INTRODUCTION
Mosquitoes are the primary vectors of several infectious diseases that have a large impact on public health (McGraw and O'Neill, Reference McGraw and O'Neill2013). They are well known as vectors of parasites (Tolle, Reference Tolle2009), viruses (Liu et al. Reference Liu, Dehning, Phipps, Swienton, Harris and Klein2016) and were recently suspected to transmit bacteria (Dieme et al. Reference Dieme, Bechah, Socolovschi, Audoly, Berenger, Faye, Raoult and Parola2015). While tropical and subtropical areas have historically been most severely affected by these diseases, mosquito-borne diseases have also emerged in temperate areas over the last decade (Rezza, Reference Rezza2014; Maria et al. Reference Maria, Maquart, Makinson, Flusin, Segondy, Leparc-Goffart, Le Moing and Foulongne2016). In addition to the growing problem mosquitoes pose as pests, attributed to the colonization of new areas, mosquito vector control has become more and more challenging for public health services (Impoinvil et al. Reference Impoinvil, Ahmad, Troyo, Keating, Githeko, Mbogo, Kibe, Githure, Gad, Hassan, Orshan, Warburg, Calderon-Arguedas, Sanchez-Loria, Velit-Suarez, Chadee, Novak and Beier2007). The absence of specific treatments and/or licensed vaccines for several mosquito-borne diseases underlines the need to improve mosquito live monitoring and subsequently, to adapt vector control measures (Benelli, Reference Benelli2016). Although mosquitoes act as vectors at the adult stage, mosquito monitoring and control of larval stages at breeding sites is the better strategy to stop their rapid spread and to limit diseases outbreaks. Mosquitoes at aquatic stages are confined to their breeding sites, making these immature stages easier to monitor and control than dispersed, flying adults (Becker et al. Reference Becker, Petrić, Zgomba, Boase, Dahl, Madon and Kaiser2010).
Generally, various mosquito species may cohabit the same area, and the species distribution can change over time (Becker et al. Reference Becker, Petrić, Zgomba, Boase, Dahl, Madon and Kaiser2010). In order to justify larvicide treatment of mosquito breeding sites, an accurate identification of the mosquito larvae present is necessary. The traditional identification method is based on detection of morphological features (Yssouf et al. Reference Yssouf, Almeras, Raoult and Parola2016). Despite the popularity of this method, largely used even today, it is time-consuming and requires an entomologist's expertise (Wang et al. Reference Wang, Li, Guo, Xing, Dong, Wang, Zhang, Liu, Zheng, Zhang, Zhu, Wu and Zhao2012). Furthermore, morphological characteristics used for identification with dichotomous keys are present only in late instar (L3/L4) larvae. Therefore, first and second instar larvae require laboratory breeding until they reach a developmental stage at which they can be morphologically identified. Rearing mosquitoes to emergence is not only time- and space-intensive, but also increases the risk of exotic mosquitoes escaping and being introduced to a new area. Moreover, the specimens at the pupal stage are generally not identifiable morphologically due to the unavailability of dichotomous keys for this stage. To circumvent these limitations, molecular tools for mosquito identification were developed (Sim et al. Reference Sim, Ramirez and Dimopoulos2009). However, these methods remain expensive and time consuming and thus are also not suitable for live monitoring of mosquito larvae.
A promising proteomic tool, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) biotyping, was recently succesfully applied to the identification of several arthropod families (Yssouf et al. Reference Yssouf, Almeras, Raoult and Parola2016). The use of this innovative tool for mosquito species identification was demonstrated at the adult (Muller et al. Reference Muller, Pfluger, Wittwer, Ziegler, Chandre, Simard and Lengeler2013; Yssouf et al. Reference Yssouf, Socolovschi, Flaudrops, Ndiath, Sougoufara, Dehecq, Lacour, Berenger, Sokhna, Raoult and Parola2013, Reference Yssouf, Parola, Lindstrom, Lilja, L'Ambert, Bondesson, Berenger, Raoult and Almeras2014) and immature stages (Steinmann et al. Reference Steinmann, Pfluger, Schaffner, Mathis and Kaufmann2013; Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014; Schaffner et al. Reference Schaffner, Kaufmann, Pfluger and Mathis2014). The distinction of species within a species complex at the adult and larval stages demonstrates the accuracy of this tool for specimen identification (Yssouf et al. Reference Yssouf, Socolovschi, Flaudrops, Ndiath, Sougoufara, Dehecq, Lacour, Berenger, Sokhna, Raoult and Parola2013; Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014). Moreover, this proteomic tool can identify mosquitoes at both the adult and immature stages and does not require laboratory-breeding of larvae, reducing the time and space needed. Recently, guidelines were established for the preparation of mosquito larvae samples for MS analysis (Nebbak et al. Reference Nebbak, Willcox, Bitam, Raoult, Parola and Almeras2016). MS identification of mosquito larvae has been tested only on laboratory-reared colonies using whole larva homogenates (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014). Although varying the diet provided to lab-reared larvae did not change MS profiles, to justify the application of this proteomic tool for live mosquito larvae monitoring, it was important to first test the reliability of MALDI-TOF MS for identification of mosquito larva specimens collected in the field.
To this aim, mosquitoes larvae collected in the spring of 2015 in Marseille, France were submitted to MALDI-TOF MS for species identification.
MATERIALS AND METHODS
Study sites and mosquito larvae collection
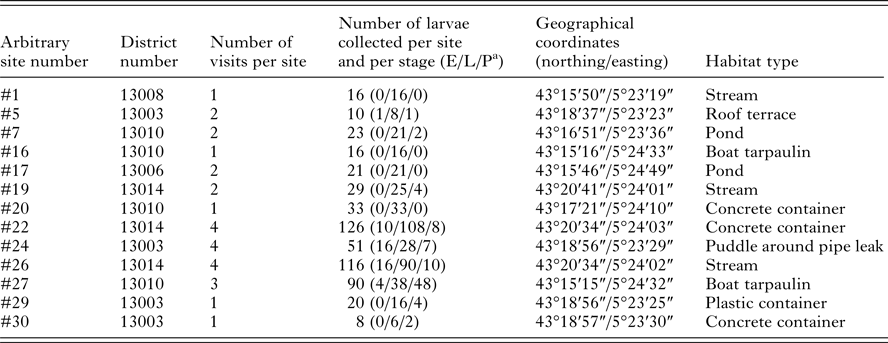
This study was conducted in the urban area of Marseille, South of France. Different habitat types containing stagnant water were selected for sampling mosquito larvae and pupae, in order to obtain a representative sample of mosquito fauna found in this area. Areas of interest were chosen based on registered complaints of mosquito nuisances provided by the Marseille city healthcare department, and Google Maps (http://www.google.fr/maps) was used to identify specific stagnant water sites in these areas. Mosquito larvae and pupae sampling were performed in April and May 2015, using standard dippers of 350 mL (Bioquip, USA). One to three dips were taken per site. Specimens were numbered prior to being transferred with habitat water into plastic bottles and labelled with the site name and date of collection. Each breeding site was visited between one and four times at a frequency of every other week. Details about the sites and mosquito collection are available in Table 1.
