (Vm2, Vm5 and Vm6) and one (Vm1) at 0.80].

Fig. 3. Dendrogram of 20 Vigna genotypes/varieties based on the UPGMA method of cluster analysis by using Jaccard's similarity coefficient obtained from four SSR marker data.
PCoA was also performed to determine the genetic relationships among 20 Vigna genotypes. Two-dimensional scatter-plot of PCoA ordination for Vigna genotypes is presented in Fig. 4. The value of variance accounted by PCoA from first three most informative principal coordinates was 85.9%’ which revealed the amount of genetic variation present in SSR molecular data derived from 20 Vigna genotypes under study. First, second and third principal coordinates explained the genetic variation among genotypes as 37.2, 27.5 and 21.2%, respectively, with a cumulative value of 85.93% as shown in online Supplementary Table S2.

Fig. 4. Two-dimensional scatter plot of 20 Vigna genotypes/varieties using principal coordination analysis of molecular data obtained from four SSR markers which confirmed the pattern obtained in the cluster analysis.
Discussion
The cultivated Vigna species are mostly grown under marginal/degraded lands, including salt-affected soils by resource-poor farmers in different regions of the world. This situation is further evidenced from green gram and black gram cultivation in seawater-intruded areas of Ayeyawaddy and Bago in Myanmar (Win et al., Reference Win, Oo, Hirasawa, Ookawa and Yutaka2011), cowpea cultivation in the high-salinity areas of Brazil (Lobato et al., Reference Lobato, Santos Filho, Costa, Goncalva-Vidigal, Moraes, Oliveira Neto, Rodriges, Cruz, Ferreira, Pita and Barreto2009), mung bean in the salinity-affected lands of Pakistan (Rao et al., Reference Rao, Nair and Nayyar2016) and desert areas of USA (Wilson et al., Reference Wilson, Liu, Lesch and Suarez2009). Although productivity of these crops is low under such unfavourable conditions, breeding for salt tolerance is a good option for improving Vigna crops in several countries. Beachpea (V. marina) offers to be a genetic donor of salt-tolerance genes for several cultivated Vigna species.
Our study reports the genetic variation for salt tolerance among V. marina accessions from different islands of Andaman and Nicobar which are vulnerable to the effects of sea level rise and climate change as evidenced by the occurrence of tsunami, 2004. Field surveys conducted for the occurrence of V. marina also revealed agro-morphological variation among the collections which were found growing under natural habitats across the islands. Among 15 leaf samples and stem cuttings of V. marina collected, we found 14 samples from the South Andaman district and one from (Lamia bay in Diglipur) North and Middle Andaman district. In a subsequent survey, it was also found in Nicobar Islands (Car Nicobar and Campbell bay).
Generally, the pH of 8.5–9.1 is considered as non-stress, 9.2–9.8 as moderate stress and >9.8 as high alkali stress (Hopmans et al., Reference Hopmans, Qureshi, Kisekka, Munns, Grattan, Rengasamy, Ben-Gal, Assouline, Javaux, Minhas, Raats, Skaggs, Wang, De Jong van Lier, Jiao, Lavado, Lazarovitch, Lil and Taleisni2021). Extremely high salt stress conditions damage the plants, but moderate to low salt stress affects plant growth rate and manifests symptoms that could be associated with morphological, physiological or biochemical changes (Hasegawa, Reference Hasegawa2013). Although upper values of pH range (7.65–9.21) indicated moderate stress in the current study, the higher EC values (9.92–13.30 dS/m) of surveyed habitats confirmed the robust salt tolerance of V. marina (Keating and Lawn, Reference Keating and Lawn1985) and its adaptation to soil-specific electrical conductances of as high as 13 dS/m (Lawn and Cottrell, Reference Lawn and Cottrell2016).
Salt tolerance is governed by several minor QTLs in Medicago truncatula (Arraouadi et al., Reference Arraouadi, Badri, Abdelly, Huguet and Aoudi2012), single major QTL in soybean [Glycine max (L.) Merr.] (Lee et al., Reference Lee, Boerma, Villagarcia, Zhou, Carter, Li and Gibbs2004; Tuyen et al., Reference Tuyen, Lal and Xu2010), two minor QTLs in field pea (Pisum sativum L.) (Leonforte et al., Reference Leonforte, Sudheesh, Cogan, Salisbury, Nicolas, Materne, Forster and Kaur2013) and a minor QTL in chickpea (Cicer arietinum L.) (Samineni, Reference Samineni2010). Nevertheless, these QTL studies used different degrees of salinity treatments and also measured different traits for assessing their tolerance levels. Therefore, in the absence of any uniform criteria and comparative approach, it is difficult to arrive at a conclusive stage. QTLs mapping for salt tolerance in V. marina, a halophytic wild leguminous plant, was first reported by Chankaew et al. (Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014) by studying the F2 population of 120 plants derived from an inter-specific cross between V. luteola and V. marina subsp. oblonga.
Based upon the results of QTL mapping by Chankaew et al. (Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014), seven SSR markers were chosen for the molecular characterization of 20 Vigna genotypes under the current study. Among them, only four SSR primers (CEDG007, CEDG087, CEDG102 and cp04220) got amplified, in which three (CEDG007, CEDG087 and cp04220) showed higher polymorphism. The SSR marker ‘CEDG087’ was earlier found to be associated with the percentage of seedlings survival (Saltol1.1) and leaf wilt score at the vegetative stage (Saltol1.2) whereas, SSR primers ‘CEDG007’ and ‘cp04220’ were found to be related to the plant-recovering ability (Saltol1.3) which got expressed at the stage of development and production of new leaves and branches under salt stress (300 mM NaCl) after some or all leaves had dropped off. An estimate of discriminatory power of SSR markers is provided by the PIC value of concerned marker, depending on the number of alleles detected and the relative frequencies of those alleles. The PIC and H e values of SSR primers CEDG087, CEDG007 and cp04220 indicated their discriminatory power among Vigna genotypes studied.
The expected product size (bp) of SSR primer ‘CEDG087’ related to seedling survival percentage and leaf wilt score at vegetative stage in our study was higher (140–180) compared to earlier study by Chankaew et al. (Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014) who obtained this value as 120. Although all V. marina and other Vigna species got amplified by this primer, only green gram varieties showed the product size of 140. Only V. marina genotypes gave amplified product (125–150 bp) for SSR primer ‘CEDG007’ related to plant-recovering ability whereas, other Vigna varieties did not exhibit any amplified product with exception of one black gram variety IPU-02-43 (175 bp). This is clearly indicating the adaptability and survivability of V. marina under salt-affected conditions due to the presence of plant-recovering ability of QTL ‘Saltol1.3’ (Chankaew et al., Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014). The banding pattern obtained under study also confirmed that the location of QTL ‘Saltol1.3’ controlling plant-recovering ability, is different from the QTLs controlling plant survival at the seedling stage and leaf wilt at the vegetative stage (Chankaew et al., Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014). Furthermore, a comparative study on understanding the salt-tolerance mechanisms in three Vigna species by Yoshida et al. (Reference Yoshida, Tomooka, Khaing, Shantha, Naito, Matsuda and Ehara2020) revealed that the V. vexillata and V. luteola were ‘Na+ excluder’ types, whereas V. marina was found as ‘Na+ includer’ type which could be due to the presence of QTL, ‘Saltol1.3’ for the plant-recovering mechanism in this halophytic species (Chankaew et al., Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014).
Cluster analysis of 20 Vigna genotypes on the basis of molecular data clearly distinguished all V. marina accessions collected from Andaman and Nicobar Islands by grouping them in cluster III from the check varieties of green gram and black gram (clusters I and II). This further confirmed the effectiveness of SSR markers in separating cultivated Vigna species from wild V. marina on the basis of genetic salt tolerance. At the germination stage, green gram cultivars can tolerate salt content up to 9–18 m mhos/cm (Rao et al., Reference Rao, Nair and Nayyar2016). Paliwal and Maliwal (Reference Paliwal and Maliwal1980) reported that green gram seeds could tolerate salinity up to 6 m mhos/cm compared to 3 m mhos/cm for black gram. Therefore, green/ black gram varieties used in our study (VBN 7, LGG 544 and TARM 1) were grouped together in sub-cluster Ib which might be due to their low tolerance level compared to V. marina (pH ranged from 7.65 to 9.21 and EC ranged from 9.9 to 13.3 dS/m). This might be due to their variation for salt tolerance at the genetic and molecular level as indicated by difference in their allele size for the SSR primer ‘CEDG087’ associated with survival (%) and leaf wilt score when subjected to salt stress at the vegetative stage.