Table 1. Details about mosquito larvae breeding sites selected in Marseille
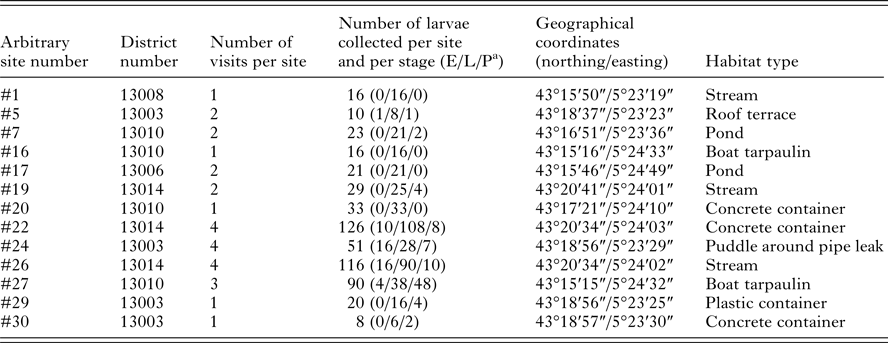
a Larva grouped in early (E, first and second instar), late (L, third and fourth instar) and pupal (P) developmental stages.
Morphological identification of mosquito larvae
At the laboratory, larvae from each collection site were rinsed successively in 70% ethanol v/v for 1 min and in sterile water for 1 min. Larvae were then classified according to their developmental stage into early instar (L1/L2), late instar (L3/L4) and pupae groups. Larvae from the late group were then identified to the genera level [Culex (Cx.), Aedes (Ae.), Ochlerotatus (Oc.), Culiseta (Cs.) or Anopheles (An.)] according to morphological criteria (Schaffner et al. Reference Schaffner, Angel, Geoffroy, Hervy, Rhaiem and Brunhes2001). Accurate morphological identification was performed on two to 30 specimens per genus depending on their availability, using specimens collected during the first site visit. Species identification was performed under the microscope (Zeiss Axio Zoom.V16, Zeiss, Marly le Roi, France) to observe details of the siphon and other body parts that are important for identification using the standard keys (Schaffner et al. Reference Schaffner, Angel, Geoffroy, Hervy, Rhaiem and Brunhes2001; Rueda, Reference Rueda2004; Becker et al. Reference Becker, Petrić, Zgomba, Boase, Dahl, Madon and Kaiser2010). Of the specimens morphologically identified, 2 to 10 larvae per species were used for MS reference database creation. For some specimens, the whole larva was treated for submission to MALDI-TOF MS and for others, only the abdomen was used. The head and thorax, stored at -20 °C, were reserved for molecular species identification.
Molecular identification
DNA was individually extracted from the head and thorax of each selected larva (n = 31) using the QIAamp DNA tissue extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Molecular identification of mosquito larvae at the species level was performed by sequencing the polymerase chain reaction (PCR) product of a fragment of the cytochrome c oxidase I gene (COI) as described previously (LCO1490 (forward): 5′-GGTCAACAAATCATAAAGATATTGG-3′; HC02198 (reverse): 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Folmer et al. Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994). Amplifications were assessed by gel electrophoresis, using 2% agarose/0·5% TBE stained with ethidium bromide. COI positive PCR products were purified using the NucleoFast 96 PCR plate (Machery-Nagel EURL, Hoerdt, France) as recommended by the manufacturer and sequenced using the same primers with the Big Dye version 1–1 Cycle Ready Reaction Sequencing mix (Applied Biosystems, Foster City, CA) and an ABI 3100 automated sequencer (Applied Biosystems). Data were collected with an ABI Prism 3130xl Genetic Analyser capillary sequencer (ABI PRISM, PE Applied Biosystems, USA). The sequences were assembled and analysed using the ChromasPro software version 1.7.7 (Technelysium Pty. Ltd., Tewantin, Australia) and blasted against GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Sample homogenization for MALDI-TOF MS
All collected larvae (whole or abdomens) and pupae were placed individually in microtubes (collection microtubes × 96, Qiagen, Hilden, Germany). The larvae were either stored at −20 °C or homogenized immediately according to the standardized protocols (Nebbak et al. Reference Nebbak, Willcox, Bitam, Raoult, Parola and Almeras2016). Briefly, a pinch of glass powder (Sigma, Lyon, France) was added to each tube in addition to 20 µL 70% (v/v) formic acid (Sigma) plus 20 µL of 50% (v/v) acetonitrile (Fluka, Buchs, Switzerland). Homogenization was carried out with a TissueLyserII instrument (Qiagen, Germany) with the optimized parameters (3 cycles of 1 min at a frequency of 30 Hertz with a quick spin centrifugation at 200 g for 1 min between each cycle).
Sample loading on MALDI-TOF target plate
After homogenization, debris was pelleted by a quick spin centrifugation (2000 rpm for 1 min), and 1·5 µL of supernatant of each sample was deposited in duplicate on the MALDI-TOF target plate (Bruker Daltonics, Wissembourg, France). Each spot was covered with 1·5 µL of CHCA matrix solution composed of saturated α-cyano-4-hydroxycynnamic acid (Sigma, Lyon, France), 50% acetonitrile (v/v), 2·5% trifluoroacetic acid (v/v) (Aldrich, Dorset, UK) and HPLC-grade water. After drying for several minutes at room temperature (RT), the target plate was introduced into a Microflex LT MALDI-TOF Mass Spectrometer (Bruker Daltonics) for analysis. To control for differences onto the target plate, matrix quality and MALDI-TOF apparatus performance, matrix solution was loaded in duplicate onto each MALDI-TOF plate with and without a bacterial test standard (Bruker Bacterial Test Standard, ref: #8255343). Moreover, two An. gambiae Giles larvae (L3) reared at the laboratory and stored at -20 °C were included on each plate and were used as homogenization positive controls.
MALDI-TOF MS parameters
Protein mass profiles were obtained using a Microflex LT MALDI-TOF Mass Spectrometer (Bruker Daltonics, Germany), with detection in the linear positive-ion mode at a laser frequency of 50 Hz within a mass range of 2–20 kDa. The acceleration voltage was 20 kV, and the extraction delay time was 200 ns. Each spectrum corresponded to ions obtained from 240 laser shots performed in six regions of the same spot and automatically acquired using the AutoXecute of the Flex Control v.2.4 software (Bruker Daltonics). The spectrum profiles obtained were visualized with Flex analysis v.3.3 software and exported to ClinProTools version v.2.2 and MALDI-Biotyper v.3.0 (Bruker Daltonics, Germany) for data processing (smoothing, baseline subtraction, peak picking) and evaluation with cluster analysis.
Spectra analysis
The reproducibility of MALDI-TOF MS spectra for each species was evaluated by visual comparison of the average spectra (MSP, Main Spectrum Profile) obtained from each late instar larva sample using the whole body or abdomen. The spectra were tested using flexAnalysis v3.3 and ClinProTools 2.2 (Bruker Daltonics) and the Composite Correlation Index (CCI) tool was used to assess spectra variations within and between each mosquito species, as previously described (Nebbak et al. Reference Nebbak, Willcox, Bitam, Raoult, Parola and Almeras2016). CCI was computed using the standard settings of mass range 3000–12 000 Da, resolution 4, 8 intervals and autocorrelation off. Correlation values [expressed by mean ± standard deviation (s.d.)] reflect the reproducibility of MS spectra. Cluster analysis (MS dendrogram) was performed by comparing the main spectra given by MALDI-Biotyper v3.0. software (Bruker Daltonics) and clustering according to protein mass profile ( m/z signals and intensities). Clustering analyses were performed to visualize the reproducibility of MS spectra from specimens of the same species, but also to examine the homogeneity of MS spectra of the same species but different origins (field or laboratory-reared). The resulting MS dendrogram illustrates how samples are related to one another.
Reference MS database expansion
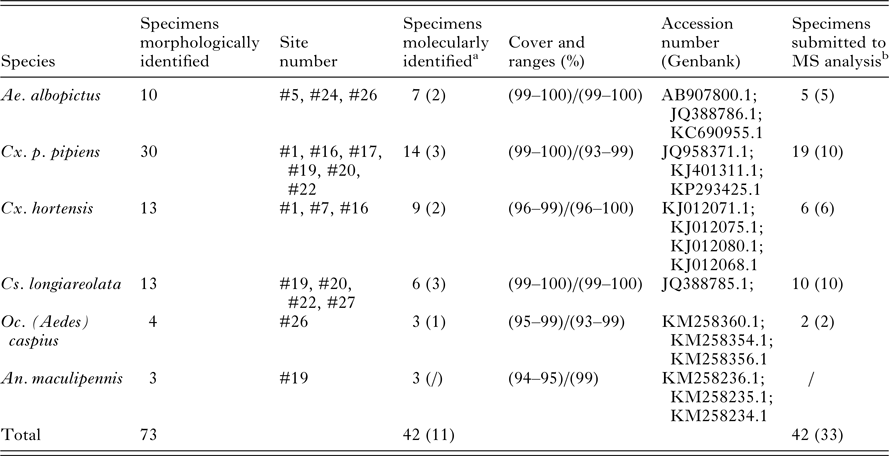
The existing homemade database initially contained, among other arthropods reference spectra, 140 MS reference spectra from six mosquito larvae species including Aedes aegypti, Aedes albopictus, Anopheles gambiae Giles, Anopheles coluzzii, Culex molestus and Culex p. pipiens (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014). As manual homogenization using pestles had been employed to prepare these samples, additional mass spectra from these species were added to the database using specimens that were homogenized automatically with glass powder, in order to be consistent with the method used in the current study. This MS reference database was further upgraded with 2–10 MS spectra of each species collected in the field, using late instar larvae from the first visit to each site that had been morphologically and molecularly identified (Table 2) (Lafri et al. Reference Lafri, Almeras, Bitam, Caputo, Yssouf, Forestier, Izri, Raoult and Parola2016). MS spectra were created with an unbiased algorithm using information on the peak position, intensity and frequency, using MALDI Biotyper software v3.0 (Broker Daltonics).
Table 2. Mosquito species collected in the field at late instar (L3/L4) aquatic stages used for morphological, molecular and MS reference database expansion
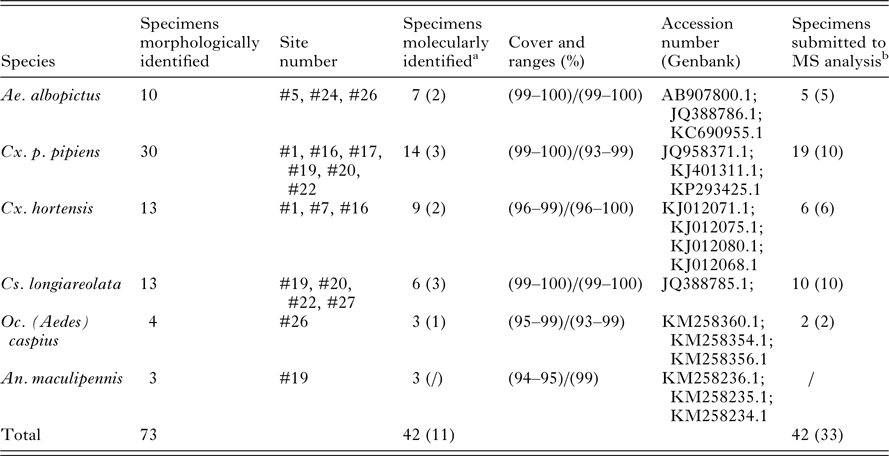
a In parentheses are indicated the number of larvae specimens submitted to molecular and mass spectrometry identification.
b In parentheses are indicated the number of larvae specimens used for MS reference database expansion.
Blind tests
Blind tests were performed with the remaining specimens collected in the field. A total of 486 MS spectra from L1 to pupal stages were tested against the expanded homemade MS reference spectra database. The reliability of species identification was estimated using the log score values (LSVs) obtained from MALDI-Biotyper software v.3.3, which ranged from 0 to 3. An LSV was obtained for each spectrum of the samples that underwent blind testing. These LSVs correspond to the degree of similarity between spectra being tested and the MS reference spectra database. Three situations were defined based on LSVs: (i) LSVs >1·8 were considered reliable for species identification (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014; Yssouf et al. Reference Yssouf, Parola, Lindstrom, Lilja, L'Ambert, Bondesson, Berenger, Raoult and Almeras2014). (ii) LSVs between 1·6 and 1·8 were subjected to additional manual controls. To be considered reliable, LSVs from duplicate tests must be included in the range 1·6–1·8 with identical first top hits for species identification. Additionally, clustering analyses were performed for some spectra to confirm species classification. (iii) Identifications from MS spectra query obtaining LSVs lower than 1·6 were not considered.