Furthermore, these cultivated Vigna varieties could not produce amplified product of the SSR primer ‘CEDG007’ (related to plant-recovering ability). On the contrary, ‘CEDG007’ produced the bands in all V. marina genotypes except for IPU-02-43. It further substantiated the interesting feature of salt tolerance i.e. plant-recovering ability in V. marina as found by Chankaew et al. (Reference Chankaew, Isemura, Naito, Ogiso-Tanaka, Tomooka, Somta, Kaga, Vaughan and Srinives2014) that the location of this QTL Saltol1.3 is relatively far (>25 cM) from the QTLs controlling plant survival at the seedling stage and leaf wilt at the vegetative stage. This QTL might be instrumental in enabling V. marina to adapt and survive under salt-affected conditions as also reported earlier (Tomooka et al., Reference Tomooka, Kaga, Isemura, Vaughan and Kole2011) stating that its young plants could survive for at least 1 month under submerged conditions in 400 mM concentration of NaCl. The group clustering formed for V. marina accessions under our study could be owing to variability in banding pattern for both SSR primers, ‘CEDG087’ and ‘CEDG007’ on the basis of allele size. The pattern of grouping obtained under cluster analysis for 20 Vigna genotypes was also confirmed by the PCoA which explained about 85.9% of genetic variation among different collections. Future research study is being planned to attempt inter-specific crosses between V. marina and cultivated Vigna species for salt-tolerance trait transfer in latter types through biotechnological tools. It is further mentioned that the seed samples of V. marina genotypes collected from Andaman and Nicobar Islands will be deposited to ICAR-National Bureau of Plant Genetic Resources (NBPGR), New Delhi for obtaining IC numbers in 2021–22. In this way, the accessions will also be available to the international and national research communities for their crop-breeding studies through submitting their requisition proforma (indent) along with material transfer agreement (MTA) to ICAR-NBPGR, New Delhi.
Conclusion
The current study on molecular profiling and cluster analysis of 20 Vigna genotypes/varieties using SSR primers associated with salt-tolerance separated V. marina (cluster III) from other Vigna species viz. green gram and black gram (clusters I and II). Two SSR markers viz. CEDG087 and CEDG007 were found very important in the current study, since they were specific to salt-tolerant QTLs Saltol1.1, Saltol1.2 and Saltol1.3. The PCoA substantiated the cluster analysis and also explained about 85.9% of genetic variation among collections. Furthermore, the results of soil sample analysis in terms of higher values of electrical conductance of the surveyed habitats also confirmed the halophytic nature of V. marina. This study indicated the effectiveness of SSR markers linked with salinity tolerance in separating cultivated Vigna species from wild V. marina. In conclusion, the findings will be useful for planning and executing the genetic transfer of the trait of robust salt tolerance of V. marina into cultivated Vigna species through novel breeding approaches for enabling pulses’ adaptation to inland and coastal saline conditions.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262121000514.
Acknowledgements
We thank The Director, ICAR-Central Island Agricultural Research Institute (CIARI), Port Blair, Andaman and Nicobar Islands, India for the financial and logistics support.
Authorship contributions
DNA isolation and gel electrophoresis: M. Prithivi; molecular data analysis and writing – original draft: K. Venkatesan; conceptualization and guidance: D.R. Manimaran; conceptualization, writing – review and editing: R.K. Gautam; supervision of all works: P.K. Singh; molecular characterization: K. Sakthivel; field survey and exploration: S.K. Zamir Ahmed; collection of genotype from Car Nicobar: S.K. Pandey and collection of genotypes and lab assistance: Shyam Sunder Rao. All authors have read and approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that there is no ethical issue(s) in this study.