RESULTS
Larvae collection and classification
Of the standing water sites considered for inclusion in the study, 13 were selected and were numbered arbitrarily. Sites were selected to include a variety of habitat types and locations, and larvae were present at all sites at the first visit. For eight sites, larvae were also found during successive visits performed every other week in April–May 2015. A total of 559 larvae were collected: 47, 426 and 86 larvae at early instar (L1/L2), late instar (L3/L4) and pupal stages, respectively (Table 1). During the first visit, a quick morphological classification of late instar larvae to the genus level was performed at each site. Based on this classification, two to five specimens per genus and per site were selected for collection and confirmatory morphological identification, and a subset of these were used to supplement the MS spectra database, excluding larvae from sites #29 and #30. No larvae collected at these two sites were included in the database supplementation; rather, specimens from these sites were only used in the blind tests and thus served as additional controls. Overall, 73 late instar larvae from the first site visit were morphologically classified into six mosquito species coming from five genera [Aedes albopictus, Culex p. pipiens, Culex hortensis, Culiseta longiareolata, Ochlerotatus (Ae.) caspius and Anopheles maculipennis; Table 2]. The COI sequencing for 42 of them, using either whole larva (n = 31) or head and thorax (n = 11) as samples for DNA extraction, corroborated the morphological identification. Excluding Oc. (Ae.) caspius and An. malucipennis larvae, specimens of the same species were found at various sites. Moreover, multiple species were found at the same site.
MALDI-TOF MS analysis
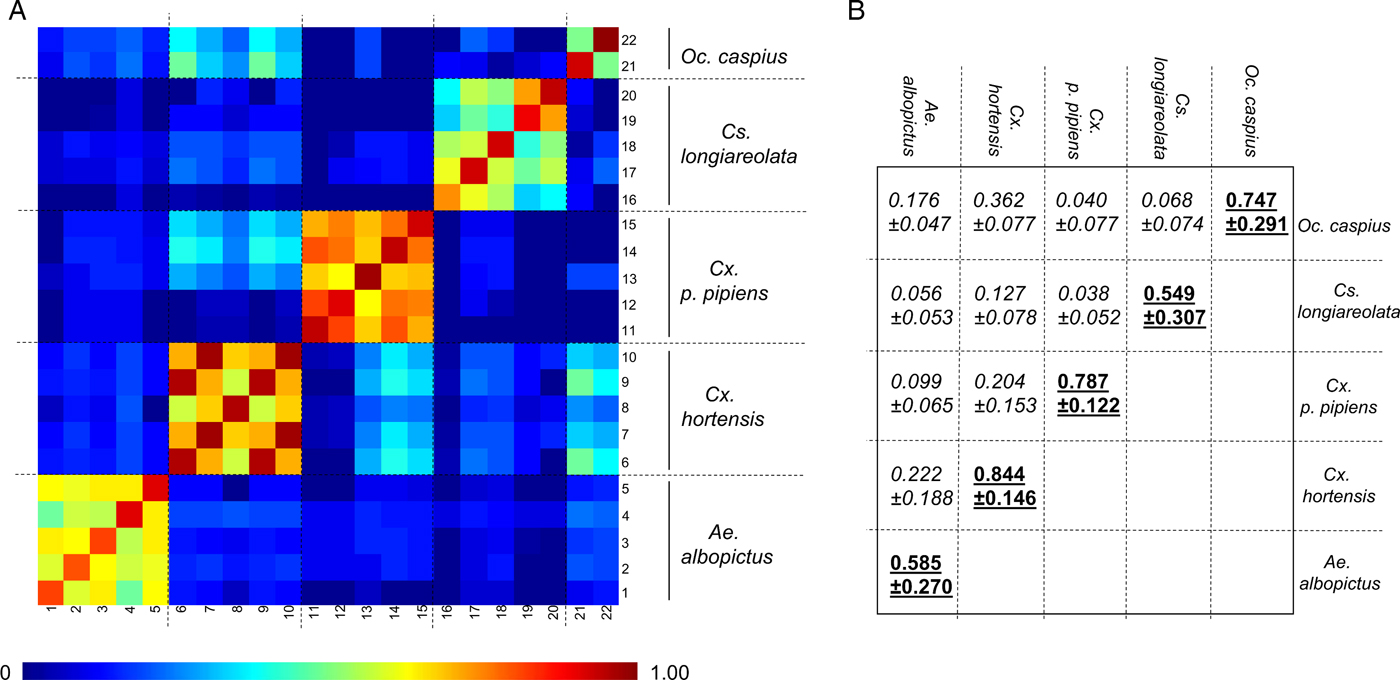
As the number of An. maculipennis larvae collected was limited (n = 3), specimens of this species were not submitted to MS analysis. Forty-two late instar larvae (L3/L4) were used to assess intra-species reproducibility and inter-species specificity of MS spectra (Table 2). For 31 of these specimens, the whole larva was used. For the remaining 11, encompassing five species, the abdomen was used for MS analysis and the head and thorax were used for molecular confirmation. Visually reproducible MS spectra were obtained for each species regardless of whether whole larvae or abdomens were used as the template (Fig. 1(i), Supplementary Fig. 1), as previously described (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014). However, because the MS profiles of abdomens had lower intensities and higher background noise, whole larvae were preferred for subsequent analyses. The CCI tool was used to analyse MS spectra from two to five specimens per species in order to objectify the correctness of morphological species identification. A heat map and the CCI values highlight the homogeneity of MS spectra for each species (Fig. 2A). Higher CCI values were obtained for comparisons of MS spectra from specimens of the same species [range of intra-species CCI means (0·549–0·844)] (Fig. 2B). The low CCI values obtained for inter-species MS spectra comparisons [range of inter-species CCI means (0·040–0·362)] support the high species-specificity of these protein profiles.

Fig. 1. Species-specific MS spectra for mosquito larvae at late instar (L3/L4) stage from field or laboratory origins. (i) Representative spectra of Ae. albopictus (A, B), Cx. p. pipiens (C, D), Cx. hortensis (E, F), Cs. longiareolata (G, H) and Oc. caspius (I, J). a.u., arbitrary units; m/z, mass-to-charge ratio. (ii) MSP dendrograms of MALDI-TOF MS spectra from late instar larvae specimens, both laboratory-reared (normal text) and collected in the field (bold). Two specimens per species and origin were used to construct the dendrogram. The dendrogram was created using Biotyper v3.0 software and distance units correspond to the relative similarity of MS spectra. The same colour code is used for each mosquito species in (i) and (ii).
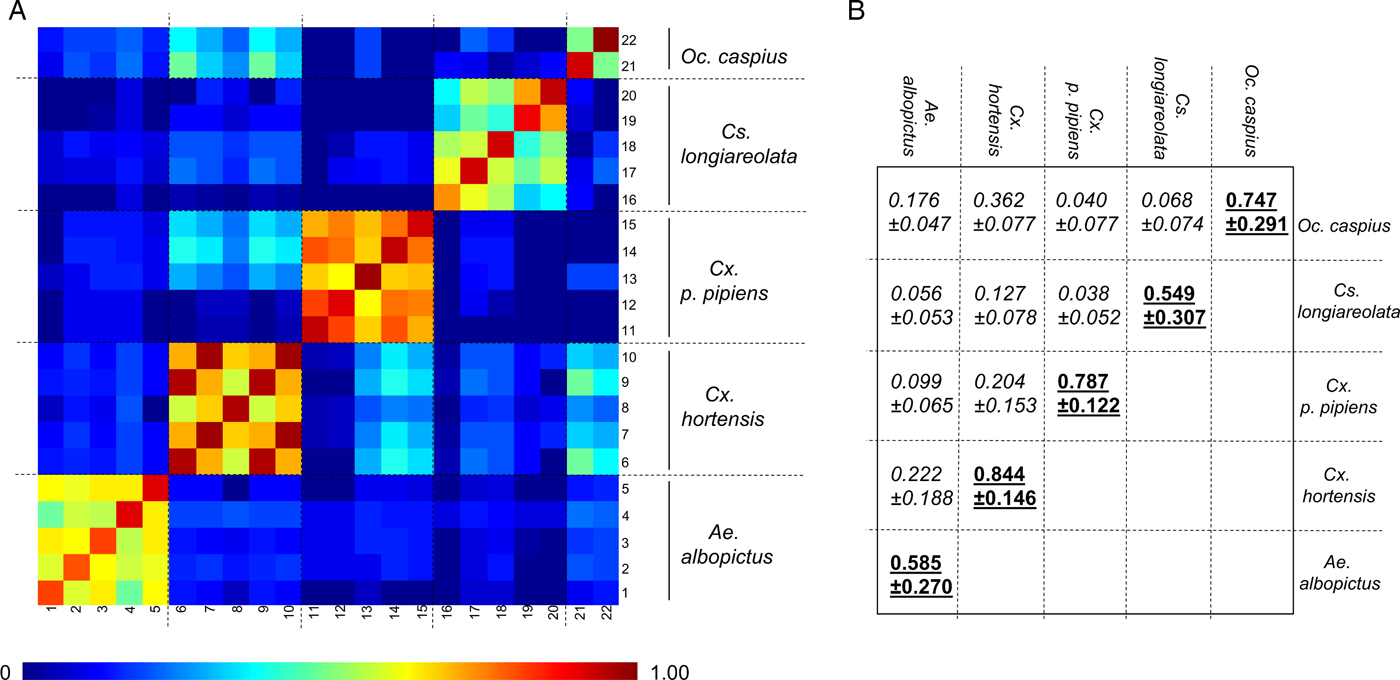
Fig. 2. Assessment of mosquito larvae MS spectra reproducibility for each species using composite correlation index (CCI). (A) MS spectra from 22 late instar larvae belonging to five different species were analysed using the CCI tool. Species are indicated to the right side of the heat map. Levels of MS spectra reproducibility are indicated in red and blue, revealing relatedness and incongruence between spectra, respectively. (B) CCI matrix was calculated using MALDI-Biotyper v3.0 software with default settings (mass range 3·0–12·0 kDa; resolution 4; 8 intervals; auto-correction off). The values correspond to the mean coefficient correlation and respective standard deviations obtained for larvae from the same species (intra-species comparisons, underlined) or between larvae from different species (inter-species comparisons, italic). CCI reflecting reproducible MS spectra are indicated in bold. CCI values are expressed as mean ± s.d.
Clustering analyses were performed to estimate the distances between MS profiles of conspecific larvae from different sites [Fig. 1(ii)]. Specimens of the same species, collected from various sites in Marseille or laboratory-reared, clustered on the same branch, supporting the species-specificity of MS protein fingerprinting profiles. However, specimens within the Culex and Aedes genera do not cluster. These results are concordant with previous studies (Yssouf et al. Reference Yssouf, Socolovschi, Flaudrops, Ndiath, Sougoufara, Dehecq, Lacour, Berenger, Sokhna, Raoult and Parola2013; Lafri et al. Reference Lafri, Almeras, Bitam, Caputo, Yssouf, Forestier, Izri, Raoult and Parola2016), which report that MALDI-TOF MS biotyping does not seem suitable for correct hierarchical taxonomic classification. Collectively, these data underline the high specificity of mosquito larvae MS spectra according to species.
Reference MS database expansion and blind tests
Of the five mosquito species submitted to MALDI-TOF MS at the larval stage, no reference MS spectra were present in the database for three of them, Cx. hortensis, Culiseta longiareolata and Oc. (Ae.) caspius [Fig. 1(ii)]. The database was therefore supplemented with MS spectra of 2–10 specimens for each of the five species, collected during the first site visits in order to be consistent with the automation and standardization method of the sample preparation used in the current study (Table 2). The remaining 486 specimens collected in the field, including larvae from the two supplementary sites (#29 and #30), were submitted to MALDI-TOF MS.
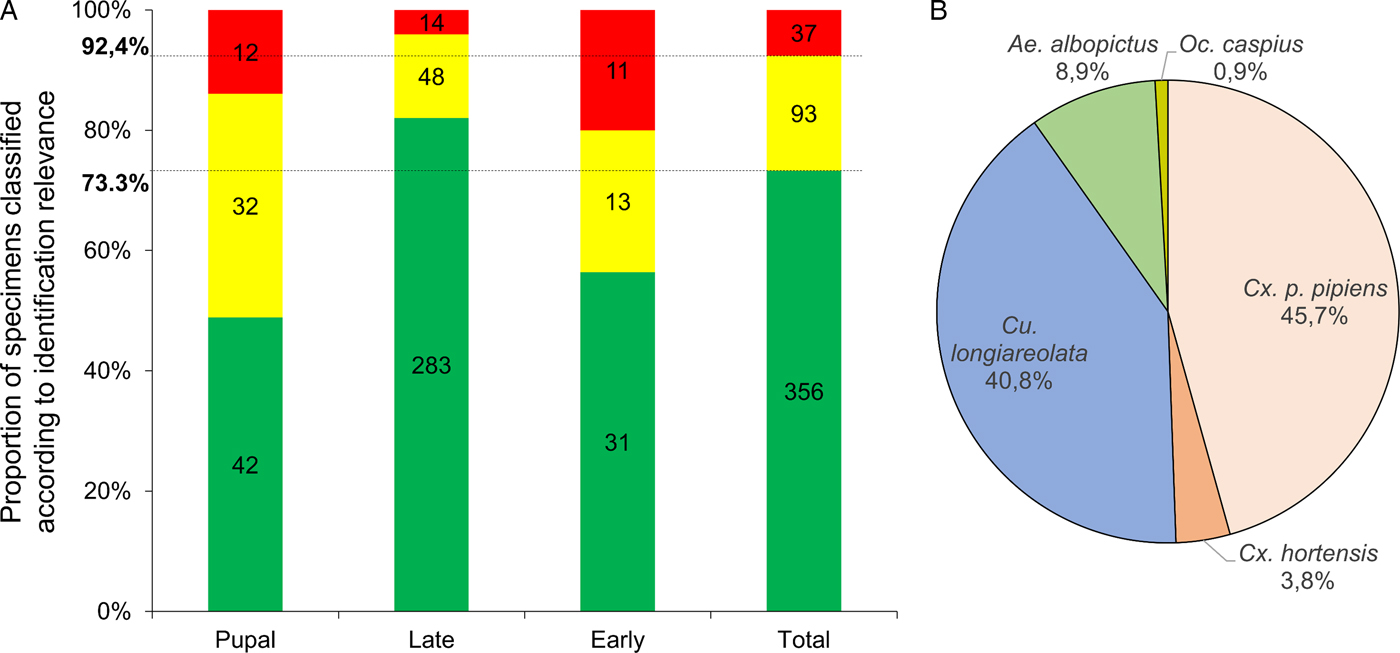
Overall, more than 92% (n = 449) of the MS spectra queried against the database were reliably identified based on the criteria detailed in Materials & Methods (Fig. 3A). Of these, 356 had LSVs above the threshold value (>1·8) and were immediately considered reliable. Ninety-three spectra required manual analysis to clarify their identification. Finally, <8% (n = 37) of the MS spectra queried against the database were not reliably identified.
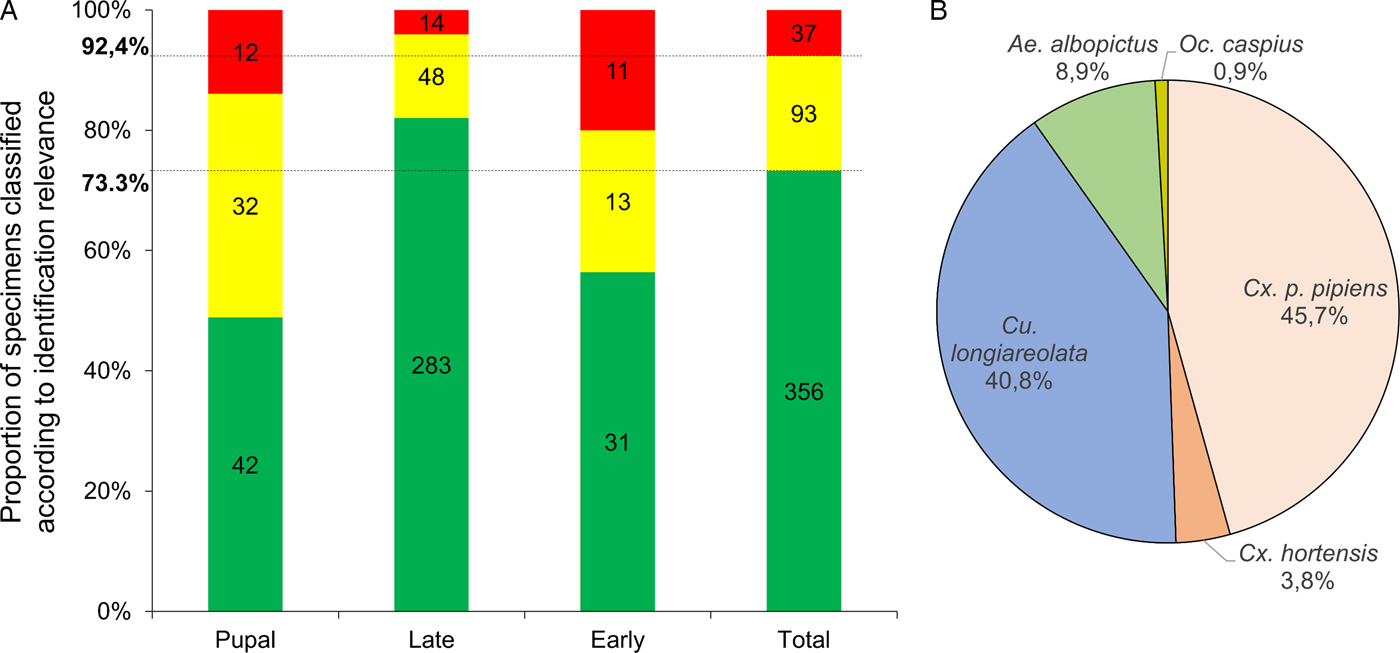
Fig. 3. Classification of mosquito aquatic stages based on LSV ranges and species identification from blind tests against MS reference database. (A) Results of MS spectra query for larvae tested blindly against the MS database were classified according to identification relevance based on LSVs. LSVs > 1·8, reliable identification (green); 1·8 > LSVs > 1·6, identification requiring manual validation (yellow) (see Materials & Methods section for details); 1·6 > LSVs; corresponding to unreliable identification and considered as non-identified (red). Results are presented for all specimens tested and for each aquatic stage. Horizontal lines indicate mean proportions obtained for all larvae submitted to MS. (B) Pie chart representing proportions of larvae identified per species. Only MS spectra from identified larvae (n = 449) were included. The percentages of specimens identified per species are indicated in brackets.
Interestingly, the proportion of larvae with the most reliable identification (LSVs > 1·8) was higher for late instar (82·0%) than for early instar (56·4%) and pupal (48·8%) stages (Fig. 3A). A visual comparison of conspecific MS spectra at distinct stages shows that MS profiles change according to stage (Supplementary Fig. 2). For early instar larvae, the intensity of MS profiles is lower than for other stages and an increase in baseline background noise is observed. The MS profile changes at the pupal stage could likely be attributed to metamorphosis (Steinmann et al. Reference Steinmann, Pfluger, Schaffner, Mathis and Kaufmann2013). Nevertheless, high LSVs (>2·0) obtained for some pupae can be explained by the conservation of several MS peaks from late instar larvae of the same species.
It is interesting to note that four larvae collected at site #17 were a striking green colour, probably due to intake of algae. These larvae were morphologically identified as Cx. p. pipiens and submission to MALDI-TOF MS confirmed morphological identification with LSVs over 2.0 (Supplementary Fig. 3). MS spectra from other specimens collected in the same site without green colouring and identified as Cx. p. pipiens presented similar MS profiles (Supplementary Fig. 3). The correct MS identification of these ‘green’ larvae confirms the robustness of MALDI-TOF MS for identification of larvae from the field.
The 449 larvae identified by MALDI-TOF MS were classified according to species (Fig. 3B). Specimens from the five species used to supplement the MS reference database were found. Culex p. pipiens and Cs. longiareolata were the most abundant species collected at the sites, representing more than 86% (n = 388) of the larvae identified. Ochlerotatus caspius was present at very low abundance at these sites (n = 4). The totality (n = 28; 100%) of the larvae from sites #29 and #30, which were only included in blind tests and not in initial database expansion, were confidently identified (LSVs > 1·8). The sites #29 and #30 were composed of Cx. p. pipiens (95%) and Cs. longiareolata (100%), respectively.
DISCUSSION
Although the application of MALDI-TOF MS biotyping tool for field mosquito monitoring of the adult and egg stages was previously demonstrated (Yssouf et al. Reference Yssouf, Parola, Lindstrom, Lilja, L'Ambert, Bondesson, Berenger, Raoult and Almeras2014; Suter et al. Reference Suter, Flacio, Farina, Engeler, Tonolla and Muller2015), larval identification using MS was limited to laboratory-reared specimens (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014). Therefore, prior to applying this tool to mosquito larvae monitoring in the field, it was indispensable to test the accuracy of MALDI-TOF MS for identification of field larvae.
In the present study, whole specimens were used for identification of larvae with MS, which presented numerous advantages but also some constraints. The larvae did not need to be dissected prior to analysis, which saved time, avoided MS profile changes due to incorrect dissection and improved identification rate by maximizing the protein quantity, which is especially important for early instar larvae. Nevertheless, if the whole larva is used for MS analysis, it cannot also be analysed using molecular tools because sample homogenization with organic buffers for MS analysis is incompatible with molecular analysis (Bisanti et al. Reference Bisanti, Ganassi and Mandrioli2008). Therefore, to confirm morphological identifications, the abdomens were submitted to MS and the heads and thoraces were reserved for molecular analysis in the present study. COI sequencing confirmed morphological identification and MS spectra from the abdomens resembled spectra from whole larvae of the same species. Moreover, the correlation analysis showed a reproducibility of MS spectra for specimens classified morphologically from the same species and an absence of MS spectra cross-recognition between species. Therefore, it was possible to corroborate morphological, molecular and MS analysis results for a subgroup of samples. This step is essential for the addition of validated MS spectra to the reference database.
Although it was previously demonstrated that MS protein profiles from laboratory-reared mosquito larvae were weakly affected by diet (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014), it was deemed important to assess the influence of habitat on MS protein profiles from larvae of the same species using field-collected specimens from different sites. The diet of mosquito larvae in the field consists of microorganisms, algae, protozoa, invertebrates and detritus intakes (Becker et al. Reference Becker, Petrić, Zgomba, Boase, Dahl, Madon and Kaiser2010). It was thought that this complex diet might have an effect on MS profiles. However, larvae from the same species collected in the field and reared in the laboratory cluster on the same branch in dendrogram construction, confirming the existence of a specific protein pattern associated with each species. Additionally, the absence of MS profile alterations and the correct identification of Cx. p. pipiens larvae with green colouring collected at site #17 confirms the robustness of this tool for mosquito larvae identification. The species-specificity of MS spectra appears well preserved for larvae of the same species collected at distinct sites and at the same site at a different collection time. The species singularity of MS spectra, essential for arthropod monitoring in the field, was previously reported for sandflies from distinct geographical origins (Lafri et al. Reference Lafri, Almeras, Bitam, Caputo, Yssouf, Forestier, Izri, Raoult and Parola2016). These results are in agreement with previous studies reporting the species-specificity of MS spectra for specimens in other arthropod families (Feltens et al. Reference Feltens, Gorner, Kalkhof, Groger-Arndt and von Bergen2010; Hoppenheit et al. Reference Hoppenheit, Murugaiyan, Bauer, Steuber, Clausen and Roesler2013; Kumsa et al. Reference Kumsa, Laroche, Almeras, Mediannikov, Raoult and Parola2016).
Interestingly, for ceratopogonid larvae, a gut dissection is recommended to increase MS spectra reproducibility (Steinmann et al. Reference Steinmann, Pfluger, Schaffner, Mathis and Kaufmann2013). Similarly, for adult mosquitoes, the abdomen is generally cut off for MS identification, because the heterogeneity of blood meals alters species-specificity of MS profiles (Kaufmann et al. Reference Kaufmann, Ziegler, Schaffner, Carpenter, Pfluger and Mathis2011; Yssouf et al. Reference Yssouf, Socolovschi, Flaudrops, Ndiath, Sougoufara, Dehecq, Lacour, Berenger, Sokhna, Raoult and Parola2013). On the other hand, using abdomens of freshly engorged mosquitoes as the MS template could be useful for determining blood sources (Niare et al. Reference Niare, Berenger, Dieme, Doumbo, Raoult, Parola and Almeras2016).
It is interesting to note that the majority of larvae submitted to MALDI-TOF MS in this study were stored at −20 °C after collection. This mode of storage was previously reported to be the best conservation method for correct and reliable identification of specimens for up to 6 months without alteration of MS spectra profiles (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014; Nebbak et al. Reference Nebbak, Willcox, Bitam, Raoult, Parola and Almeras2016). Then, in the case of larval samples could not be treated rapidly for MS submission, the frozen mode appeared as the best storing method to preserve larvae.
Blind tests against the expanded MS reference database yielded correct identifications for more than 92% of the MS spectra queried independently of developmental stage. Of these, more than 73% were confidently identified with LSVs above the previously established threshold value (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014; Nebbak et al. Reference Nebbak, Willcox, Bitam, Raoult, Parola and Almeras2016). The poor MS spectra from specimens that required manual validation or were not identified are generally attributed to lower MS spectra quality (lower peak intensity and diversity). This phenomenon could be the result of differences in sample size, storage mode, homogenization method, loading onto the MALDI plate, or the quality of the reagents, among other things.
A limiting factor frequently observed in previous studies using MALDI-TOF MS for arthropod identification is the sample homogenization method (Yssouf et al. Reference Yssouf, Parola, Lindstrom, Lilja, L'Ambert, Bondesson, Berenger, Raoult and Almeras2014; Nebbak et al. Reference Nebbak, Willcox, Bitam, Raoult, Parola and Almeras2016). The homogenization of arthropod specimens in preparing the samples for submission to MALDI-TOF MS has traditionally been carried out manually with pestles (Yssouf et al. Reference Yssouf, Almeras, Raoult and Parola2016). This homogenization method could yield different results dependent on the skill of the technician and is not ideal for preparing numerous samples. To address these shortcomings, standardized protocols for automation of sample homogenization were applied in this study as previously described (Nebbak et al. Reference Nebbak, Willcox, Bitam, Raoult, Parola and Almeras2016).
Nevertheless, identification rates were not equivalent for each immature stage. The best identification results were obtained for the MS spectra from late instar larvae (more than 80% with LSVs > 1·8). Because only MS profiles from late instar larvae were included in the MS spectra reference database, it was not surprising that matching rates were higher for this stage. Conversely, <60% of early instar larvae were correctly identified with LSVs above 1·8. The lower identification rate could be attributed to the lower protein quantity loaded onto the MALDI plate due to the smaller size of early stages. Indeed, a link has been reported between protein concentration and peak detection by MS (Steinmann et al. Reference Steinmann, Pfluger, Schaffner, Mathis and Kaufmann2013). Moreover, in contrast to the pioneering study, where volume of organic buffer used was adapted according to larva size (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014), in the current study a single volume of organic buffer was added for sample homogenization regardless of stage. Although the simplification of the protocol can decrease the rate of identification of early stages by MS, the standardization of organic buffer volume allows for the automation of buffer loading onto the MALDI plate could enable high-throughput analysis.
At the pupal stage, of the 86 pupae tested blindly, <50% obtained LSVs > 1·8. The large size of the specimens at this stage allowed extraction of sufficiently high amounts of proteins to obtain MS spectra with numerous intense peaks. The misidentification is instead attributed to protein pattern changes, which could likely be the result of metamorphosis, a process in which protein repertory evolves (Steinmann et al. Reference Steinmann, Pfluger, Schaffner, Mathis and Kaufmann2013). This phenomenon was previously mentioned (Dieme et al. Reference Dieme, Yssouf, Vega-Rua, Berenger, Failloux, Raoult, Parola and Almeras2014), and it was suggested that the MS reference database should be expanded with representative MS spectra from both larval and pupal stages. It would be interesting to test whether the pupae that were successfully identified in the current study are in the first stage of metamorphosis, while the more important proteomic changes and consequent changes in MS pattern correspond to advanced metamorphosis steps. Additional experiments could be done in the future to validate these hypotheses. Similarly, the comparison of MS spectra of ceratopogonids during metamorphosis (larval, pupal and adult stages), revealed a drastic change of MS profiles according to developmental stage (Steinmann et al. Reference Steinmann, Pfluger, Schaffner, Mathis and Kaufmann2013). These authors outlined the requirement to include several MS spectra per stage and per species in the MS database. The evolution of the MS spectra at the mosquito pupal stage observed in the present study is a limiting factor to obtaining a high identification rate with the inclusion of only MS spectra from late stage larvae of each species in the MS database.
The blind tests showed that two mosquito species were dominant, Cx. p. pipiens and Cs. longiareolata, representing more than 86% of the larvae collected. Cx. p. pipiens, belonging to the Cx. pipiens complex, is considered to be a species of public and veterinary health importance. This mosquito species is a vector of human pathogens such as viruses [West Nile Virus (WNV), St. Louis encephalitis virus (SLEV)], filarial worms (Wuchereria bancrofti) and avian malaria (Plasmodium spp.) (Farajollahi et al. Reference Farajollahi, Fonseca, Kramer and Marm2011).
Culiseta longiareolata (Macquart) is an ornithophilic species with no public health interest that is widely distributed in the Mediterranean region (Becker et al. Reference Becker, Petrić, Zgomba, Boase, Dahl, Madon and Kaiser2010). The detection of Cx. p. pipiens and Cs. longiareolata at the same site was previously reported (Becker et al. Reference Becker, Petrić, Zgomba, Boase, Dahl, Madon and Kaiser2010), and both species were previously collected in Marseille (Delaunay et al. Reference Delaunay, Jeannin, Schaffner and Marty2009; Cotteaux-Lautard et al. Reference Cotteaux-Lautard, Berenger, Fusca, Chardon, Simon and Pages2013). Very few An. maculipennis were collected at only one site and time point. A closer monitoring of more sites over a longer period of time could help to improve the known mosquito species repertoire and to analyse seasonal changes in mosquito species distribution.
Despite the diversity of habitat types and locations in the selected sites, few distinct mosquito species were found. Now that the application of this tool for field larvae identification has been verified, it should be used to monitor mosquito larvae populations across space and time in order to demonstrate the potential of this tool to survey mosquito fauna. The use of MALDI-TOF MS requires an initial investment to purchase the high-cost equipment. However, the low cost of reagents, the rapidity of MS analysis (around 1 min per spot), the accuracy of species identification and the simplicity of the protocols have made this technology a standard tool for the systematic identification of micro-organisms as part of routine analyses in clinical microbiology laboratories (Bizzini et al. Reference Bizzini, Durussel, Bille, Greub and Prod'hom2010; Welker and Moore, Reference Welker and Moore2011). It is likely that the same revolution will occur for medical entomology studies and that this tool will become a standard method for live monitoring of arthropod populations.
Concluding remarks
The emergence of mosquito-borne diseases worldwide underlines the urgency to improve entomological surveillance at the adult and immature stages using tools compatible with real-time monitoring. The present study confirms the suitability and applicability of MALDI-TOF MS for mosquito larvae identification using specimens collected in the field. The demonstration of the low influence of diet on MS spectra of larvae further confirms the species-specificity of protein patterns. Despite the limitations of this tool for the identification of larvae at early instar and pupal stages, it remains highly competitive at the economic and throughput levels. The next step is the application of this tool to establish seasonal variations in mosquito larvae density and species diversity according to habitat type. MALDI-TOF MS appears to be a promising tool for mosquito species identification of the adult and now larval stages.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTION
Conceived and designed the experiments: AL, JMB. Performed the experiments: NA, KS, ACW, AL. Analysed the data: AL, NA. Contributed reagents/materials/analysis tools: RD, PP. Wrote the paper: NA, AL. Contributed to the paper redaction: ACW, PP.
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182017001354
ACKNOWLEDGEMENTS
We would like to thank the City of Marseille and, in particular, the ‘Service de la Santé Publique et des Handicapés (SSPH)’, for the assistance provided to this research through the expertise of its territory, which was represented by Mr. Patrick PADOVANI, Deputy Mayor, Delegate for Health, Hygiene, Disabled Persons, Alzheimer, AIDS and drug addiction. We would also like to thank the ‘Régie des Transports de Marseille (RTM)’ through its president, Mr Maxime TOMMASINI, for providing travel capabilities to our students for sample collection. This participation enabled the deployment and follow-up of this study. We would like to thank the Honors Carolina Burch Fellowship for enabling ACW to carry out an internship in our laboratory and participate in this project.
FINANCIAL SUPPORT
This work has been carried out thanks to the support of the ‘Agence Régionale de Santé Provence-Alpes-Côte d'Azur (ARS-PACA)’ in the framework of 2015 health/environment projects (n° 20150668).